Key Points
HbF induction by salubrinal is not mediated through changes in globin mRNA stability, mRNA cellular localization, or HbA levels.
Translation efficiency of γ-globin mRNA is increased during stress recovery following salubrinal-enhanced eIF2α phosphorylation.
Abstract
Fetal hemoglobin (HbF) induction can ameliorate the clinical severity of sickle cell disease and β-thalassemia. We previously reported that activation of the eukaryotic initiation factor 2α (eIF2α) stress pathway increased HbF through a posttranscriptional mechanism. In this study, we explored the underlying means by which salubrinal, an activator of eIF2α signaling, enhances HbF production in primary human erythroid cells. Initial experiments eliminated changes in globin messenger RNA (mRNA) stability or cellular location and reduction of adult hemoglobin as possible salubrinal mechanisms. We then determined that salubrinal selectively increased the number of actively translating ribosomes on γ-globin mRNA. This enhanced translation efficiency occurred in the recovery phase of the stress response as phosphorylation of eIF2α and global protein synthesis returned toward baseline. These findings highlight γ-globin mRNA translation as a novel mechanism for regulating HbF production and as a pharmacologic target for induction of HbF.
Introduction
Induction of fetal hemoglobin (HbF) is an effective therapeutic strategy for β-hemoglobinopathies.1-5 Most studies have focused on understanding transcriptional regulation of hemoglobin switching to discover new mechanism-based therapeutic approaches to γ-globin gene activation.6 However, there are data indicating that HbF may also be posttranscriptionally regulated.7-10
Recently, we identified the eukaryotic initiation factor 2α (eIF2α) pathway as a critical posttranscriptional regulator of HbF.11 We found that increasing eIF2α phosphorylation (p-eIF2α) increased HbF protein without changing γ/(γ+β) messenger RNA (mRNA) levels. The most dramatic posttranscriptional HbF induction was elicited by salubrinal (Sal), a pharmacologic enhancer of p-eIF2α, which increased HbF up to 4.5-fold without altering globin mRNA levels, cellular differentiation, or total Hb content. Here, we investigate the posttranscriptional mechanism of HbF induction when eIF2α is activated by Sal.
Methods
Cell culture and chemicals
K562 cells were maintained in RPMI medium (Cellgro) with 10% fetal bovine serum and 1% penicillin-streptomycin. CD34+ cells were obtained from the University of Washington or from Dr Patrick Gallagher (Yale Medical School) using protocols approved by the institutional review board. Cultures were maintained as described in Sankaran et al.12 Salubrinal (Sal-003; Tocris Bioscience) and actinomycin-D (Sigma-Aldrich) were dissolved in dimethylsulfoxide, and puromycin (Sigma-Aldrich) was dissolved in phosphate-buffered saline.
mRNA and protein analyses
RNA isolation, complementary DNA synthesis, quantitative polymerase chain reaction, Hb high-performance liquid chromatography, and western blotting were performed as previously described (see the supplemental Methods, available on the Blood Web site).11 Transcript levels were calculated by the method of Larionov et al13 relative to glyceraldehyde-3-phosphate dehydrogenase (GAPDH) expression. Polymerase chain reaction primers are listed in supplemental Table 1.
Lentiviral infection
Vectors and virus were generated as previously described.11 The sequence targeted by hemoglobin β gene (HBB) short hairpin RNA was 5′-TGGCCCATCACTTTGGCAAAG-3′, and infections were performed on days 8 and 9. Green fluorescent protein was monitored by flow cytometry for transduction efficiency (range, 90% to 95%).
Results and discussion
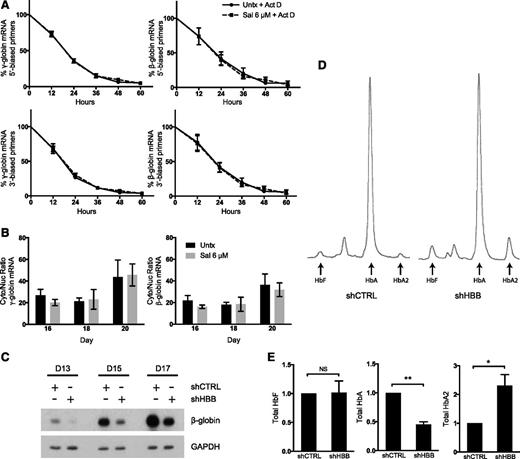
To investigate Sal’s mechanism, we used an erythroid primary cell differentiation system12 in which Sal was applied at doses previously shown to increase p-eIF2α and reduce protein translation.11 First, we assessed whether Sal changed γ- and β-globin mRNA stability. Cells were treated with Sal in combination with actinomycin D, an inhibitor of transcription, and compared with actinomycin D treatment alone. During a 60-hour time course, Sal did not change the relative half-life of γ- or β-globin mRNA (Figure 1A). Next, we determined whether changes in γ- or β-globin mRNA cellular localization could explain Sal’s ability to increase HbF. Cytoplasmic and nuclear RNA fractions were compared as cytoplasmic mRNA:nuclear mRNA ratios. Sal treatment did not alter the cellular location of γ- or β-globin when compared with the control (Figure 1B), indicating that changes in mRNA transport were not sufficient to account for the difference in HbF after Sal treatment.
