Key Points
The H3K27me3 demethylase UTX is recurrently mutated in male T-ALL and escapes X-inactivation in female T-ALL blasts and normal T cells.
The loss of Utx contributes to T-ALL formation in vivo and UTX inactivation confers sensitivity to H3K27me3 inhibition.
Abstract
T-cell acute lymphoblastic leukemia (T-ALL) is an aggressive form of leukemia that is mainly diagnosed in children and shows a skewed gender distribution toward males. In this study, we report somatic loss-of-function mutations in the X-linked histone H3K27me3 demethylase ubiquitously transcribed X (UTX) chromosome, in human T-ALL. Interestingly, UTX mutations were exclusively present in male T-ALL patients and allelic expression analysis revealed that UTX escapes X-inactivation in female T-ALL lymphoblasts and normal T cells. Notably, we demonstrate in vitro and in vivo that the H3K27me3 demethylase UTX functions as a bona fide tumor suppressor in T-ALL. Moreover, T-ALL driven by UTX inactivation exhibits collateral sensitivity to pharmacologic H3K27me3 inhibition. All together, our results show how a gender-specific and therapeutically relevant defect in balancing H3K27 methylation contributes to T-cell leukemogenesis.
Introduction
T-cell acute lymphoblastic leukemia (T-ALL) is an aggressive hematologic malignancy that occurs in children and adolescents and is diagnosed more frequently in males than females. Current treatment schedules consist of intensified chemotherapy and are often associated with considerable side effects.1,2 T-ALL arises from a multistep oncogenic process in which different genetic alterations drive malignant transformation of immature T-cell progenitors. Several key oncogenic drivers mark particular molecular-genetic subgroups as demonstrated by genome-wide transcriptome studies on large cohorts of primary T-ALL samples.3-5 Activation of NOTCH signaling has been recognized as an oncogenic hallmark of T-ALL driven by activating NOTCH1 mutations6 and loss-of-function mutations targeting the E3-ubiquitin ligase FBXW7.7,8 Furthermore, a plethora of additional mechanisms of T-cell transformation have been elucidated involving an increasing number of T-ALL oncogenes and tumor suppressors.9
Recent sequencing studies have identified the core components EZH2, EED, and SUZ12 of the polycomb repressive complex 2 (PRC2), which mediates gene silencing through trimethylation of H3K27,10 as tumor suppressors in the pathogenesis of T-ALL.9,11,12 Moreover, conditional ablation of Ezh2 in mouse hematopoietic stem cells was shown to be sufficient for murine T-ALL development.12 The histone demethylase ubiquitously transcribed X (UTX) chromosome tetratricopeptide repeat protein counters the enzymatic activity of PRC2 by removing di- and trimethyl groups from H3K27.13,14 In 2009, somatic loss-of-function mutations targeting the UTX gene were identified in a variety of human tumors, including multiple myeloma, esophageal, and renal cancer.15 Recently, a general role for UTX as tumor suppressor in human cancer was further supported by the identification of recurrent inactivating UTX mutations in several leukemia and solid tumor cancer types.15-17
In this study, we identified somatic loss-of-function mutations targeting the histone demethylase UTX in human T-ALL and provide in vitro evidence for its tumor suppressor function. Notably, our study reveals that the histone demethylase UTX can serve in vivo as a bona fide tumor suppressor in the molecular pathogenesis of T-ALL. Finally, we show that UTX mutant leukemias are more sensitive to treatment with an H3K27me3 inhibitor, providing new opportunities for epigenetically targeted therapy in T-ALL.
Methods
Collection of T-ALL patient and normal T-cell samples
A cohort of 35 bone marrow samples from primary T-ALL patients was collected from different medical institutes (Universitair Ziekenhuis Ghent, Ghent, Belgium; Universitair Ziekenhuis Leuven, Leuven, Belgium; and Hôpital Purpan, Toulouse, France). The study was approved by the Medical Ethical Commission of Ghent University Hospital (Ghent, Belgium; B67020084745). Normal T cells were obtained from human thymus tissue derived from female pediatric patients undergoing cardiac surgery. The thymic T cells were obtained and used according to the guidelines of the Medical Ethical Commission of Ghent University Hospital, and the study was performed in accordance with the Declaration of Helsinki.
