Key Points
HOXA9 parallels blood development, but is restricted to HEP, and diminishes as they differentiate into blood cells.
Functional assays reveal how HOXA9 enhances blood formation by promoting commitment of HEP to CD45+ cells with higher clonogenic potential.
Abstract
The molecular determinants regulating the specification of human embryonic stem cells (hESCs) into hematopoietic cells remain elusive. HOXA9 plays a relevant role in leukemogenesis and hematopoiesis. It is highly expressed in hematopoietic stem and progenitor cells (HSPCs) and is downregulated upon differentiation. Hoxa9-deficient mice display impaired hematopoietic development, and deregulation of HOXA9 expression is frequently associated with acute leukemia. Analysis of the genes differentially expressed in cord blood HSPCs vs hESC-derived HSPCs identified HOXA9 as the most downregulated gene in hESC-derived HSPCs, suggesting that expression levels of HOXA9 may be crucial for hematopoietic differentiation of hESC. Here we show that during hematopoietic differentiation of hESCs, HOXA9 expression parallels hematopoietic development, but is restricted to the hemogenic precursors (HEP) (CD31+CD34+CD45−), and diminishes as HEPs differentiate into blood cells (CD45+). Different gain-of-function and loss-of-function studies reveal that HOXA9 enhances hematopoietic differentiation of hESCs by specifically promoting the commitment of HEPs into primitive and total CD45+ blood cells. Gene expression analysis suggests that nuclear factor-κB signaling could be collaborating with HOXA9 to increase hematopoietic commitment. However, HOXA9 on its own is not sufficient to confer in vivo long-term engraftment potential to hESC-hematopoietic derivatives, reinforcing the idea that additional molecular regulators are needed for the generation of definitive in vivo functional HSPCs from hESC.
Introduction
The great regenerative potential of human stem cells was first revealed by the successful accomplishment of bone marrow transplantation in human patients.1 Since then, hematopoietic stem and progenitor cells (HSPCs) from different human sources are transplanted as a rescue therapy approach into patients suffering from autoimmune and hematologic disorders.2 However, HSPC availability is limited and immunologic compatibility between the donor and recipient remains a major barrier.2 Human embryonic stem cells (hESCs) are characterized by their unlimited proliferation ability and pluripotency (ie, their capacity to produce cells of multiple lineages/tissues representing all 3 embryonic germ layers).3,4 They represent a unique in vitro model for human developmental biology, disease modeling, drug screening, and a potential source for cell replacement strategies.5 The hESCs have been used to study the developmental and cellular mechanisms underlying the emergence of human hematopoietic cells. hESCs generate hematopoietic cells through an intermediate bipotent hemogenic precursor (HEP), capable of differentiating into both endothelial and hematopoietic cells.6-8 Although hESC-derived hematopoietic cells possesses the developmental potential to become HSPCs,6,7,9-11 the efficient generation of adequate numbers of hematopoietic cells capable of in vivo long-term multilineage reconstitution remains a challenge.12-17 Therefore, a better understanding of the intrinsic regulators and signaling pathways driving hematopoietic differentiation of hESC is required. In this context, is key study the role of early hematopoietic regulators that closely mimic the in vivo developmental program of human hematopoietic specification.
A comparative gene expression profile analysis between hESC-derived CD34+ and cord blood (CB)-derived CD34+ cells revealed that HOXA9 is the most differentially expressed gene, being highly expressed in definitive CB-CD34+ cells, but having a low expression in hESC-derived CD34+ cells.12,18 HOXA9 is a homeodomain transcription factor key in hematopoiesis and leukemogenesis.19 It is highly expressed in HSPCs, and is downregulated during differentiation.20 Hoxa9 knockout mice are viable, but they display reduced spleen and thymus size and decreased numbers of granulocytes, lymphocytes, and committed progenitors.21 Hoxa9−/− hematopoietic stem cells (HSCs) show a defective ability to repopulate irradiated recipients in competitive transplantation assays,22 whereas enforced expression of Hoxa9 in bone marrow HSPCs enhances expansion of HSCs and myeloid precursors.23 Furthermore, HOXA9 is frequently overexpressed in human acute myeloid leukemia (AML) and MLL-rearranged acute lymphoblastic leukemia.24-26 Recently, it has been shown that enforced expression of HOXA9, along with ERG2, RORA, SOX4, and MYB provided human pluripotent stem cell (hPSC)-derived hematopoietic cells with short-term engraftment capacity.15 Although these findings establish that HOXA9 plays a key role in hematopoiesis, its function during normal human hematopoietic development remains unknown. Here we show that HOXA9 expression parallels hematopoietic development. Using genetic gain-of-function and loss-of-function studies, we demonstrate that HOXA9, rather than affecting proliferation or survival, robustly enhances hematopoietic differentiation of hESCs by promoting the commitment of HEPs into primitive and total CD45+ blood cells with higher clonogenic potential biased to granulocyte–colony-forming unit (CFU) phenotype.
Material and methods
The hESC culture
H9 (Wicell, Madison, WI) and AND-1 (Spanish Stem Cell Bank) hESC lines were maintained undifferentiated in a feeder-free culture, as previously described27 in Matrigel (BD Biosciences, MA)-coated T25 flasks. HESCs were fed daily with human mesenchymal stem cell-conditioned medium supplemented with 8 ng/mL basic fibroblast growth factor (Miltenyi, Germany). Media was changed daily and cells were split weekly by dissociation with 200 U/mL of collagenase IV (Invitrogen). Approval from the Spanish National Embryo Ethical Committee was obtained to work with hESCs.
Plasmid construction and lentiviral transduction
The Hoxa9 complementary DNA (cDNA) was subcloned in the pRRL-EF1α-PGK-green fluorescent protein (GFP) vector. The following lentivectors were generated: pRRL-EF1α-PGK-GFP (empty vector [EV]) and pRRL-EF1α-Hoxa9-PGK-GFP (HOXA9). Viral particles pseudotyped with VSV-G were generated on 293T cells by calcium-phosphate transfection protocol and concentrated by ultracentrifugation.16 The hESCs were infected overnight on the day of passage with concentrated virus in the presence of polybrene at 8 µg/mL (Sigma-Aldrich). The following day, the viral supernanant was removed and infected hESCs were washed with fresh media and maintained in culture. HOXA9 expression was confirmed at the RNA and protein level. Similarly, pLKO.1 vectors containing the scramble or HOXA9 specific sequences (short-hairpin HOXA9 [shHOXA9]; mission short hairpin RNA (shRNA), Sigma-Adrich) were used to produce viral particles on 293T cells. The hESCs were infected with concentrated viral supernatant as described above. The following day, the viral supernanant was removed and the infected hESCs were washed with fresh media and maintained in culture. Transduced cells were selected with Puromycin (Sigma-Adrich) at 0.5 µg/mL for 15 days.
Hematopoietic differentiation of hESCs
Embryoid body (EB) system.
