Key Points
PIK3CA and NRAS mutations are recurrent in BRAFV600E wild-type ECD patients.
57.5% (46/80) of ECD patients have a BRAFV600E mutation, and an additional 10.9% and 3.7% have PIK3CA and NRAS mutations, respectively.
Abstract
Erdheim-Chester disease (ECD) is a rare histiocytic disorder that is challenging to diagnose and treat. We performed molecular analysis of BRAF in the largest cohort of ECD patients studied to date followed by N/KRAS, PIK3CA, and AKT1 mutational analysis in BRAF wild-type patients. Forty-six of 80 (57.5%) of patients were BRAFV600E-mutant. NRAS mutations were detected in 3 of 17 ECD BRAFV600E wild-type patients. PIK3CA mutations (p.E542K, p.E545K, p.A1046T, and p.H1047R) were detected in 7 of 55 patients, 4 of whom also had BRAF mutations. Mutant NRAS was present in peripheral blood CD14+ cells, but not lymphoid cells, from an NRASQ61R mutant patient. Our results underscore the central role of RAS-RAF-MEK-ERK activation in ECD and identify an important role of activation of RAS-PI3K-AKT signaling in ECD. These results provide a rationale for targeting mutant RAS or PI3K/AKT/mTOR signaling in the subset of ECD patients with NRAS or PIK3CA mutations.
Introduction
Erdheim-Chester disease (ECD) is a histiocyte proliferation with frequent multiorgan involvement,1 and aggressive phenotypes in ECD may lead to death despite treatment.2 Recently, ECD and the related disorder Langerhans cell histiocytosis (LCH) have been identified to have BRAFV600E mutations in 40% to 70% of patients.3-6 The discovery of BRAFV600E mutations in ECD and LCH has provided an important therapeutic target, and treatment of BRAFV600E-mutant ECD and LCH patients with vemurafenib has demonstrated dramatic therapeutic efficacy in pilot studies.7 Therefore, accurate identification of BRAFV600E mutations in ECD and LCH is critical. However, the heterogeneous nature of ECD and LCH lesions frequently presents a challenge to the accurate identification of BRAFV600E mutations in lesional tissue.8 In addition, several groups have noted that a larger proportion of LCH and ECD lesions have activation of ERK signaling than that demonstrated to have the BRAFV600E mutation.3,8,9 Interestingly, Cangi et al recently identified that 18 of 18 ECD patients had a BRAFV600E mutation in DNA from whole lesional tissue if an ultrasensitive methodology was used, suggesting that current data underestimate the true mutational frequency of BRAFV600E mutations in ECD.8 Concurrently, we recently identified an NRASQ61R mutation in an ECD patient who was definitively BRAF–wild-type.10
Given this, we analyzed 80 ECD patients for the BRAFV600E mutation followed by interrogation of 25 ECD thought to be BRAF–wild-type using a variety of sensitive techniques for repeat BRAF mutational analysis. In parallel, we analyzed these samples for NRAS, KRAS, PIK3CA, and AKT1 mutations.
Methods
Patients
Eighty ECD patients were included in this study, approved by the Ethics Committee Ile de France III (#2011-A00447-34) and the Institutional Review Board at Memorial Sloan-Kettering Cancer Center. Informed consent was provided according to the Declaration of Helsinki.
Genetic analyses
Genomic DNA was extracted from formalin-fixed, paraffin-embedded samples after histologic review and enrichment by macrodissection to ≥10% histiocytes. All samples derived from patients before any therapy. Workflow of genetic analysis is depicted in supplemental Figure 1 (available on the Blood Web site). Detection of BRAFV600 and NRASQ61 mutations was performed by pyrosequencing.11 BRAF status of 41 of the 80 patients was already reported (BRAFV600E mutational frequency was 51% in this initial series).5,6,12 Fifty-eight cases, in which 25 initially did not have BRAF p.V600 mutation detected, were further analyzed for BRAF mutations with other methods including multiplex picodroplet digital polymerase chain reaction (PCR)13 (Raindance Technologies; details in the supplemental Methods). Screening for mutations in other genes was performed with Sequenom mass spectrometric–based genotyping assays as previously described14 (NRAS, KRAS, PIK3CA, and AKT1 hotspot mutations), and next-generation targeted sequencing analysis for regions of mutations in BRAF, N/KRAS, and PIK3CA by Illumina MiSeq as described in the supplemental Methods.