Murine and human T-ALL cell lines
The MOHITO T-ALL mouse cell line was cultured in RPMI-1640 medium (Gibco, Life Technologies, Carlsbad, CA) supplemented with 20% fetal calf serum, glutamine (2 mM), penicillin (100 U/mL)-streptomycin (100 μg/mL), IL-7 (10 ng/mL), and IL-2 (5 ng/mL) (PeproTech, Rocky Hill, NJ).18
Human T-ALL cell lines PEER, TALL-1, and PF-382 were obtained from the German Collection of Microorganisms and Cell Cultures (Braunschweig, Germany). The human ovarian adenocarcinoma cell line OVCAR-3 and the human colon adenocarcinoma cell line HT-29 were obtained from the American Type Culture Collection repository (Manassas, VA). The cell lines were cultured in RPMI-1640 medium supplemented with 10% or 20% fetal calf serum, glutamine (2 mM), penicillin (100 U/mL), and streptomycin (100 μg/mL) under controlled conditions (37°C, 5% CO2).
DNA and RNA isolation
DNA isolation was performed using the QIAamp DNA Mini Kit (Qiagen, Hilden, Germany). RNA isolation was performed using the miRNeasy Mini Kit with DNA digestion on-column (Qiagen). DNA and RNA concentration was measured on the NanoDrop 1000 Spectrophotometer. RNA quality was assessed using the Experion Automated Electrophoresis System according to the manufacturer’s instructions (Bio-Rad Laboratories, Hercules, CA). After RNA quality assessment, complementary DNA (cDNA) synthesis was performed using the iScript cDNA Synthesis Kit (Bio-Rad Laboratories).
Sequencing analysis
We sequenced all coding exons of PTEN, PHF6, and UTX, and mutation hotspot regions for NOTCH1 and FBXW7 in 35 T-ALL patient samples by Sanger sequencing. Primer sequences are noted in supplemental Table 1 on the Blood Web site. The PCRx Enhancer System (Invitrogen, Life Technologies, Carlsbad, CA) was used for NOTCH1 polymerase chain reactions (PCR). FBXW7, PTEN, PHF6, and UTX amplification was performed using the KAPA Taq HotStart PCR Kit (Kapa Biosystems, Wilmington, MA). For all reactions, the following PCR protocol was used: 95°C for 10 minutes (96°C for 15 seconds, 57°C for 1 minute, followed by 72°C for 1 minute) for 40 cycles, then 72°C for 10 minutes. PCR products were purified and analyzed using the Applied Biosystems 3730xl DNA Analyzer (Applied Biosystems, Life Technologies).
Sanger sequencing of Utx, Ezh2, Suz12, and Eed in the MOHITO cell line, and Sanger sequencing of UTX, EZH2, SUZ12, and EED on human T-ALL cell lines was performed at the Geoffrey Beene Translational Oncology Core Facility at Memorial Sloan-Kettering Cancer Center (MSKCC).
Array comparative genomic hybridization (CGH)
T-ALL patient samples were profiled for copy number analysis on SurePrint G3 Human 4×180K CGH Microarrays (Agilent Technologies, Santa Clara, CA). In brief, patient and control genomic DNAs (gDNAs) were labeled using random prime labeling with Cy3 and Cy5 dyes (PerkinElmer, Waltham, MA), respectively. Next, hybridization was performed according to the manufacturer’s instructions (Agilent Technologies), followed by data-analysis using the in-house developed analysis tool arrayCGHbase.19
Single nucleotide polymorphism (SNP) genotyping
Genotyping and allelic expression analysis of SNPs, rs181547731 and rs20539, located in the 3′ untranslated region and exon 20 of UTX, respectively, was performed in both gDNA and cDNA samples of 3 female T-ALL patients, 3 female normal T-cell donors, and 2 female cancer cell lines (OVCAR-3 and HT-29). The standard PCR and sequencing protocols were used as described above. Primer sequences are listed in supplemental Table 1.
Real-time quantitative PCR (qPCR)
The qPCR reaction was performed using the LightCycler 480 (Roche, Basel, Switzerland) and qPCR data were analyzed by the ΔΔCt method using the in-house developed qPCR analysis program, qBasePLUS (Biogazelle, Ghent, Belgium). Primers used to measure the expression of the genes of interest and reference genes are provided in supplemental Table 2.