Undifferentiated hESCs were treated with collagenase IV for 5 minutes, and scraped off from the matrigel. The hESCs were transferred to low-attachment plates and incubated overnight in media composed by knockout-Dulbecco’s modified Eagles medium (Invitrogen) supplemented with 20% non–heat-inactivated fetal bovine serum (FBS) for hESCs (Gibco), 1 mM glutamine, 0.1 mM nonessential amino acids, and 0.1 mM β-mercaptoethanol. The next day (day 1), the EBs were centrifuged and the medium was changed for the same media supplemented with bone morphogenetic protein 4 (BMP-4) (25 ng/mL), Fms-related tyrosine kinase 3 ligand (Flt-3L) (300 ng/mL), stem cell factor (SCF) (300 ng/mL), IL-3 (10 ng/mL), IL-6 (10 ng/mL), and granulocyte-colony stimulating factor (G-CSF) (50 ng/mL),10,27 with medium changes every 4 days.
OP9 coculture system.
OP9 stroma was prepared by plating OP9 cells in gelatin-coated dishes in α-MEM basal medium supplemented with 20% non–heat-inactivated FBS (Gibco). OP9 cells were grown for 8 days to form an overgrown monolayer. The hESC lines grown in Matrigel-coated flasks were prepared as a suspension of small aggregates using collagenase IV followed by gentle scraping in differentiation medium ([DM]; α-Minimum Essential Medium [MEM] basal medium, 10% non–heat-inactivated FBS, 100 mM monothioglycerol, and 50 mg/mL ascorbic acid). One-tenth of this suspension was plated on 8-day overgrown OP9 stroma in 10 mL of DM. On the next day, the media was replaced by 20 mL of DM to remove unattached cells. From day 3 of coculture, a half-volume media change was performed every other day.11,17
Real-time reverse transcriptase-polymerase chain reaction
Total RNA was extracted from hESCs-OP9 cocultures, differentiating EBs, HEPs, and CD45+ hematopoietic cells using the total RNA Purification Kit (Norgen Biotek, Canada). After DNAase treatment, first-strand cDNA synthesis was performed using the First-Strand cDNA Synthesis Kit (Amersham). The resulting cDNA was analyzed for HOXA9 expression by using Brilliant II SYBR Green QPCR master mix on an Mx3005P System (Stratagene). The primers used were: HOXA9:5′-CCACGCTTGACACTCACACT-3′ and 5′-TCGCTGGGTTGTTTTTCTCT-3′, glyceraldehyde-3-phosphate dehydrogenase:5′-TGCACCACCAACTGCTTAGC-3′ and 5′-GGCATGGACTGTGGTCATGAG-3′. Brachyury:5′-ATGAGCCTCGAATCCACATAGT-3′ and 5′-TCCTCGTTCTGATAAGCAGTCA-3′; MIXL1:5′-GGATCCAGGTATGGTTCCAG-3′ and 5′-GGAGCACAGTGGTTGAGGAT-3′; OCT4:5′-TCTGCAGAAAGAACTCGAGCAA-3′, and 5′-AGATGGTCGTTTGGCTGAACAC-3′; NANOG:5′-TGCAGTTCCAGCCAAATTCTC-3′, and 5′-CCTAGTGGTCTGCTGTATTACATTAAGG-3′; SOX2:5′-CCCCCGGCGGCAATAGCA-3′, and 5′-TCGGCGCCGGGGAGATACAT-3′.
Western blot analysis
The hESC cultures (5 × 105 cells) were dissociated with trypsin-EDTA for 10 minutes and the single cell suspension was subsequently lysed in 50 µL of radioimmunoprecipitation assay (RIPA) buffer containing protease inhibitors for 30 minutes. There was 30 µL of the whole-cell lysate mixed with 10 µL of loading buffer, resolved on 8% sodium dodecyl sulfate polyacrylamide gel electrophoresis and transferred to polyvinylidene difluoride (PVDF) membranes using a semi-dry transfer system. The membrane was blocked for 30 minutes with the blocking reagent chemiluminiscence western blotting kit (Roche). For HOX9 detection, the membrane was incubated overnight with anti-HOXA9 antibody (1:100 dilution; Upstate) followed by 2 hours incubation with anti-mouse horseradish peroxidase (HRP) (Millipore). The HOXA9 protein (36 KDa) was detected as above. Anti-β-actin antibody detection (clone AC-15; Sigma-Aldrich) was used as a loading control.
FACS analysis
The hESC colonies were disassociated with trypsin-EDTA (Invitrogen) and the cell suspension was stained with phycoerythrin-conjugated TRA-1-60, TRA-1-81, SSEA3, and SSEA4 antibodies (BD Biosciences).28 After washing, cells were stained with 7-amino-actinomycin D (7AAD; BD Biosciences). Differentiating EBs at days 7, 10, 15, and 22 of development were dissociated using collagenase B (Roche) for 2 hours at 37°C followed by 10 minutes incubation at 37°C with Cell Dissociation Buffer (Invitrogen). A single cell suspension was stained with anti-CD34−PECy7, anti-CD31−PE, anti-CD45−APC (Miltenyi), and 7-AAD. The hESC-OP9 cocultures were treated for 1 hour with collagenase IV followed by 20 minutes with Tryple, dissociated by pipetting, and filtered through a 70 μm strainer. Single cell suspensions were stained as above. Live cells identified by 7-AAD exclusion were analyzed using a fluorescence-activated cell sorter (FACS)-Canto II cytometer equipped with the FACS Diva analysis software (BD Biosciences). The proportion of HEPs (CD31+CD34+CD45−), primitive blood cells (CD34+CD45+), and total blood cells (CD45+) was analyzed within the GFP positive (GFP+) hESC-derived cells.
Hemogenic progenitors OP9 cocultures
Day 10 EBs were dissociated as described above, and the HEP population was purified by magnetic bead separation using the human CD34 Microbead kit and the AutoMACS Pro separator (Miltenyi) by following the manufacturer’s instructions. Purity of the CD34 fraction was assessed by flow cytometry and only purities >95% were used. Purified HEPs (3 × 104) were then plated on OP9 stroma on 6-well plates using DM supplemented with hematopoietic cytokines (300 ng/mL stem cell factor, 300 ng/mL Flt3L, 10 ng/mL IL-3, 10 ng/mL IL-6, and 50 ng/mL granulocyte–stem cell factor). At day 4 of coculture, the cells were harvested and analyzed by flow cytometry as described above. The proportion of CD41a+CD235a early hematopoietic progenitors within the HEP population was analyzed using CD41a−PECy7 and CD235a eFuor450 antibodies using an FACSCanto cytometer.