Immunohistochemistry
Details of immunohistochemical methods and analyses are provided in the supplemental Methods.
Results and discussion
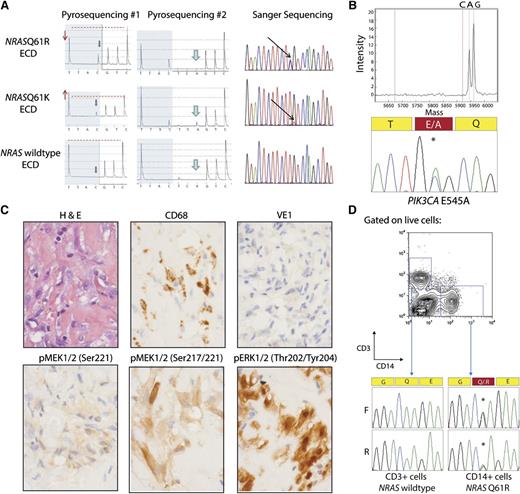
A BRAF c.1799T>A, p.V600E mutation was initially detected in 38 of 80 (47.5%) ECD patients using direct pyrosequencing of DNA obtained from lesional tissue enriched for histiocytes. Of note, there was no correlation between BRAFV600E allele burden from pyrosequencing and percent of histiocytes in these samples. As was mentioned earlier, recent work from our group identified a single ECD patient as having an NRASQ61R mutation without any evidence of BRAFV600E mutation based on pyrosequencing, Sanger sequencing, or locked nucleic acid PCR followed by pyrosequencing.10 Given this result, we screened with pyrosequencing all patients for mutations in NRASQ61 and detected 2 of 80 NRASQ61-mutated cases (Figure 1 and Table 1).
NRAS and PIK3CA mutations in ECD histiocytes and CD14+ cells from peripheral blood. (A) Detection of NRAS p.Q61 mutation by pyrosequencing (first 2 columns) and confirmation by Sanger sequencing (third column). The lower pyrogram corresponds to an NRAS–wild-type ECD case, and the 2 others correspond to patients with NRAS mutations. Mutants are detectable with the appearance of a new peak at the first C injection (gray arrows), and size variation of the peak at the first T injection compared with the second C injection (red arrows and dashed line). Pyrosequencing with another sequence of injection (ESGTTACTCAGTCAGCT) was used to further identify the mutations (middle column) as c.181C>A mutations. (B) Detection of PIK3CA E545A mutation by Sequenom and Sanger sequencing in an NRAS/KRAS/BRAF–wild-type ECD patient. (C) Phosphorylated MEK (pMEK) and ERK (pERK) detected by immunohistochemistry in BRAFV600E–wild-type, NRAS-mutant ECD. Histiocytes (noted by hematoxylin and eosin and CD68 stain) failed to stain for BRAFV600E VE1 monoclonal antibody but had high cytoplasmic expression of pMEK1/2 with 2 different antibodies as well as cytoplasmic and nuclear expression of pERK1/2 (original magnification ×600). (D) Genotyping of CD14+ monocytes and CD3+ T cells purified from the peripheral blood of an NRASQ61R-mutant ECD patient with double-FACS sorting reveals the presence of NRAS mutation in CD14+ cells but not in T cells.
NRAS and PIK3CA mutations in ECD histiocytes and CD14+ cells from peripheral blood. (A) Detection of NRAS p.Q61 mutation by pyrosequencing (first 2 columns) and confirmation by Sanger sequencing (third column). The lower pyrogram corresponds to an NRAS–wild-type ECD case, and the 2 others correspond to patients with NRAS mutations. Mutants are detectable with the appearance of a new peak at the first C injection (gray arrows), and size variation of the peak at the first T injection compared with the second C injection (red arrows and dashed line). Pyrosequencing with another sequence of injection (ESGTTACTCAGTCAGCT) was used to further identify the mutations (middle column) as c.181C>A mutations. (B) Detection of PIK3CA E545A mutation by Sequenom and Sanger sequencing in an NRAS/KRAS/BRAF–wild-type ECD patient. (C) Phosphorylated MEK (pMEK) and ERK (pERK) detected by immunohistochemistry in BRAFV600E–wild-type, NRAS-mutant ECD. Histiocytes (noted by hematoxylin and eosin and CD68 stain) failed to stain for BRAFV600E VE1 monoclonal antibody but had high cytoplasmic expression of pMEK1/2 with 2 different antibodies as well as cytoplasmic and nuclear expression of pERK1/2 (original magnification ×600). (D) Genotyping of CD14+ monocytes and CD3+ T cells purified from the peripheral blood of an NRASQ61R-mutant ECD patient with double-FACS sorting reveals the presence of NRAS mutation in CD14+ cells but not in T cells.