Interleukin depletion assay
Short hairpin RNAs (shRNAs) were designed using the online program Designer of Small Interfering RNA, followed by cloning into the mouse stem cell virus (MSCV)-based retroviral backbonce, which encodes the shRNA along with green fluorescent protein (GFP) under the control of the SV40 promoter (MLS). Next, the interleukin (IL-2/IL-7) dependent MOHITO cell line was transduced with shRNAs Utx, and GFP percentage was evaluated using the Guava Flow Cytometer (Millipore, Billerica, MA). Subsequently, MOHITO cells were depleted of IL-2 and IL-7 until less than 10% of viable cells were detected, followed by rescue through supplementing IL-2 and IL-7 back in the media. After each round of interleukin depletion, the GFP percentage was measured. The sequences of the shRNAs are listed in supplemental Table 3.
T-ALL mouse model
Fetal liver cells were isolated at embryonic days 13 to 14, followed by retroviral transduction of shRNAs or empty control labeled with GFP (MLS backbone) and NOTCH1 labeled with mCherry (MSCV IRES GFP backbone).20 Next, the transduced fetal liver cells were tail vein injected after lethal irradiation of mouse recipients. Subsequently, mouse recipients were monitored for leukemia onset by analysis of lymphoblast counts in blood smears and by physical appearance. By the time of leukemia formation, various tissues including the spleen, lungs, liver, kidneys, and lymph nodes were fixed for histologic evaluation. Furthermore, the lymphoblasts from the spleen and thymus were mashed to single-cell suspensions and frozen in 10% dimethylsulfoxide. Survival data were analyzed using the Kaplan-Meier method and statistical significance was calculated using the log-rank (Mantel-Cox) test.
Immunophenotypic analysis
Surface marker analysis of murine tumor cells was performed using a BD LSR II flow cytometer with monoclonal antibodies CD3-APC (BD Biosciences, San Jose, CA), CD8a-PECY7 (eBioscience, San Diego, CA), and CD4-biotin (BD Biosciences). Data were analyzed with FACSDiva 6.1.2 software (BD Biosciences) and compensations were set using OneComp eBeads (eBioscience).
Histology
Histologic analysis including hematoxylin and eosin staining, Ki67 staining, H3K27me3 staining, and terminal deoxynucleotidyltransferase-mediated dUTP nick end labeling assay of the different mouse leukemias was performed at the Laboratory of Comparative Pathology of MSKCC.
Western blotting
Cells were lysed using a lysis buffer containing 2% sodium dodecyl sulfate, 62.5 mM Tris pH 6.8, 10% glycerol, and 5% β-mercaptoethanol supplemented with EDTA free protease inhibitor cocktail (Roche). Denaturated samples were loaded on 10% or 7.5% gels (for 1 hour 15 minutes, 250V) (Bio-Rad Laboratories) for electrophoresis, followed by immunoblotting on polyvinylidene flouride membranes using a wet blotting system (for 1 hour, 250V) (Bio-Rad Laboratories). Membranes were incubated with primary and secondary antibody dilutions in 5% milk/Tris-buffered saline with Tween solutions, developed using SuperSignal West Dura Chemiluminescent Substrate (Pierce, Thermo Scientific, Rockford, IL), and visualized on ChemiDoc-It Imaging System 500 (UVP, Upland, CA). The following antibodies were used: Utx (155 kDa and A302-374A; Bethyl Laboratories, Montgomery, TX), H3K27me3 (17 kDa and 9733; Cell Signaling, Danvers, MA), EZH2 (95-100 kDA and 612666; BD Biosciences), and tubulin (50 kDa and T5168; Sigma-Aldrich, St. Louis, MO).
Gene expression arrays of murine T-ALL samples
RNA samples were profiled for gene expression analysis on SurePrint G3 Mouse 8×60K Microarrays according to the manufacturer’s instructions (Agilent Technologies). Normalization of gene expression data were done by quantile normalization using R. Background correction was done based on the dark corner probe group of the Agilent slides. Differential gene expression analysis was performed using fold change analysis and P-value calculation was based on unpaired Student t test. Biased gene set enrichment analysis (GSEA) was performed using a gene set consisting of differential probes between the Utx sh#1 and control murine leukemias, and evaluated against gene expression data of the Utx-deficient MOHITO samples. Unbiased GSEA was executed against annotated gene sets of the Molecular Signatures Database. Gene expression data are accessible on ArrayExpress (accession number E-MTAB-2921).