Sal does not change mRNA stability, mRNA cellular localization, or total HbA to induce HbF. (A) Neither γ-globin nor β-globin relative mRNA half-life is changed in cells treated with Sal in combination with 5 μg/mL actinomycin D (Act D) versus actinomycin D alone. Expression is reported as fold change relative to the untreated control (Untx) on day 15 prior to treatment. Actinomycin D and 6 μM Sal were simultaneously applied on day 15. mRNA expression was quantified by using primers located at either the 5′ or 3′ end of each mRNA. Error bars represent ± standard error of the mean (SEM) of 3 independent experiments. (B) Sal does not change the cytoplasmic (Cyto):nuclear (Nuc) ratio of γ- or β-globin mRNA compared with the untreated control. Cells were treated with 6 μM Sal on days 15 and 18. Cytoplasmic and nuclear RNA were isolated on days 15, 18, and 20, mRNA expression for γ- and β-globin was quantified in each compartment by using primers that spanned at least 1 exon-exon junction, and the cytoplasmic mRNA:nuclear mRNA ratio was compared. Error bars represent ± SEM of 4 independent experiments. (C) Western blotting shows that short hairpin HBB (shHBB) causes 50% knockdown of β-globin protein compared with short hairpin control (shCTRL). Protein lysates were taken on days 13, 15, and 17 of differentiation after infections on days 8 and 9. glyceraldehyde-3-phosphate dehydrogenase was used as a loading control. (D) High-performance liquid chromatography (HPLC) was performed on day 20 of culture to assess the proportions of HbF, HbA, and HbA2 in shCTRL- and shHBB-infected cells. These HPLC traces from a representative experiment reveal that shHBB enhances the percentage of HbF and percentage of HbA2 compared with shCTRL. (E) β-globin knockdown does not increase total HbF, but it does enhance total HbA2. After analysis by HPLC, the area under the curve for HbF, HbA, and HbA2 was corrected for the total Hb concentration in 2 × 106 cells. Total amounts of HbF, HbA, and HbA2 are depicted as fold change relative to the shCTRL. Error bars represent ± SEM of 3 independent experiments. P values were determined by using an unpaired 2-tailed Student t test. NS, not significant. *P < .05; **P < .001.
Sal does not change mRNA stability, mRNA cellular localization, or total HbA to induce HbF. (A) Neither γ-globin nor β-globin relative mRNA half-life is changed in cells treated with Sal in combination with 5 μg/mL actinomycin D (Act D) versus actinomycin D alone. Expression is reported as fold change relative to the untreated control (Untx) on day 15 prior to treatment. Actinomycin D and 6 μM Sal were simultaneously applied on day 15. mRNA expression was quantified by using primers located at either the 5′ or 3′ end of each mRNA. Error bars represent ± standard error of the mean (SEM) of 3 independent experiments. (B) Sal does not change the cytoplasmic (Cyto):nuclear (Nuc) ratio of γ- or β-globin mRNA compared with the untreated control. Cells were treated with 6 μM Sal on days 15 and 18. Cytoplasmic and nuclear RNA were isolated on days 15, 18, and 20, mRNA expression for γ- and β-globin was quantified in each compartment by using primers that spanned at least 1 exon-exon junction, and the cytoplasmic mRNA:nuclear mRNA ratio was compared. Error bars represent ± SEM of 4 independent experiments. (C) Western blotting shows that short hairpin HBB (shHBB) causes 50% knockdown of β-globin protein compared with short hairpin control (shCTRL). Protein lysates were taken on days 13, 15, and 17 of differentiation after infections on days 8 and 9. glyceraldehyde-3-phosphate dehydrogenase was used as a loading control. (D) High-performance liquid chromatography (HPLC) was performed on day 20 of culture to assess the proportions of HbF, HbA, and HbA2 in shCTRL- and shHBB-infected cells. These HPLC traces from a representative experiment reveal that shHBB enhances the percentage of HbF and percentage of HbA2 compared with shCTRL. (E) β-globin knockdown does not increase total HbF, but it does enhance total HbA2. After analysis by HPLC, the area under the curve for HbF, HbA, and HbA2 was corrected for the total Hb concentration in 2 × 106 cells. Total amounts of HbF, HbA, and HbA2 are depicted as fold change relative to the shCTRL. Error bars represent ± SEM of 3 independent experiments. P values were determined by using an unpaired 2-tailed Student t test. NS, not significant. *P < .05; **P < .001.