Characteristics of NRAS- and PIK3CA-mutant ECD patients and those not identified as having BRAF, NRAS, or PIK3CA mutations*
| Age† (y/Gender) . | Principal organs involved by histiocytosis . | Tissue biopsy site . | % Histiocytes in biopsy . | BRAF p.V600 . | Other genes . | Follow up (mo, status at last follow-up) . |
|---|---|---|---|---|---|---|
| 67/M | Heart, aorta, RPF, pleura | Peritoneum | 70 | Wild-type | NRAS p. G12D | 43, DoD |
| 56/M | Aorta, RPF, pleura, bone | Pleura | 70 | Wild-type | NRAS p. Q61K | 108, AwD |
| 65/M | Aorta, pleura | Pleura | 40 | Wild-type | NRAS p.Q61R | 26, AwD |
| 32/M | Aorta, paraparesis | Paravertebral | 10 | Wild-type | PIK3CA p.E542K | 100, AwD |
| 32/M | CNS, visual loss, bone | Bone | 10 | Wild-type | PIK3CA p.E545K | Not available |
| 29/M | RPF, bone | Bone | 20 | Wild-type | PIK3CA p.H1047R | 50, AwD |
| 62/M | CNS, aorta, bone, sinus | Perirenal | 30 | p.V600E | PIK3CA p.A1046T | 60, AwD |
| 52/M | DI, bone, RPF | Skin | 60 | p.V600E | PIK3CA p.H1047L | 24, AwD |
| 58/M | Lung, CNS, X, aorta | Skin | 50 | p.V600E | PIK3CA p.H1047L | 15, AwD |
| 42/M | E, bone, aorta, RPF | Orbital | 60 | p.V600E | PIK3CA p.H1047L | 30, AwD |
| 53/M | CNS, bone | Bone | 10 | Wild-type | None detected | 17, AwD |
| 49/M | RPF, DI, sclerosing cholangitis, sinus | Perirenal | 20 | Wild-type | None detected | 48, AwD |
| 56/M | Lung, bone | Bone | 40 | Wild-type | None detected | 44, AwD |
| 81/F | Aorta, X, skin | Skin | 50 | Wild-type | None detected | 52, AwD |
| 70/M | Aorta, RPF, pleura, skin, arthritis | Skin | 30 | Wild-type | None detected | 18, AwD |
| 65/M | Lung, bone | Bone | 40 | Wild-type | None detected | 72, AwD |
| 55/M | Heart, RPF, lung, bone, X | Skin | 70 | Wild-type | None detected | 33, AwD |
| 54/M | Mesenteric | Mesentery | 40 | Wild-type | None detected | 27, AwD |
| 39/M | CNS, E, skin | Skin | 90 | Wild-type | None detected | 134, AwD |
| 42/F | Not available | Bone | 40 | Wild-type | None detected | Not available |
| 30/M | Heart, aorta, paraparesis, liver | Paravertebral | 30 | Wild-type | None detected | 20, AwD |
| Age† (y/Gender) . | Principal organs involved by histiocytosis . | Tissue biopsy site . | % Histiocytes in biopsy . | BRAF p.V600 . | Other genes . | Follow up (mo, status at last follow-up) . |
|---|---|---|---|---|---|---|
| 67/M | Heart, aorta, RPF, pleura | Peritoneum | 70 | Wild-type | NRAS p. G12D | 43, DoD |
| 56/M | Aorta, RPF, pleura, bone | Pleura | 70 | Wild-type | NRAS p. Q61K | 108, AwD |
| 65/M | Aorta, pleura | Pleura | 40 | Wild-type | NRAS p.Q61R | 26, AwD |
| 32/M | Aorta, paraparesis | Paravertebral | 10 | Wild-type | PIK3CA p.E542K | 100, AwD |
| 32/M | CNS, visual loss, bone | Bone | 10 | Wild-type | PIK3CA p.E545K | Not available |
| 29/M | RPF, bone | Bone | 20 | Wild-type | PIK3CA p.H1047R | 50, AwD |
| 62/M | CNS, aorta, bone, sinus | Perirenal | 30 | p.V600E | PIK3CA p.A1046T | 60, AwD |
| 52/M | DI, bone, RPF | Skin | 60 | p.V600E | PIK3CA p.H1047L | 24, AwD |
| 58/M | Lung, CNS, X, aorta | Skin | 50 | p.V600E | PIK3CA p.H1047L | 15, AwD |
| 42/M | E, bone, aorta, RPF | Orbital | 60 | p.V600E | PIK3CA p.H1047L | 30, AwD |