Gene expression arrays of human T-ALL samples
Differential UTX expression analysis (unpaired Student t test) was performed between female and male T-ALL patient samples on published gene expression data from Clappier et al.21 Patient genders was defined based on karyotype information. Gene expression data are accessible on ArrayExpress (accession number E-MTAB-593).21
Chromatin immunoprecipitation (ChIP)-sequencing
Lymphoblasts from murine leukemias were crosslinked in 1% formaldehyde for 10 minutes on a rotating platform in a 37°C oven. Next, 2.5 M glycine (0.122 M final concentration) was added for 5 minutes at 37°C to quench the crosslinking reaction. The cells were subsequently pelleted at 4°C for 5 minutes at 1750 rpm, followed by 3 washing steps in cold phosphate-buffered saline. The final pellet was stored at −80°C. The following H3K27me3 ChIP-procedure (H3K27me3: 17 kDa, Cat #6002; Abcam, Cambridge, United Kingdom) was performed as previously described (Béguelin et al22 ): H3K27me3 ChIP-seq libraries were prepared using the Illumina ChIP-Seq sample kits based on the manufacturer’s instructions. Libraries were validated using an Agilent Technologies 2100 Bioanalyzer and 8 to 10 pM was sequenced on the HiSeq2000 sequencer (Illumina, San Diego, CA). The raw data were aligned to mm9 genome using ELAND. Reads were normalized for total ChIP-seq reads and quantified in 1 kb bins genome-wide. The enriched regions were identified as consecutive bins with read counts >1 standard deviation of the genome-wide mean.
Cell viability assay
Murine and human T-ALL cell lines were treated with 3-deazaneplanocin A (DZNep) hydrochloride (Sigma-Aldrich), using a dilution series ranging from 50 nM to 100 μM. After 24 hours, cell viability was measured using the luminescence detection kit CellTiter-Glo Cell Viability Assay (Promega, Madison, WI).
Results
Mutations recurrently target the H3K27me3 demethylase UTX in male T-ALL patients
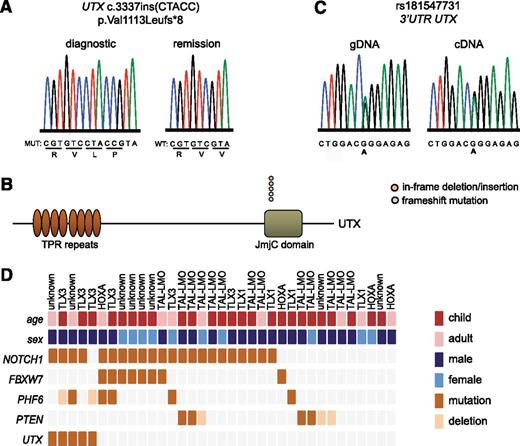
To identify a potential role for the H3K27me3 demethylase UTX in the molecular pathogenesis of T-ALL, we performed sequencing and copy number analysis in a series of 35 primary leukemia samples (10 female vs 25 male), including 25 pediatric and 10 adult T-ALL cases. We identified loss-of-function mutations in UTX in 5 out of 35 (14.3%) primary T-ALL patient samples (Figure 1A and supplemental Table 4), including 3 frameshift and 2 inframe insertion/deletion mutations. UTX mutations were localized in a hotspot region within the catalytic Jumonji-C domain of the protein (Figure 1B) and sequencing of available remission material confirmed the somatic origin of these mutations (Figure 1A). The somatic UTX mutations in our patient population were exclusively identified in samples of male origin.
UTX mutations in human T-ALL. (A) DNA sequencing chromatogram showing a UTX mutation in the gDNA of a male primary T-ALL patient sample. The mutation is absent in remission material of the same patient. (B) Graphical representation of the localization of genetic lesions in the UTX protein structure. In-frame deletion/insertion mutations are depicted in orange circles and frameshift mutations in blue circles. (C) Genotyping and allelic expression analysis of SNP rs181547731 in gDNA and cDNA derived from female T-ALL lymphoblasts. (D) Graphical representation of the different mutations (dark orange rectangles) and deletions (light orange rectangles) present in a set of T-ALL oncogenes and tumor suppressor genes in 35 primary T-ALL patient samples. The different T-ALL subgroups include TAL-LMO, TLX3, TLX1, HOXA, and patients for whom the subgroup is unknown. The age subgroups include children (age ≤15 years; dark red rectangles) and adults (age >15 years; light red rectangles). Male and female T-ALL patient samples are presented in dark blue and light blue rectangles, respectively. JmjC, Jumonji C; TPR, tetratricopeptide repeat.