Previously, we observed that Sal treatment not only increased HbF, but concomitantly reduced adult hemoglobin (HbA).11 We questioned whether reducing HbA was sufficient to increase HbF. Decreased β-globin translation or inhibition of β-globin chain association with α-globin could reduce HbA, thereby allowing α-globin chains to preferentially complex with γ-globin chains. To test this hypothesis, we used a short-hairpin RNA targeting β-globin that reduced protein levels by ∼50% (Figure 1C). This was an appropriate level of knockdown because Sal only modestly reduced HbA in previous experiments. When equal amounts of Hb were analyzed by HPLC, β-globin knockdown increased the percentage of HbF by twofold (4% to 8%) and the percentage of HbA2 by fivefold (2% to 10%) and decreased HbA from 94% to 82% (Figure 1D). However, the absolute Hb content per cell was reduced by ∼50% with β-globin knockdown. When total Hb levels per cell were calculated, HbF was unchanged (Figure 1E). In contrast, total HbA2 was increased, a finding consistent with elevated HbA2 levels seen in patients with β-thalassemia intermedia.14 These results confirm that reducing HbA is not sufficient for increasing HbF but it does increase HbA2.
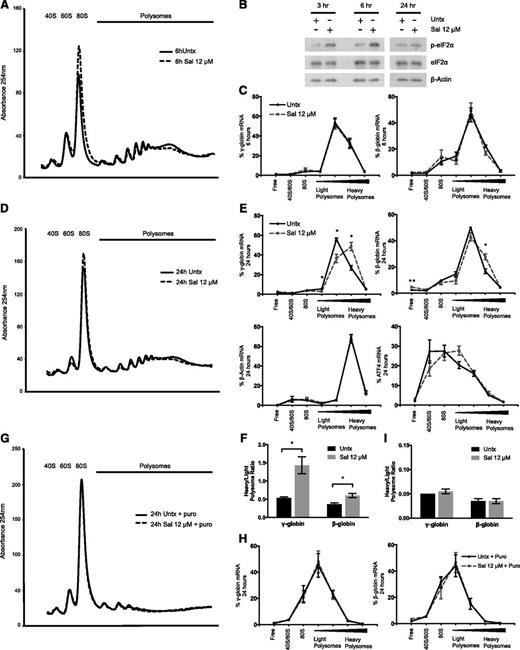
We next tested whether Sal affected globin translation. By using polysome profiling, we confirmed that Sal treatment of 6 hours reduced protein translation, as indicated by a shift in mRNA from the polysomes to the 80S peak (Figure 2A). This coincided with an increase in p-eIF2α, which reduces translation initiation15 (Figure 2B). However, quantification of γ- and β-globin mRNA isolated from pooled polysome fractions revealed that Sal did not significantly change the translation efficiency of either mRNA after 6 hours (Figure 2C).