CFU assays
Human clonogenic progenitor assays were performed by plating 5 × 104 cells from differentiating EBs or OP9-hESC cocultures into methylcellulose H4436 (Stem Cell Technologies, Canada). Localized clusters of >50 cells were counted between days 8 and 14 using standard morphological criteria.29
Cell cycle analysis of hemogenic progenitors
Day 10 EV- and HOXA9-EBs were dissociated, harvested, fixed in 70% ice-cold ethanol, and stored overnight at −20°C. The next day, the cells were washed with phosphate-buffered saline and incubated with anti-CD31− fluorescein isothiocyanate and anti-CD34− fluorescein isothiocyanate (Miltenyi) for 15 minutes. After washing, the cells were resuspended in buffer containing 50 μg/mL of propidium iodide and 100 μg/mL of RNase. Cell cycle distribution was acquired using an FACSCanto II cytometer and analyzed with Modfit software (Verity Software, Topsham, ME).16
Apoptosis assay
The apoptotic status of EV- and HOXA9-CD34+CD45− HEPs was assessed using the Annexin-V apoptosis detection kit (BD Biosciences), according to the manufacturer’s instructions. Briefly, day 10 EBs were dissociated and washed twice before staining with anti-CD34−PECy7 (Miltenyi), Annexin-V-PE, and 7-AAD. Apoptotic cells were detected by gating the Annexin V+/7-AAD+ fraction.
In vivo teratoma formation
In vivo pluripotency was analyzed as previously described.30 In brief, hESCs were resuspended in phosphate-buffered saline plus 30% Matrigel (BD Biosciences) and implanted subcutaneously in the flank of 8-week-old nonobese diabetic/severe combined immunodeficiency IL2Rγ−/− mice (NOD scid gamma mice; Jackson Laboratories, Bar Harbor, MA). Teratoma growth was monitored by palpation and mice were killed 8 weeks after implantation. Teratomas were fixed and embedded in paraffin. Hematoxylin and eosin staining was performed on paraffin-embedded teratoma sections.
Mice transplantation and analysis of engraftment
CB-derived CD34+ cells (3 × 104 cells/50 µL), as well as EV-EB or HOXA9-EB day 15 hematopoietic cells were transplanted intrahepatically (5 × 105 cells/50 µL) into newborn irradiated (2.4 Gy) NOD scid gamma mice, housed under sterile conditions, as described in detail.31 Mice health was monitored throughout the entire experiment. Mice were killed 6 to 8 weeks after transplantation, and BM, spleen, liver, and peripheral blood were collected and analyzed for human hematopoietic engraftment using anti-HLA-ABC-PE and anti-CD45−APC (BD Biosciences) as previously described.31,32 The Animal Care Committee of the University of Granada approved all mouse protocols.
Gene expression profiling and data analysis
CD31+CD45− HEPs were purified by FACS from day 15 differentiating EV- and HOXA9-EBs. The RNA was isolated and its quality checked in the Agilent 2100 Bioanalyzer. RNA was labeled (Quick-Amp Labeling kit, Agilent) with Cy3 and hybridized with the Gene Expression Hybridization kit to a Whole Human Genome Microarray (G4112F; Agilent) following the manufacturer’s instructions. Two independent samples per condition were labeled and hybridized. A gene was considered differentially expressed in HOXA9-EPs when it was greater than twofold upregulated/downregulated (P value < .01) compared with EV-HEPs. Analysis of gene functions was performed using the Ingenuity Pathway Analysis software (Ingenuity Systems Inc., Redwood City, CA). Data are available at Gene Expression Omnibus (GEO) under accession number GSE61017.
Statistical analysis
All data are expressed as mean ± standard error of the mean. Statistical comparisons were performed (GraphPad Prism program) with nonparametric test (Mann-Whitney U test), 2-tailed t test P value (95% confidence interval). Statistical significance was defined as a P value <0.05, where *P < .05, **P < .01 and ***P < .001, where applicable.
Results
HOXA9 is expressed in HEPs during hematopoietic differentiation of hESCs
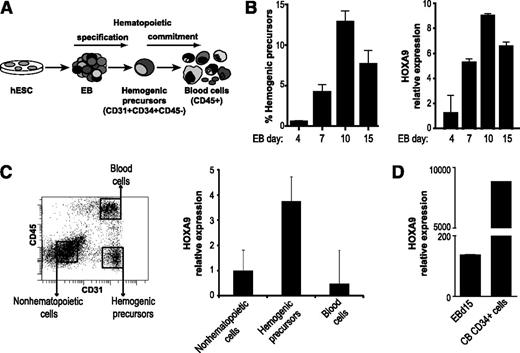
To dissect the role of HOXA9 during human embryonic hematopoiesis, we first analyzed HOXA9 expression kinetics during EB hematopoietic differentiation of the hESC lines H9 and AND1. During hematopoietic development, hESCs are first specified into HEPs (CD45−CD31+), which subsequently commit into CD45+ hematopoietic cells (Figure 1A). HOXA9 is detected by day 4 of EB development and its expression gradually increases, paralleling the emergence of the HEPs population (Figure 1B). To further determine which population within the differentiating EBs express HOXA9, its expression was analyzed in HEPs, hematopoietic cells (CD45+), and nonhematopoietic/remaining cells (CD45−CD31−) FACS-purified at day 15 of EB differentiation (Figure 1C). Using the nonhematopoietic cells population as a reference, we found that HOXA9 expression is highly enriched in HEPs and its expression diminished as the HEPs undergo blood differentiation (Figure 1C). A previous transcriptome analysis identified HOXA9 as the most upregulated gene in CB-derived CD45+CD34+ HSPCs as compared with hESC-derived CD45+CD34+ cells.12 Similarly, we confirmed that HOXA9 expression is 60-fold lower in differentiating day 15 EBs than in CB-CD34+ cells (Figure 1D). These results suggest that HOXA9 may be a regulator of the hematopoietic differentiation of hESCs.
HOXA9 is expressed in the HEPs during differentiation of hESC. (A) Schema of hESC hematopoietic differentiation system based on EB formation. (B) Kinetics of the emergence of HEPs during the hematopoietic differentiation protocol (left). Time course expression of HOXA9 during EB hematopoietic development (right). The expression of endogenous HOXA9 correlates with the HEPs emergence throughout EB differentiation. (C) Quantitative polymerase chain reaction analysis in isolated cell populations demonstrating that HOXA9 expression is enriched in the HEP population. Gene expression is shown relative to nonhematopoietic cells. (D) A 60-fold reduction of HOXA9 expression on day 15 EB in comparison with definitive CB-CD34+ hematopoietic cells. Relative expression is shown normalized to undifferentiated hESCs. Data represent mean ± standard error of the mean for 3 independent experiments.
HOXA9 is expressed in the HEPs during differentiation of hESC. (A) Schema of hESC hematopoietic differentiation system based on EB formation. (B) Kinetics of the emergence of HEPs during the hematopoietic differentiation protocol (left). Time course expression of HOXA9 during EB hematopoietic development (right). The expression of endogenous HOXA9 correlates with the HEPs emergence throughout EB differentiation. (C) Quantitative polymerase chain reaction analysis in isolated cell populations demonstrating that HOXA9 expression is enriched in the HEP population. Gene expression is shown relative to nonhematopoietic cells. (D) A 60-fold reduction of HOXA9 expression on day 15 EB in comparison with definitive CB-CD34+ hematopoietic cells. Relative expression is shown normalized to undifferentiated hESCs. Data represent mean ± standard error of the mean for 3 independent experiments.