UTX mutations in human T-ALL. (A) DNA sequencing chromatogram showing a UTX mutation in the gDNA of a male primary T-ALL patient sample. The mutation is absent in remission material of the same patient. (B) Graphical representation of the localization of genetic lesions in the UTX protein structure. In-frame deletion/insertion mutations are depicted in orange circles and frameshift mutations in blue circles. (C) Genotyping and allelic expression analysis of SNP rs181547731 in gDNA and cDNA derived from female T-ALL lymphoblasts. (D) Graphical representation of the different mutations (dark orange rectangles) and deletions (light orange rectangles) present in a set of T-ALL oncogenes and tumor suppressor genes in 35 primary T-ALL patient samples. The different T-ALL subgroups include TAL-LMO, TLX3, TLX1, HOXA, and patients for whom the subgroup is unknown. The age subgroups include children (age ≤15 years; dark red rectangles) and adults (age >15 years; light red rectangles). Male and female T-ALL patient samples are presented in dark blue and light blue rectangles, respectively. JmjC, Jumonji C; TPR, tetratricopeptide repeat.
Equal dosage of chromosome X genes between male and female cells is regulated by random inactivation of one copy of the X chromosome in female cells. However, previous studies have shown that some genes, including UTX,23 can escape chromosome X-inactivation in certain tissues. Notably, we analyzed two silent SNPs (rs181547731 situated in the 3′ untranslated region and rs20539 situated in exon 20 of UTX) in lymphoblasts from 3 female T-ALL patients, and confirmed bi-allelic expression of UTX in female T-ALL lymphoblasts (Figure 1C and supplemental Figure 1A). In agreement, UTX expression is elevated in female T-ALL patient samples in comparison with male T-ALL patient cases (Student t test, P = .001; female: n = 5, male: n = 20) (supplemental Figure 1B).21 In addition, we were able to demonstrate that UTX is also able to escape X-inactivation in normal T cells derived from female donors (supplemental Figure 1C) and in 2 female cancer cell lines, the ovarian adenocarcinoma cell line OVCAR-3 and the colon adenocarcinoma cell line HT-29 (supplemental Figure 1D).
In T-ALL, a wide variety of oncogenic lesions cooperate to induce T-cell transformation. To identify genetic lesions that co-occur with the loss of UTX in T-ALL, we classified our cohort into the known molecular genetic subgroups based upon the presence of genetic defects and/or aberrant expression of transcription factor oncogenes (Figure 1D).3-5 Furthermore, we screened our patient population for alterations targeting T-ALL–specific oncogenes and tumor suppressors (Figure 1D). This analysis showed that UTX mutations co-occur with aberrant expression of the TLX3 oncogene, activating NOTCH1 mutations, and mutations or deletions targeting the putative chromatin reading factor PHF6 (Figure 1D). Of note, no deletions were identified in the second H3K27me3 eraser and UTX family member Jumonji D3.
Utx acts as a tumor suppressor in T-ALL
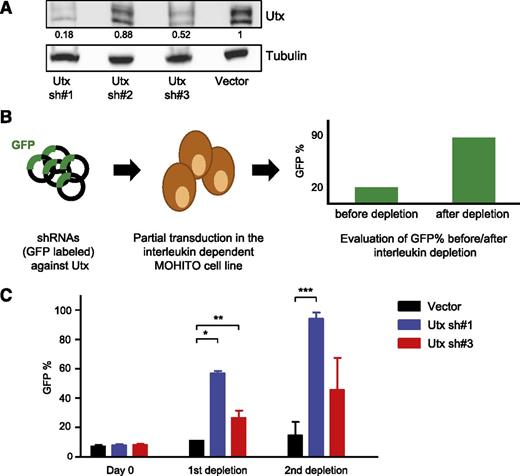
To functionally validate the role of Utx as a putative tumor suppressor in the context of malignant T-cell transformation, we used MOHITO, the interleukin (IL2/IL7) dependent murine T-ALL cell line.18 We transduced MOHITO cells with shRNAs against Utx and an empty vector control, and assessed knockdown efficiency using western blot analysis (Figure 2A). Both Utx shRNA#1 and Utx shRNA#3 mediate a clear decrease in Utx protein levels. Next, we examined if knockdown of Utx can provide an oncogenic advantage in vitro to the MOHITO cells. First, MOHITO cells were partially transduced with Utx shRNA#1, Utx shRNA#3, or empty vector control (GFP co-expression). After successive rounds of interleukin depletion that led to a drop in cell viability, GFP enrichment was assessed as an indicator of tumor promoting activity (Figure 2B). Notably, murine tumor cells infected with functional Utx shRNA#1 or Utx shRNA#3 showed significant GFP enrichment over MOHITO cells infected with an empty vector control (Figure 2C), indicating that Utx loss grants oncogenic properties to the MOHITO cells.