Sal selectively increases the translation efficiency of γ-globin mRNA. (A) Representative polysome profile on day 15 after 6 hours of 12 μM Sal treatment reveals a polysome-to-monosome shift, indicative of halted translation initiation and reduced translation. (B) Western blotting shows that p-eIF2α is increased at 3 and 6 hours after 12 μM Sal treatment on day 15 relative to an untreated control. At 24 hours after treatment, p-eIF2α levels are similar between the 2 conditions. Total eIF2α and β-actin are used as loading controls. (C) After 6 hours of treatment with 12 μM Sal, γ- and β-globin translation efficiency is unchanged relative to the untreated control. Polysome profiling fractions were pooled into 7 larger fractions containing ribosome-free 40S/60S or 80S, or polysome-bound RNA ranging from lower to higher ribosome occupancy. γ- and β-globin mRNA were quantified in each pooled fraction and normalized to an exogenous luciferase mRNA control. The percent of total mRNA for each fraction was calculated. Error bars represent ± SEM of 3 independent experiments. (D) After 24 hours of 12 μM Sal treatment, the representative polysome profile on day 16 shows a slight polysome-to-monosome shift. (E) After 24 hours, 12 μM Sal significantly increases the translation efficiency of γ- and β-globin mRNA, whereas β-actin and activating transcription factor 4 translation is not significantly changed. mRNA was quantified in each pooled fraction and normalized to an exogenous luciferase control, and the percentage of total mRNA found in each fraction was calculated. Error bars represent ± SEM of 3 independent experiments. P values were determined by using an unpaired 2-tailed Student t test. *P < .05; **P < .01. (F) At 24 hours, Sal increases translation efficiency of γ-globin to a greater extent than β-globin. Percentage of mRNA in the lightest 2 polysome fractions was compared with the percentage of mRNA in the heaviest 2 polysome fractions by using a heavy-to-light polysome ratio. Error bars represent ± SEM of 3 independent experiments. P values were determined by using an unpaired 2-tailed Student t test. *P < .05. (G) The representative polysome profile at 24 hours shows ribosome dissociation and a dramatic shift to the monosome peak when 12 μM Sal and the untreated control are incubated with 1 mM puromycin (Puro). (H) Puromycin treatment after 24 hours of 12 μM Sal shifts the percentage of γ- and β-globin mRNA to lighter polysome fractions to the same extent as in the untreated control. Error bars represent ± SEM of 2 independent experiments. (I) Puromycin reduces the heavy:light polysome ratio to the same extent in 12 μM Sal-treated and untreated cells. Error bars represent ± SEM of 2 independent experiments.
Sal selectively increases the translation efficiency of γ-globin mRNA. (A) Representative polysome profile on day 15 after 6 hours of 12 μM Sal treatment reveals a polysome-to-monosome shift, indicative of halted translation initiation and reduced translation. (B) Western blotting shows that p-eIF2α is increased at 3 and 6 hours after 12 μM Sal treatment on day 15 relative to an untreated control. At 24 hours after treatment, p-eIF2α levels are similar between the 2 conditions. Total eIF2α and β-actin are used as loading controls. (C) After 6 hours of treatment with 12 μM Sal, γ- and β-globin translation efficiency is unchanged relative to the untreated control. Polysome profiling fractions were pooled into 7 larger fractions containing ribosome-free 40S/60S or 80S, or polysome-bound RNA ranging from lower to higher ribosome occupancy. γ- and β-globin mRNA were quantified in each pooled fraction and normalized to an exogenous luciferase mRNA control. The percent of total mRNA for each fraction was calculated. Error bars represent ± SEM of 3 independent experiments. (D) After 24 hours of 12 μM Sal treatment, the representative polysome profile on day 16 shows a slight polysome-to-monosome shift. (E) After 24 hours, 12 μM Sal significantly increases the translation efficiency of γ- and β-globin mRNA, whereas β-actin and activating transcription factor 4 translation is not significantly changed. mRNA was quantified in each pooled fraction and normalized to an exogenous luciferase control, and the percentage of total mRNA found in each fraction was calculated. Error bars represent ± SEM of 3 independent experiments. P values were determined by using an unpaired 2-tailed Student t test. *P < .05; **P < .01. (F) At 24 hours, Sal increases translation efficiency of γ-globin to a greater extent than β-globin. Percentage of mRNA in the lightest 2 polysome fractions was compared with the percentage of mRNA in the heaviest 2 polysome fractions by using a heavy-to-light polysome ratio. Error bars represent ± SEM of 3 independent experiments. P values were determined by using an unpaired 2-tailed Student t test. *P < .05. (G) The representative polysome profile at 24 hours shows ribosome dissociation and a dramatic shift to the monosome peak when 12 μM Sal and the untreated control are incubated with 1 mM puromycin (Puro). (H) Puromycin treatment after 24 hours of 12 μM Sal shifts the percentage of γ- and β-globin mRNA to lighter polysome fractions to the same extent as in the untreated control. Error bars represent ± SEM of 2 independent experiments. (I) Puromycin reduces the heavy:light polysome ratio to the same extent in 12 μM Sal-treated and untreated cells. Error bars represent ± SEM of 2 independent experiments.