Enforced expression of HOXA9 preserves hESC pluripotency
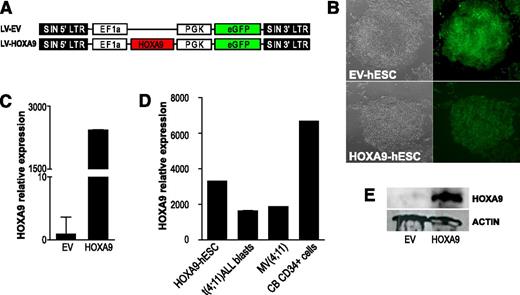
To investigate the effect of HOXA9 in the hematopoietic differentiation of hESCs, transgenic HOXA9-expressing H9 and AND1 hESC lines were generated. HoxA9 cDNA was subcloned in a lentiviral vector expressing GFP as reporter (Figure 2A). The hESCs were transduced with either the EV or the HoxA9-expressing vector (HOXA9). More than 90% of the cells were transduced as assayed by fluorescence microscopy (Figure 2B) and FACS for GFP expression (Figure 4A). Stable ectopic expression of Hoxa9 was confirmed by quantitative reverse-transcription polymerase chain reaction (Figure 2C-D) and WB (Figure 2E). More than 20 weeks after transduction, EV- and HOXA9-hESC cultures were analyzed for pluripotency markers and functional assays. HOXA9-hESCs retained the expression of both the hESC-associated surface markers SSEA-3, SSEA-4, TRA-1-60, and TRA-1-81 (supplemental Figure 1A), and the pluripotency transcription factors OCT4, NANOG, SOX2, and REX-1 (supplemental Figure 1B). Functionally, EV- and HOXA9-hESCs formed teratomas with identical efficiency (100%), latency (50-55 days), and histologic composition (supplemental Figure 1C). Together, these data indicate successful and stable ectopic expression of Hoxa9 in hESCs, which does not interfere with hESC pluripotency.
Enforced expression of Hoxa9 in hESCs. (A) Schematic representation of the lentiviral vectors used. (B) Bright field and fluorescence images of colonies of empty vector (EV)-expressing and HOXA9-expressing hESCs. (C) Quantitative polymerase chain reaction showing Hoxa9 transcript overexpression in hESCs. (D) Relative Hoxa9 expression levels reached in HOXA9-hESCs as compared with the endogenous HOXA9 expression in CB-CD34+, t(4;11) leukemic blasts and the leukemic cell line MV(4;11). Relative expression is shown normalized to EV-hESCs. (E) Western blot analysis demonstrating ectopic Hoxa9 protein expression in hESCs.
Enforced expression of Hoxa9 in hESCs. (A) Schematic representation of the lentiviral vectors used. (B) Bright field and fluorescence images of colonies of empty vector (EV)-expressing and HOXA9-expressing hESCs. (C) Quantitative polymerase chain reaction showing Hoxa9 transcript overexpression in hESCs. (D) Relative Hoxa9 expression levels reached in HOXA9-hESCs as compared with the endogenous HOXA9 expression in CB-CD34+, t(4;11) leukemic blasts and the leukemic cell line MV(4;11). Relative expression is shown normalized to EV-hESCs. (E) Western blot analysis demonstrating ectopic Hoxa9 protein expression in hESCs.
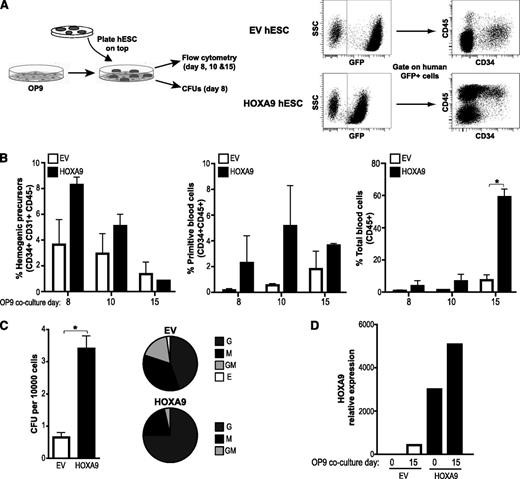
HOXA9 enhances hematopoietic differentiation of hESCs in OP9 coculture. (A) Schema of hESC hematopoietic differentiation system based on OP9 coculture and endpoint analysis (left panel). Representative flow cytometry displaying how HEPs (CD45−CD31+CD34+) and blood cells (CD45+/CD45+CD34+) are identified within the human GFP+ population (right panel). (B) Enforced expression of Hoxa9 enhances the differentiation into primitive blood cells (CD34+CD45+) and total blood cells (CD45+). (C) CFU read out from day 15 EBs confirms a significantly increase in hematopoietic clonogenic potential of the HOXA9-hESC blood derivatives. Scoring of CFU reveals a skew toward G-CFU in Hoxa9-transduced progenitors (right pie charts). (D) Ectopic Hoxa9 remains highly expressed throughout the differentiation. Relative expression is shown normalized to undifferentiated hESCs. Data are presented as mean ± standard error of the mean for 3 independent experiments. G, granulocyte; M, monocyte; GM, granulocyte-macrophage; E, erythroid.
HOXA9 enhances hematopoietic differentiation of hESCs in OP9 coculture. (A) Schema of hESC hematopoietic differentiation system based on OP9 coculture and endpoint analysis (left panel). Representative flow cytometry displaying how HEPs (CD45−CD31+CD34+) and blood cells (CD45+/CD45+CD34+) are identified within the human GFP+ population (right panel). (B) Enforced expression of Hoxa9 enhances the differentiation into primitive blood cells (CD34+CD45+) and total blood cells (CD45+). (C) CFU read out from day 15 EBs confirms a significantly increase in hematopoietic clonogenic potential of the HOXA9-hESC blood derivatives. Scoring of CFU reveals a skew toward G-CFU in Hoxa9-transduced progenitors (right pie charts). (D) Ectopic Hoxa9 remains highly expressed throughout the differentiation. Relative expression is shown normalized to undifferentiated hESCs. Data are presented as mean ± standard error of the mean for 3 independent experiments. G, granulocyte; M, monocyte; GM, granulocyte-macrophage; E, erythroid.