Knockdown of Utx augments the oncogenic activity in the murine T-ALL cell line MOHITO. (A) Western blot analysis of GFP sorted MOHITO knockdown samples containing 3 different hairpins against Utx compared with empty vector control. Blots were incubated with antibodies against Utx and tubulin. Expression levels were normalized to tubulin levels and compared with the vector control. (B) Graphical illustration of the interleukin depletion assay in the IL2-IL7–dependent MOHITO cell line. MOHITO cells are partially transduced with vectors encoding shRNAs against Utx and coexpressing GFP, after which the GFP percentage is measured by fluorescence-activated cell sorter analysis before and after successive rounds of interleukin depletion. (C) GFP percentage measured at Day 0, and after 1st and 2nd depletion rounds in Utx sh#1 and Utx sh#3 knockdown samples and controls. The depletion assay was repeated twice. The GFP enrichment was statistically significant after the 1st depletion for Utx sh#1* and Utx sh#3**, and after the 2nd depletion for Utx sh#1*** compared with empty vector control respectively (unpaired Student t test: *P = .005; **P = .049; ***P = .008).
Knockdown of Utx augments the oncogenic activity in the murine T-ALL cell line MOHITO. (A) Western blot analysis of GFP sorted MOHITO knockdown samples containing 3 different hairpins against Utx compared with empty vector control. Blots were incubated with antibodies against Utx and tubulin. Expression levels were normalized to tubulin levels and compared with the vector control. (B) Graphical illustration of the interleukin depletion assay in the IL2-IL7–dependent MOHITO cell line. MOHITO cells are partially transduced with vectors encoding shRNAs against Utx and coexpressing GFP, after which the GFP percentage is measured by fluorescence-activated cell sorter analysis before and after successive rounds of interleukin depletion. (C) GFP percentage measured at Day 0, and after 1st and 2nd depletion rounds in Utx sh#1 and Utx sh#3 knockdown samples and controls. The depletion assay was repeated twice. The GFP enrichment was statistically significant after the 1st depletion for Utx sh#1* and Utx sh#3**, and after the 2nd depletion for Utx sh#1*** compared with empty vector control respectively (unpaired Student t test: *P = .005; **P = .049; ***P = .008).
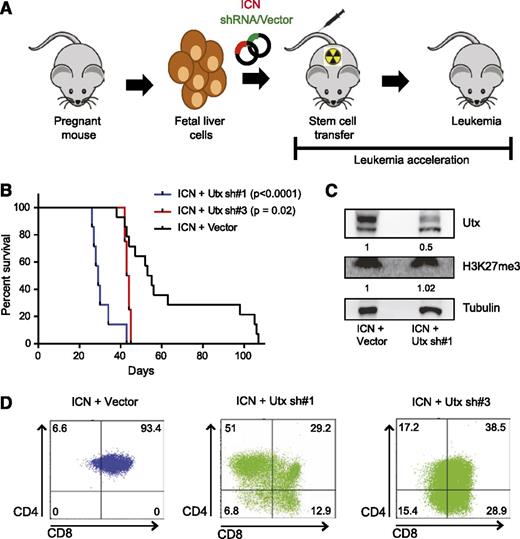
Next, we used a bone marrow transplant mouse model of NOTCH1-induced T-ALL20 (Figure 3A) to study the in vivo role of Utx in the pathogenesis of T-ALL. Leukemic onset in mouse recipients receiving fetal liver cells with enforced NOTCH1 expression occurred around 55 days (n = 14, mean latency = 54 days) (Figure 3B). Infection of fetal liver cells with Utx shRNA#1 together with activated NOTCH1 resulted in an acceleration of T-ALL onset (n = 7, P < .0001; mean latency = 29 days) (Figure 3B), confirming the in vitro oncogenic properties observed in the T-ALL MOHITO cells. Introduction of Utx shRNA#3 provided a milder effect on T-ALL latency (n = 4, P = .02; mean latency = 43.5 days) (Figure 3B).