We then questioned whether translation was changed during stress recovery, which has been reported for other mRNAs.16,17 After 24 hours of Sal treatment, p-eIF2α recovered to control levels (Figure 2B) and the polysome profile from Sal-treated cells was shifted less to the 80S fraction (Figure 2D). During this recovery phase, the translation efficiency of γ- and β-globin was increased as indicated by a significant shift to higher ribosome occupancy (Figure 2E). In contrast, β-actin translation efficiency remained unchanged and activating transcription factor 4 (ATF4) mRNA shifted slightly, but not significantly, to the polysome fractions. Together, these results suggest that globin translation efficiency is specifically increased 24 hours after Sal treatment. When the ratio of heavy-to-light polysomes was compared, Sal increased the γ-globin ratio by 2.9-fold (P = .019) but increased the β-globin ratio by only 1.6-fold (P = .025) (Figure 2F), indicating that γ-globin mRNA was preferentially translated. Because our previous results showed that 2 Sal treatments over 5 days increased HbF up to 4.5-fold,11 smaller changes in translation efficiency would be enhanced by multiple treatments to account for the observed increase in HbF.
However, the steady-state number of ribosomes on a transcript is not necessarily a measure of active translation.18 To confirm that changes in globin translation efficiency were the result of the addition of functional ribosomes, we cotreated cells with Sal and puromycin. Puromycin requires peptidyl-transferase activity to be incorporated into nascent peptides and causes premature ribosome release, which differentiates stalled from active ribosomes.19 At 24 hours after Sal treatment, puromycin caused disruption of polyribosomes and increased the 80S peak in Sal-treated and control cells (Figure 2G). Puromycin also produced a shift to lower ribosome occupancy of γ- and β-globin transcripts (Figure 2H). Notably, puromycin eliminated the difference in translation efficiency observed between Sal-treated and control cells (Figure 2H-I). These results confirm that Sal enhances globin translation efficiency at 24 hours, as evidenced by a greater number of actively translating ribosomes occupying the transcripts.
To further implicate enhanced protein translation as a mechanism responsible for increased HbF after Sal treatment, we used the K562 cell line. We first confirmed that Sal increased γ-globin protein levels without changing γ-globin mRNA at 24 hours as seen in primary cells (supplemental Figure 1A-B). Next, we used the nonradioactive SUnSET method to analyze protein synthesis.20 In this method, low doses of puromycin are incorporated into neosynthesized peptides20-22 and are monitored by flow cytometry to deduce protein translation rates. The puromycin fluorescence was not significantly changed after 24 hours of Sal treatment, confirming similar levels of total protein synthesis (supplemental Figure 1C). In control experiments, the fluorescence of cells preincubated with the translation inhibitor cycloheximide was reduced by 70%. We then analyzed cells that were dual stained for puromycin and HbF and found that Sal significantly increased HbF fluorescence by 22% (supplemental Figure 1D). Additionally, immunocytochemistry indicated a similar subcellular localization of puromycin and HbF (supplemental Figure 1E), suggesting that nascent γ-globin translation enhances HbF production by Sal.
In summary, we have determined that posttranscriptional induction of HbF by Sal is the result of enhanced γ-globin mRNA translation that occurs during the recovery phase of the p-eIF2α stress response. This provides an explanation for previous observations in which HbF induction exceeded increases in γ-globin mRNA.9,10 Our results suggest that control of γ-globin mRNA translation is an important mechanism for regulating HbF production that could be used to benefit patients with β-hemoglobinopathies.
The online version of this article contains a data supplement.
There is an Inside Blood Commentary on this article in this issue.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank Dr Patrick Gallagher for kindly supplying CD34+ cells, Dr Lionel Lewis and Bernie Beaulieu for assistance with Hb HPLC, Dr Elizabeth McCoy and Dr Alexei Kisselev for guidance regarding polysome profiling, and Dr Edwin Hahn, Dr Rodwell Mabaera, Dr Rachel West, and Dr Emily Schaeffer for valuable discussions.
This work was supported by the National Institutes of Health (National Heart, Lung and Blood Institute grant HL73442 [C.H.L.] and the National Institute of Diabetes and Digestive and Kidney Diseases grant F30DK094540 [C.K.H.]) and by the Knights of the York Cross of Honour and the Royal Order of Scotland (Masonic charitable organizations).
Authorship
Contribution: C.K.H. designed and performed the research, analyzed and interpreted the data, and wrote the manuscript; and C.H.L. designed and oversaw the research, analyzed and interpreted the data, and wrote the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Christopher H. Lowrey, Section of Hematology/Oncology, Dartmouth-Hitchcock Medical Center, One Medical Center Dr, Lebanon, NH 03756, e-mail: christopher.h.lowrey@dartmouth.edu.