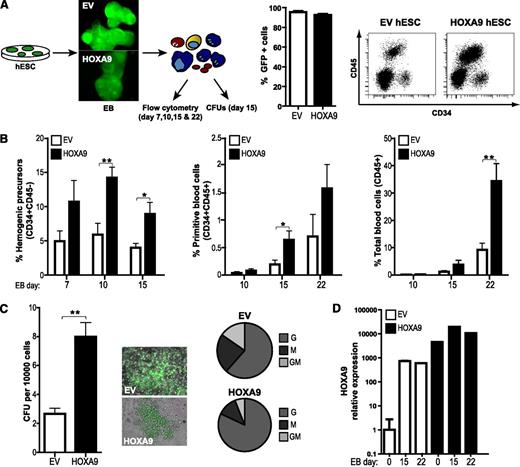
Overexpression of HOXA9 during EB development leads to an increased hematopoietic differentiation. (A) Schematic of the EB-based hematopoietic differentiation and endpoint analyses (left panel). More than 90% of the cells within both empty vector (EV)-hESC/EB and HOXA9-hESC/EBs cultures were transduced (GFP+; middle panel). Representative flow cytometry displaying how HEPs (CD34+CD31+CD45−) and blood cells (CD45+) are identified (right panel). (B) HOXA9 robustly promotes differentiation into HEPs, primitive blood cells (CD34+CD45+), and total blood cells (CD45+). (C) Hoxa9-expressing blood progeny displays threefold higher clonogenic potential (left panel). Colonies retained GFP expression after the CFU assay, and scoring of CFU reveals a skew toward G-CFU in Hoxa9-transduced progenitors (middle and right panels). (D) Quantitative PCR analysis demonstrating that exogenous levels of Hoxa9 remain high throughout EB differentiation. Data represent mean ± standard error of the mean for 6 independent experiments.
Overexpression of HOXA9 during EB development leads to an increased hematopoietic differentiation. (A) Schematic of the EB-based hematopoietic differentiation and endpoint analyses (left panel). More than 90% of the cells within both empty vector (EV)-hESC/EB and HOXA9-hESC/EBs cultures were transduced (GFP+; middle panel). Representative flow cytometry displaying how HEPs (CD34+CD31+CD45−) and blood cells (CD45+) are identified (right panel). (B) HOXA9 robustly promotes differentiation into HEPs, primitive blood cells (CD34+CD45+), and total blood cells (CD45+). (C) Hoxa9-expressing blood progeny displays threefold higher clonogenic potential (left panel). Colonies retained GFP expression after the CFU assay, and scoring of CFU reveals a skew toward G-CFU in Hoxa9-transduced progenitors (middle and right panels). (D) Quantitative PCR analysis demonstrating that exogenous levels of Hoxa9 remain high throughout EB differentiation. Data represent mean ± standard error of the mean for 6 independent experiments.
Augmented hematopoietic differentiation from HOXA9 hESCs
Once we confirmed that enforced Hoxa9 expression in hESCs is compatible with pluripotency and homeostasis, we studied its developmental impact throughout embryonic hematopoiesis. Hematopoietic differentiation from the EV- and HOXA9-hESC lines H9 and AND1 was assessed using both OP9 coculture11,17 (Figure 3A) and EB formation10,27 (Figure 4A and supplemental Figure 2). Regardless of the hematopoietic differentiation protocol used (Figures 3B and 4B and supplemental Figure 2), enforced expression of Hoxa9 consistently enhanced the production of HEPs (CD45−CD31+; between twofold and fivefold), and more robustly, primitive blood cells (CD45+CD34+; between twofold and 15-fold) and total blood cells (CD45+; between twofold and eightfold) (Figures 3B and 4B and supplemental Figure 2A). It has been described the existence of CD45−CD34+CD41a+CD235a+ primitive hematopoietic progenitors within the HEP population.33 As shown in supplemental Figure 2B, HOXA9 overexpression also increased (up to 12-fold) the proportion of primitive CD45−CD34+CD41a+CD235+cells. Functionally, we tested the clonogenic potential of hematopoietic progenitors derived from either day 8 of OP9-hESC cocultures (Figure 3C) or day 15 EB development (Figure 4C), measured by the ability to form CFUs in semisolid cultures. Hematopoietic cells generated from HOXA9-hESCs display a significantly increased (fivefold and threefold, respectively) clonogenic potential. Importantly, CFU scoring revealed a HOXA9-mediated skew toward granulocyte colony forming units (G-CFU) phenotype (Figures 3C and 4C).
To ensure that the observed developmental effect is related to Hoxa9 overexpression, we confirmed by quantitative reverse-transcription polymerase chain reaction stable transgene expression upon differentiation of HOXA9-hESCs in OP9 (Figure 3D) and EBs (Figure 4D). Together, these data confirm that HOXA9 positively regulates human embryonic hematopoiesis.
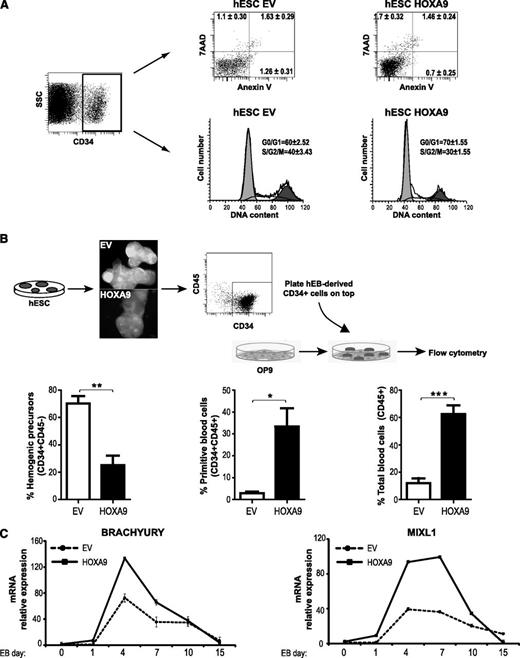
The increased hematopoietic outcome of HOXA9-hESCs may be the consequence of HOXA9-mediated proliferation/survival of the emerging HEPs. In fact, HOXA9 has been associated to cell survival and proliferation of leukemic cells.34 To address this, apoptosis and cell cycle distribution were analyzed within the HEP population. No differences in the proportion of apoptotic HEPs (measured by 7AAD exclusion and Annexin V staining) were detected between HOXA9 and EV HEPs (2.2% vs 2.9%; Figure 5A), and HOXA9-HEPs cycled slightly less than EV-HEPs (30% vs 40%; Figure 5A). Therefore, next, we wanted to elucidate whether HOXA9 could be mediating the commitment of HEPs toward hematopoietic cells. For this, HEPs were magnetic-activated cell sorting–purified from day 10 EV- and HOXA9-EBs and plated on OP9 stroma for hematopoietic differentiation. After 4 days, ∼70% of the EV-cells remained as HEPs in the OP9 culture. In contrast, only ∼20% of the HOXA9-expressing HEPs remained in the OP9 coculture, while the majority of them differentiated into CD45+ and CD45+CD34+ cells (Figure 5B). In addition, the expression kinetics during EB development of the master early mesodermal transcription factors, Brachyury and MIXL1, indicate that HoxA9 contributes to hematopoietic commitment by inducing mesodermal specification from very early-stage (Figure 5C). Together, these data indicate that HOXA9 induces mesodermal and HEP specification and subsequent promotion of blood commitment, rather than selective proliferation or survival of HEPs.