Loss of Utx accelerates leukemia development in a NOTCH1-induced T-ALL mouse model. (A) Graphical illustration of NOTCH1-induced T-ALL mouse model. Fetal liver cells are partially transduced with vectors encoding NOTCH1 (ICN) (coexpressing mCherry), and the shRNA or empty vector control (coexpressing GFP), followed by tail vein injection in lethally irradiated mouse recipients and monitoring of leukemia onset. (B) Kaplan-Meier curves and log-rank (Mantel-Cox) analysis show accelerated leukemia onset in Utx sh#1 (n = 7, P < .0001) (blue) and Utx sh#3 (n = 4, P = .02) (red) mice as compared with empty vector (n = 14) (black) mouse recipients. (C) Western blot analysis of Utx and H3K27me3 in a representative Utx sh#1 mouse leukemia sample. Utx protein levels and H3K27me3 levels are quantified by Image J, normalized to tubulin levels, and compared with expression levels in control mice. (D) Immunophenotypical fluorescence-activated cell sorter analysis of CD4 and CD8 T-cell markers in the Utx sh#1, Utx sh#3, and empty vector control mouse leukemias. A representative example of each subtype is depicted.
Loss of Utx accelerates leukemia development in a NOTCH1-induced T-ALL mouse model. (A) Graphical illustration of NOTCH1-induced T-ALL mouse model. Fetal liver cells are partially transduced with vectors encoding NOTCH1 (ICN) (coexpressing mCherry), and the shRNA or empty vector control (coexpressing GFP), followed by tail vein injection in lethally irradiated mouse recipients and monitoring of leukemia onset. (B) Kaplan-Meier curves and log-rank (Mantel-Cox) analysis show accelerated leukemia onset in Utx sh#1 (n = 7, P < .0001) (blue) and Utx sh#3 (n = 4, P = .02) (red) mice as compared with empty vector (n = 14) (black) mouse recipients. (C) Western blot analysis of Utx and H3K27me3 in a representative Utx sh#1 mouse leukemia sample. Utx protein levels and H3K27me3 levels are quantified by Image J, normalized to tubulin levels, and compared with expression levels in control mice. (D) Immunophenotypical fluorescence-activated cell sorter analysis of CD4 and CD8 T-cell markers in the Utx sh#1, Utx sh#3, and empty vector control mouse leukemias. A representative example of each subtype is depicted.
In all murine tumors, histologic analysis revealed aggressive leukemia phenotypes marked by an absence of apoptosis, high proliferation, and infiltration of lymphoblasts in different organs (supplemental Figure 2A). In addition, evaluation of the shRNA-mediated Utx knockdown confirmed the loss of Utx expression in vivo in the NOTCH1 T-ALL mouse model (Figure 3C and supplemental Figure 2B). Notably, immunohistochemistry and western blot analysis of H3K27me3 revealed that the H3K27me3 levels between Utx-driven and NOTCH1-driven control murine T-cell tumors were largely comparable (Figure 3C and supplemental Figure 2C). Immunophenotypic characterization of mouse leukemias indicated an earlier T-cell maturation arrest for tumors driven by loss of Utx as compared with NOTCH1-control tumors (Figure 3D and supplemental Tables 5-7). The NOTCH1-only controls were typically CD4+CD8+ double positive. In contrast, immunophenotypic marker expression in Utx-driven tumors was more heterogeneous, including cell populations that lacked both CD8 and CD4 surface expression (Figure 3D and supplemental Tables 5-7). Hence, the demethylase Utx is able to restrain T-ALL development in vivo.
Transcriptional programs driven by loss of Utx in murine T-ALL models
To gain more insight into the oncogenic mechanisms mediated by the loss of Utx, we performed gene expression profiling of NOTCH1 control tumors and mouse leukemias driven by Utx inactivation. Supervised gene expression analysis (FC >1.5, P < .05; Figure 4A) showed that Utx-deficient mouse tumors are associated with a specific gene expression signature. Utx leukemias are characterized by downregulation of the T-ALL tumor suppressor genes Ect2l11 and Nf1,24 and activation of the T-ALL oncogene Tal125 (Figure 4A). We could confirm down and upregulation of Utx-regulated genes using qPCR analysis (representative examples are shown in supplemental Figure 4. Furthermore, GSEA26 in the mouse tumors driven by loss of Utx showed enrichment of gene sets linked to early T-cell lymphocytes27 (P < .01; Figure 4B and supplemental Figure 3A) and LYL1 neighboring genes in T-ALL3 (P = .02; supplemental Figure 3B), in line with the early immunophenotypic arrest observed in these murine leukemias (Figure 3D).