| 53/M | CNS, bone | Bone | 10 | Wild-type | None detected | 17, AwD |
| 49/M | RPF, DI, sclerosing cholangitis, sinus | Perirenal | 20 | Wild-type | None detected | 48, AwD |
| 56/M | Lung, bone | Bone | 40 | Wild-type | None detected | 44, AwD |
| 81/F | Aorta, X, skin | Skin | 50 | Wild-type | None detected | 52, AwD |
| 70/M | Aorta, RPF, pleura, skin, arthritis | Skin | 30 | Wild-type | None detected | 18, AwD |
| 65/M | Lung, bone | Bone | 40 | Wild-type | None detected | 72, AwD |
| 55/M | Heart, RPF, lung, bone, X | Skin | 70 | Wild-type | None detected | 33, AwD |
| 54/M | Mesenteric | Mesentery | 40 | Wild-type | None detected | 27, AwD |
| 39/M | CNS, E, skin | Skin | 90 | Wild-type | None detected | 134, AwD |
| 42/F | Not available | Bone | 40 | Wild-type | None detected | Not available |
| 30/M | Heart, aorta, paraparesis, liver | Paravertebral | 30 | Wild-type | None detected | 20, AwD |
AwD, alive with disease; CNS, central nervous system; DI, diabetes insipidus; DoD, dead of disease; E, exophthalmos; RPF, retroperitoneal fibrosis; X, xanthelasma.
The 42 ECD patients with BRAF p.V600E mutations, but without PIK3CA mutation, are not included in this table. For 17 other BRAFV600E/NRAS–wild-type ECD patients, PIK3CA, KRAS, and AKT1 hotspot mutations were not investigated, because biopsies and tumor DNA samples were exhausted.
Age at time of diagnosis.
Although a BRAFV600E mutational frequency of 47.5% is roughly consistent with the frequency of BRAFV600E mutation demonstrated in ECD previously,5,6 given the results of Cangi et al,8 we performed further detailed analyses for BRAFV600E mutations in the subset of ECD patients thought to be BRAF–wild-type based on direct pyrosequencing analysis. Using both allele-specific real-time PCR, next-generation sequencing of lesional DNA (mean depth of 66×), and multiplex picodroplet digital PCR in a subset of 25 samples without initial BRAFV600E mutation detection, we were able to identify an additional 8 patients as being BRAFV600E-mutant, increasing the frequency of BRAFV600E mutations in ECD to 57.5% (46/80) (supplemental Figure 1). We cannot exclude the possibility that the frequency of BRAFV600E mutations found here (46/80) differs from that of Cangi et al (18/18)8 because of technical differences in modalities used for BRAFV600E mutation detection between the 2 studies.
Because both the PI3K/AKT/mTOR and RAF/MEK/ERK pathways are downstream effectors of RAS signaling, we also screened for mutations in commonly mutated genes in the PI3K/AKT pathway using a combination of mass spectrometric–based genotyping as well as next-generation sequencing analysis of frequently mutated regions of NRAS, KRAS, PIK3CA, and AKT1. We identified a third patient as having an NRAS mutation (Table 1). Immunohistochemical analysis for phosphorylated MEK1/2 (pMEK1/2) and ERK1/2 (pERK1/2) in NRAS-mutant ECD revealed that histiocytes were clearly positive for pMEK1/2 Ser217 and Ser217/221 as well as pERK1/2 Thr202/Tyr204 (Figure 1C). Positive staining with pERK1/2 was detected in 11 cases without BRAF, NRAS, or PIK3CA mutations as well as 3 BRAFV600E and 3 NRAS mutant samples (supplemental Figure 2).