HOXA9 promotes specification rather than survival or proliferation of HEPs. (A) Apoptosis and cell cycle analysis on empty vector (EV)- or HOXA9-hESC–derived HEPs. Similar numbers of dead cells (Annexin V+/7AAD+), apoptosis-undergoing cells (Annexin V+), and cycling cells (S, G2, and M) were found in EV-HEPs versus HOXA9-HEPs. (B) HEPs were purified at day 10 of differentiation and cocultured with OP9 for 4 days. HOXA9-HEPs differentiated faster toward CD34+CD45+, and CD45+ hematopoietic cells. Data represent mean ± standard error of the mean (SEM) for 4 independent experiments. (C) Gene expression kinetics of the mesendodermal transcription factors Brachyury and MixL1 during EV- and HOXA9-EB hematopoietic differentiation. Data represent mean ± SEM for 2 independent experiments. SSC, side scatter.
HOXA9 promotes specification rather than survival or proliferation of HEPs. (A) Apoptosis and cell cycle analysis on empty vector (EV)- or HOXA9-hESC–derived HEPs. Similar numbers of dead cells (Annexin V+/7AAD+), apoptosis-undergoing cells (Annexin V+), and cycling cells (S, G2, and M) were found in EV-HEPs versus HOXA9-HEPs. (B) HEPs were purified at day 10 of differentiation and cocultured with OP9 for 4 days. HOXA9-HEPs differentiated faster toward CD34+CD45+, and CD45+ hematopoietic cells. Data represent mean ± standard error of the mean (SEM) for 4 independent experiments. (C) Gene expression kinetics of the mesendodermal transcription factors Brachyury and MixL1 during EV- and HOXA9-EB hematopoietic differentiation. Data represent mean ± SEM for 2 independent experiments. SSC, side scatter.
HOXA9 silencing abrogates hematopoietic differentiation of hESCs
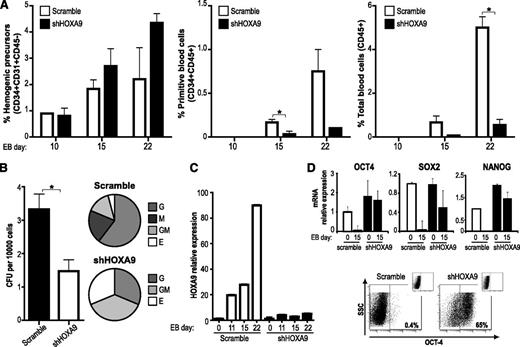
To further demonstrate that endogenous HOXA9 modulates human embryonic hematopoietic specification, we undertook a loss-of-function approach in which endogenous HOXA9 expression was stably inhibited with shHOXA9-expressing lentiviral vectors.17 HOXA9 knockdown did not affect hESC pluripotency properties (supplemental Figure 3).35 Transgenic shHOXA9-hESCs displayed a much less efficient hematopoietic differentiation as compared with control scramble-hESCs (Figure 6A). HOXA9 knockdown had no significant effect on HEPs generation, but it strongly (up to fivefold) abolished both primitive (CD45+CD34+) and total (CD45+) blood cells during EB development (Figure 6A), and by day 22 of EB differentiation shHOXA9-hESC hardly produced hematopoietic cells in comparison with scramble short hairpin RNA-hESCs (Figure 6). As a result, hematopoietic progenitors derived from shHOXA9-hESCs showed a ∼2.5-fold decrease on hematopoietic clonogenic potential as compared with the scramble control (Figure 6B). In contrast to HOXA9 overexpression, silencing of HOXA9 skewed the CFU phenotype toward erythroid and granulocyte-macrophage colony forming unit phenotype at the expense of M- and G-CFUs (Figure 6B). Quantitative reverse transcriptase-polymerase chain reaction revealed that endogenous HOXA9 levels were highly reduced (∼90%) upon shHOXA9 expression, validating the observed HOXA9 knockdown-mediated hematopoietic developmental defects (Figure 6C). Interestingly, after 15 days of hematopoietic differentiation, shHOXA9 EBs retain the expression of the pluripotent transcription factors OCT4, SOX2, and NANOG (Figure 6D). Flow cytometric analysis confirmed that shHOXA9 day15 EBs are composed by OCT4+ cells (Figure 6D). These data indicate that HOXA9 knockdown abrogates hESC hematopoietic differentiation by retaining a pluripotency identity/gene expression profile.
HOXA9 silencing abrogates hematopoietic differentiation of hESC. (A) Hematopoietic differentiation of hESCs transduced with an irrelevant short hairpin RNA sequence (scramble) or 3 different short hairpin RNA specific sequences for HOXA9 (shHOXA9). (B) CFU read out from day 15 EBs confirming greater than a twofold decrease of hematopoietic progenitor potential in short hairpin (shHOXA9) cells. Scoring of CFUs revealed a skewed differentiation toward erythroid lineage in shHOXA9 progenitors. (C) Quantitative reverse-transcription polymerase chain reaction analysis showing a highly reduced HOXA9 expression throughout the differentiation in shHOXA9 EBs. Data represent mean ± SEM for 3 independent experiments. (D) Quantitative reverse-transcription polymerase chain reaction showing the expression of the pluripotency factors OCT4, SOX2, and NANOG in scramble- and shHOXA9-hEBs after 15 days of hematopoietic differentiation (upper panel). Flow cytometry confirming that shHOXA9-EBs are composed by OCT4+ cells after 15 days of hematopoietic differentiation (lower panel). mRNA, messenger RNA.
HOXA9 silencing abrogates hematopoietic differentiation of hESC. (A) Hematopoietic differentiation of hESCs transduced with an irrelevant short hairpin RNA sequence (scramble) or 3 different short hairpin RNA specific sequences for HOXA9 (shHOXA9). (B) CFU read out from day 15 EBs confirming greater than a twofold decrease of hematopoietic progenitor potential in short hairpin (shHOXA9) cells. Scoring of CFUs revealed a skewed differentiation toward erythroid lineage in shHOXA9 progenitors. (C) Quantitative reverse-transcription polymerase chain reaction analysis showing a highly reduced HOXA9 expression throughout the differentiation in shHOXA9 EBs. Data represent mean ± SEM for 3 independent experiments. (D) Quantitative reverse-transcription polymerase chain reaction showing the expression of the pluripotency factors OCT4, SOX2, and NANOG in scramble- and shHOXA9-hEBs after 15 days of hematopoietic differentiation (upper panel). Flow cytometry confirming that shHOXA9-EBs are composed by OCT4+ cells after 15 days of hematopoietic differentiation (lower panel). mRNA, messenger RNA.
HOXA9 is not sufficient to confer hESC-hematopoietic derivatives in vivo engraftment potential
Next, we analyzed whether overexpression of HOXA9 suffices to endow with in vivo engraftment capacity to the hESC-derived HOXA9-expressing hematopoietic cells. 5 × 105 hESC-derived EV and HOXA9 hematopoietic cells were intrahepatically transplanted into newborn NSG mice.31,32 Although HOXA9 regulates hematopoietic differentiation in vitro, it did not confer on its own in vivo function/engraftment potential to hESC-derived hematopoietic cells (supplemental Figure 4). In accordance with this, a recent report described that other factors such as ERG, RORA, SOX4, and MYB, along with HOXA9 are necessary to confer short-term engraftment potential to EB-derived hematopoietic cells.15
Transcriptional changes underlie the HOXA9-mediated hematopoietic differentiation of hESC
To identify patterns of gene expression that could explain (at the molecular level) the developmental impact of HOXA9 in hematopoietic commitment of hESCs, we next performed gene expression profiles (GEP) in FACS-purified EV- and HOXA9-HEPs. There were 1855 probes differentially expressed in HOXA9- vs EV-HEPs (regulation greater than or equal to twofold up/down; P < .01); 985 probes (53%) were upregulated and 870 probes (47%) were downregulated in HOXA9-HEPs as compared with EV-HEPs (supplemental Table 1). We analyzed the altered genes using the GeneCodis public web-based tool.36 Among the top 10 most significant gene ontology biological processes enriched in the HOXA9-upregulated genes, we found “signal transduction,” “immune response,” “inflammatory response,” and interestingly, “negative regulation of proliferation” (supplemental Figure 5A), supporting the slight decrease in cycling HOXA9-HEPs (Figure 5A). In contrast, among the top 10 most significant gene ontology biological processes enriched in the HOXA9-downregulated genes they included “nervous system development” and “skeletal system development,” suggesting that HOXA9 may facilitate blood commitment by inhibiting alternative differentiation pathways (supplemental Figure 5B).
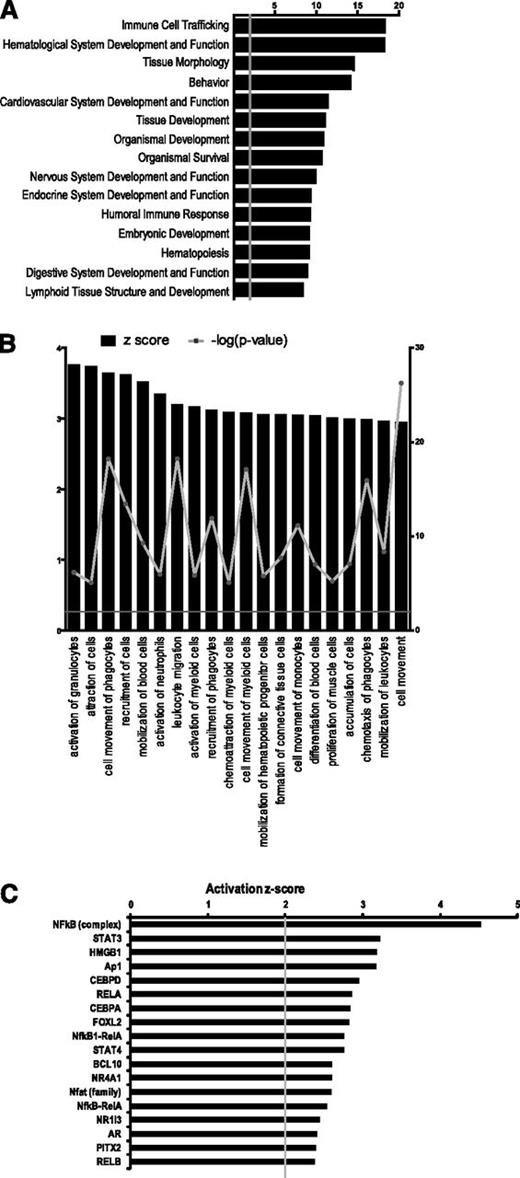
The genes differentially expressed in HOXA9- vs EV-HEPs were then analyzed at the functional level using the IPA software. IPA analysis revealed “immune cell trafficking” and “hematological system development and function” as the top 2 biological functions altered by HOXA9 expression (Figure 7A). The IPA software allows the prediction of activated and inhibited biofunctions based on z score value. We found that within the top 20 predicted activated biofunctions (z score >3) in HOXA9-HEPs, the majority were related to blood cell function and cell mobilization (Figure 7B). Interestingly, previous microarray analyses by McKinney-Freeman et al.37 also found a relevant role for cell migration in HSCs of aorta-gonad mesonephros during hematopoietic development.
GEP in HOXA9-expressing HEPs. (A) Top 15 biological functions of genes differentially expressed in HOXA9-HEPs compared with empty vector-HEPs, ranked by P value. The top 2 biofunctions in HOXA9-HEPs are “immune cell trafficking” and “hematological system development and function.” (B) The top 20 predicted activated biofunctions (z score >3) in HOXA9-HEPs. Z score: black bars, left Y-axis; -log (P value): filled gray circle with gray line, right Y-axis. (C) Upstream transcription factor regulators predicted to be activated in HOXA9-HEPs (z score >2).
GEP in HOXA9-expressing HEPs. (A) Top 15 biological functions of genes differentially expressed in HOXA9-HEPs compared with empty vector-HEPs, ranked by P value. The top 2 biofunctions in HOXA9-HEPs are “immune cell trafficking” and “hematological system development and function.” (B) The top 20 predicted activated biofunctions (z score >3) in HOXA9-HEPs. Z score: black bars, left Y-axis; -log (P value): filled gray circle with gray line, right Y-axis. (C) Upstream transcription factor regulators predicted to be activated in HOXA9-HEPs (z score >2).
To identify transcriptional regulators that could explain the gene expression pattern produced by HOXA9 in HEPs, we looked for upstream regulators using the IPA software. C/EBPα, a master regulator of granulopoiesis38,39 was predicted to be activated in HOXA9-HEPs (Figure 7C), in line with the HOXA9-mediated skew toward G-CFU phenotype (Figures 3C and 4C). It is worth mentioning that 8 of the top 20 upstream regulators predicted to be activated in HOXA9-HEPs participate in the nuclear factor (NF)-κB pathway. Five are members of the NF-κB pathway itself (NF-κB, RELA, NF-κB-RelA, NF-κB1-RelA, RELB), and 3 (STAT3, HGMB1 and BCl10) are cofactors/activators of the NF-κB pathway (Figure 7C). Regulation of NF-κB pathway is associated with hESC differentiation40 and HSC homeostasis and differentiation.41 Furthermore, high expression of components of the NF-κB pathway is a distinctive molecular feature of somatic CD34+ HSPCs42 and is related to primitive HSC state,43 suggesting that NF-κB signaling may cooperate with HOXA9 in the enhanced hematopoietic differentiation.
Discussion
Improving differentiation of hPSCs into HSPCs capable of long-term, multilineage engraftment in vivo is key for their potential use in regenerative medicine, drug screening, and disease-modeling applications. Generation of adequate numbers of functional HSPCs from hPSCs relies on our understanding of the intrinsic factors and microenvironment cues underlying early hematopoietic development. Previous gene expression profile studies identified HOXA9 as the most downregulated gene in hESC-derived HSPCs as compared with somatic (CB-derived) CD34+ HSPCs,12,18 suggesting that the absence of HOXA9 may underlie, at least in part, the poor in vitro hematopoietic differentiation of hPSCs and the lack of in vivo functional hPSC-derived hematopoiesis.