Analysis of regions of recurrent mutations in PIK3CA in a subset of 30 BRAFV600E-mutant ECD revealed 7 of 58 ECD patients overall with PIK3CA mutations, of whom 4 were BRAF-mutated (Figure 1 and Table 1). This finding is consistent with data demonstrating a frequent overlap of PIK3CA mutations with mutations in the mitogen-activated protein kinase pathway.14
The cell-of-origin for histiocytic disorders including LCH and ECD has been long debated, and recent work from both Berres et al4 and Cangi et al8 demonstrated the presence of BRAFV600E in peripheral blood CD14+ cells in a proportion of BRAFV600E-mutant LCH and ECD patients, respectively. Genetic analysis of CD14+ cells purified by double fluorescence-activated cell sorting (FACS) of peripheral blood mononuclear cells from an NRASQ61R-mutant ECD patient clearly revealed the presence of the NRAS mutation in CD14+ cells but not in CD3+ cells (Figure 1D). These data further suggest that the histiocytic proliferations in ECD are derived from genetically aberrant circulating myeloid hematopoietic cells.
The discovery of recurrent BRAFV600E mutations in ECD as well as the consistent activation of ERK regardless of BRAF mutational status confirms the central role of RAF/MEK/ERK activation in ECD. The identification of recurrent NRAS and PIK3CA mutations in ECD further affirms activation of both the PI3K-AKT and RAF/MEK/ERK pathways in a proportion of ECD patients. Although larger series of patients are necessary to determine whether the clinical characteristics of NRAS- or PIK3CA-mutated ECD patients differ from BRAF-mutated or BRAF/NRAS/PIK3CA–wild-type ECD patients, several of the NRAS- and PIK3CA-mutant patients here had multiorgan disease requiring treatment (Table 1). We suspect that ECD patients with RAS mutations may benefit from targeted anti-MEK therapy, as have benefitted RAS mutant metastatic melanoma patients.15-17 Likewise, therapeutic targeting of a PI3K/AKT/mTOR pathway may be important for BRAF–wild-type/PIK3CA-mutant ECD patients. Although pharmacologic targeting of this pathway has been pursued for advanced cancer patients14,18,19 and LCH,20 this treatment approach has only recently been initiated in ECD with a study of sirolimus for ECD patients (ACTRN12613001321730).21 Correlation of clinical response to PI3KCA mutational status may be critical to interpreting results of this ongoing study and in future studies targeting this pathway in ECD.
The online version of this article contains a data supplement.
There is an Inside Blood Commentary on this article in this issue.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank all of the 48 colleagues who sent us samples and/or clinical data from ECD patients, as well as D. Peschaud, M. Bakari, Y. Pothin, and N. Terrones, and for technical contributions to BRAF mutation analyses.
This work was supported by grants from Enfance et Santé, Histiocytose France, Association pour la Recherche et l’Enseignement en Pathologie, INSERM/Registre Maladies Rares, The Erdheim-Chester Disease Global Alliance, and The Geoffrey Beene Cancer Center of Memorial Sloan-Kettering Cancer Center.
Authorship
Contribution: J.-F.E., E.L.D., Z.H.-R., F.C.-A., O.A.-W., and J.H. designed the study, collected the data, contributed to data interpretation, wrote the manuscript, and approved the manuscript; and F.C., D.M.H., E.K., R.R., M.P., S.A., C.G., Z.A., G.F., C.L.G., K.L., M.C., J.-E.K., S.T., P.N., J.D., and V.T. collected the data, contributed to data interpretation, and approved the manuscript.
Conflict-of-interest disclosure: J.-F.E. received honoraria from Roche and GlaxoSmithKline for counseling on patients with melanomas on the diagnosis and/or treatment with BRAF inhibitors. J.H. received honoraria from GlaxoSmithKline and Roche for counseling on targeted treatments of patients with histiocytosis. The remaining authors declare no competing financial interests.
Correspondence: Jean-François Emile, EA4340 & Pathology Department, Ambroise Paré Hospital, 9 Av Ch de Gaulle, F-92104 Boulogne, France; e-mail: jean-francois.emile@uvsq.fr; and Omar Abdel-Wahab, Human Oncology and Pathogenesis Program and Leukemia Service, Department of Medicine, Memorial Sloan-Kettering Cancer Center, 1275 York Ave, New York, NY 10065; e-mail: abdelwao@mskcc.org.
References
Author notes
J.-F.E., E.L.D., O.A.-W., and J.H. contributed equally to this study.