HOXA9 has a crucial role in blood cell development.24,26Hoxa9 knockout mice display an impaired hematopoiesis21 and Hoxa9−/− HSPCs show a defective ability to repopulate irradiated recipients in competitive transplantation assays.22 However, the role of HOXA9 during early human hematopoietic development remains elusive. Here, we analyzed the contribution of HOXA9 to human embryonic hematopoietic differentiation using hESCs as a model. Endogenous HOXA9 expression parallels hESCs hematopoietic specification, as it is highly enriched in the HEP population and subsequently diminishes as they differentiate into CD45+ blood cells. This is in line with previous reports studying somatic hematopoiesis in which HOXA9 is highly expressed in HSPCs and is then downregulated upon differentiation.20 Enforced expression of Hoxa9 in hESCs robustly promoted differentiation into primitive and total blood cells with higher clonogenic potential. Importantly, these HOXA9-mediated effects were confirmed using both the EB and OP9 differentiation systems. HOX genes are potential regulators of HSC self-renewal and differentiation.44 In human embryonic hematopoiesis, HOXA9 did not confer proliferative or survival advantage to HEPs, but it promotes blood lineage commitment once HEPs are generated. In addition, the expression kinetics of Brachyury and MIXL1 indicate that HoxA9 contributes to hematopoietic commitment by inducing mesodermal specification from very early stage of hematopoietic differentiation. On the other hand, HOXA9 silencing does not affect the specification of HEPs, but it abolishes subsequent differentiation of HEPs into blood, indicating that HOXA9 may have a key role in the process of the hematopoietic transition from HEPs. Interestingly, upon hematopoietic differentiation shHOXA9-EBs retain the expression of OCT4, SOX2, and NANOG, and 65% of the shHOXA9-EB cells are OCT4+, suggesting that HOXA9 knockdown abrogates hESC hematopoietic differentiation by retaining a pluripotency identity/gene expression profile.
HOXA9 silencing induced a robust skew toward erythroid CFU phenotype. Indeed, HOXA9 has been reported to repress the erythroid differentiation program of somatic CD34+ cells,45 and therefore it is plausible that HOXA9 downregulation removes this blockade on erythroid differentiation. Conversely, HOXA9 overexpression skewed blood progenitor clonogenic potential toward a granulocyte phenotype, in line with previous work in mouse. Hoxa9−/− mice are deficient in the production of mature granulocytes,21 and Hoxa9 overexpression in mouse BM cells enhanced granulopoiesis due to a significant increase in their progenitor cell numbers.23 Interestingly, in our microarray analysis, C/EBPα was predicted to be one of the upstream regulator responsible for the changes in gene expression due to Hoxa9 overexpression. C/EBPα has been identified as master regulator of granulocytic development.38,39 It has also been recently demonstrated that C/EBPα regulates HOXA9 during leukemic initiation in AML.46 The observed skew toward granulopoiesis could be due to an increased activity of C/EBPα gene in HOXA9-hESC.
HOXA9 expression on its own is not sufficient to endow hESC-derived hematopoietic cells with in vivo reconstituting function. Doulatov et al.15 reported that enforced expression of ERG, RORA, SOX4, MYB, and HOXA9 in hPSCs-hematopoietic progenitors is necessary to confer them with short-term engraftment potential. However, additional, yet undefined intrinsic factors and/or extrinsic signals are still required for the generation of in vivo long-term hematopoietic reconstitution from either mouse or human PSCs. Despite the fact that HOXA9 overexpression is strongly linked to leukemogenesis,25,47,48 it has been demonstrated that cooperating mutations are needed for oncogenic transformation.49,50 In hESCs, ectopic HOXA9 did not provide proliferative/survival advantage to hESC-derived hematopoietic cells either in vitro or in vivo. In addition, both HOXA9 overexpression and silencing was compatible with hESC pluripotency, ruling out HOXA9-mediated transformation potential.
The GEP of aorta-gonad mesonephros progenitors was reported to be enriched in “cell movement,” “blood vessel development,” and “inflammatory response” biofunctions, which can be explained by the migratory behavior of these embryonic HSPC.37 Our GEP revealed that HOXA9-HEPs are similarly enriched in “immune cell trafficking” and “cell movement” biofunctions, suggesting that HOXA9 might confer to the hESC-derived HEPs a molecular signature that more closely resembles hemogenic progenitors found during early hematopoietic development. We also used IPA to identify transcriptional activated by Hoxa9 overexpression. We found that several members of the NF-κB signaling were predicted to be activated in HOXA9-HEPs, suggesting that NF-κB signaling may cooperate with HOXA9 to augment embryonic hematopoietic differentiation. A reciprocal regulatory relationship between HOXA9 and NF-κB has been reported in endothelial and smooth muscle cells.51,52 Notably, in leukemia cells, NF-κB signaling cooperates with MLL oncoproteins to regulate the expression of MLL target genes, such as HOXA9 and MEIS1.53 As both HOXA9 and NF-κB have been implicated in normal hematopoiesis and leukemia, the link between them during human hematopoietic differentiation deserves a closer examination. In conclusion, we provide new information regarding the role of HOXA9 function in human embryonic hematopoiesis. We demonstrate that HOXA9 promotes hematopoietic differentiation from the bipotent hemogenic progenitor; however it failed to confer with in vivo long-term engraftment to the hESC-derived HSPC. Further work should explore which alternative genes/pathways, including NF-κB, collaborate with HOXA9 during normal human hematopoiesis.
The online version of this article contains a data supplement.
There is an Inside Blood Commentary on this article in this issue.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank the Plataforma Andaluza de Bioinformática for access to Ingenuity Pathways Analysis software (Andalusian Bioinformatics Platform) (www.scbi.uma.es).
This work was funded by grants from the Ministry of Economy and Competitiveness (SAF2013-43065) (P.M.); the Fondo de Investigación Sanitaria (PI10/00449) (P.M.) and (PI11/00119) (C.B.); the European Rare Diseases Net (PI12/03112) (P.M.); the Spanish Association Against Cancer Foundation (CI110023) (P.M,), and Andalusian Health Government (CSJA2006/0030) (P.M.).
C.B. (CP07/0059), P.J.R. (CP09/0063), and V.R.-M. (CP12/03175) are supported by a Miguel Servet contracts from the Fondo de Investigación Sanitaria/Instituto Salud Carlos III. P.M acknowledges the support from “Obra Social La Caixa/Fundaciò Josep Carreras.”
Authorship
Contribution: V.R.-M. and P.M. designed the research; V.R.-M., O.N.-M., T.R., P.J.R., V.A., and C.B. performed research and analyzed data; V.R.-M. and P.M. wrote the paper; and all authors have read and approved the manuscript in its present form.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Pablo Menendez, Institució Catalana de Recerca i Estudis Avançats, Josep Carreras Leukaemia Research Institute. Facultat de Medicina, University of Barcelona, Carrer Casanova 143, Barcelona 08036, Spain; e-mail: pmenendez@carrerasresearch.org; and Verónica Ramos-Mejía, Centre for Genomics and Oncological Research, PTS Granada, Av. de la Ilustración 114, Granada 18016, Spain; e-mail: veronica.ramos@genyo.es.