Gene networks regulated by Utx in T-ALL. (A) Differentially expressed genes (FC >1.5; P < .05) between Utx knockdown and control murine leukemias are represented in a heat map. A selection of genes is shown in rows and each column represents 1 individual mouse leukemia sample. The scale bar shows color-coded differential expression from the mean in standard deviation units with red indicating higher levels and blue lower levels of expression. (B) Unbiased GSEA of gene expression signatures associated with murine leukemias driven by loss of Utx or NOTCH1 only vector controls. Gene sets involving early T-lymphocytes (P < .01) are significantly enriched in Utx-driven leukemias. (C) H3K27me3 ChIP-seq profiles at 2 specific gene loci (Lzts2 and Pcgf2) in murine leukemias driven by loss of Utx or NOTCH1 only vector controls. (D) GSEA of transcripts significantly downregulated upon Utx knockdown in murine leukemias (FC >1.5, P < .05; 252 probes) in gene expression signatures obtained from MOHITO samples driven by loss of Utx. Heatmap displays the TOP 25 leading edge of this gene set in Utx-driven MOHITO samples. FDR, false discovery rate; NES, negative enrichment score.
Gene networks regulated by Utx in T-ALL. (A) Differentially expressed genes (FC >1.5; P < .05) between Utx knockdown and control murine leukemias are represented in a heat map. A selection of genes is shown in rows and each column represents 1 individual mouse leukemia sample. The scale bar shows color-coded differential expression from the mean in standard deviation units with red indicating higher levels and blue lower levels of expression. (B) Unbiased GSEA of gene expression signatures associated with murine leukemias driven by loss of Utx or NOTCH1 only vector controls. Gene sets involving early T-lymphocytes (P < .01) are significantly enriched in Utx-driven leukemias. (C) H3K27me3 ChIP-seq profiles at 2 specific gene loci (Lzts2 and Pcgf2) in murine leukemias driven by loss of Utx or NOTCH1 only vector controls. (D) GSEA of transcripts significantly downregulated upon Utx knockdown in murine leukemias (FC >1.5, P < .05; 252 probes) in gene expression signatures obtained from MOHITO samples driven by loss of Utx. Heatmap displays the TOP 25 leading edge of this gene set in Utx-driven MOHITO samples. FDR, false discovery rate; NES, negative enrichment score.
Next, we performed H3K27me3 ChIP sequencing analysis on murine tumor material to evaluate if genes downregulated in Utx-driven mouse leukemias showing differential H3K27me3 levels at their promoters. In total, 51 out of 258 downregulated genes (19.7%) showed an enhanced H3K27 trimethylation at their promoter regions in at least 2 Utx sh#1 samples compared with 2 control samples. In comparison, 1801 out of 39 430 genes (4.5%) on the gene expression array demonstrated enrichment of H3K27me3 at their promoter region (hypergeometric test, P = 6.9058e-19, log = −41.8168). Hence, the genes that were downregulated after Utx knockdown had statistically more H3K27me3 enrichment as compared with the general number of H3K27me3 enrichment over all genes. Notably, the ChIP-sequencing analysis revealed an accumulation of H3K27me3 at promoter regions of genes with putative tumor suppressor activity such as Pcgf2,28 Lzts2,29 Dock4,30 Pura,31 Col2a1,32 and Slc26a433 in the Utx tumors (Figure 4C and supplemental Figure 3C).
To further explore the relationship between the genetic program mediated by the in vivo transplant model and the in vitro MOHITO model, we performed GSEA26 of gene expression signatures associated with Utx knockdown in MOHITO cells. Interestingly, this analysis revealed a significant enrichment of genes downregulated upon the loss of Utx in the mouse leukemias in Utx-deficient MOHITO cells (P = .007; 82 of 252 probes in core enrichment) (Figure 4D).
Loss of Utx provides a selective pressure toward DZNep treatment
Because the loss of UTX affects the genome-wide distribution of H3K27me3, we hypothesized that decreased UTX levels would render leukemia cells more vulnerable to treatment with 3-DZNep, an epigenetic compound that specifically targets H3K27me3.34 Treatment of 3 T-ALL cell lines with DZNep demonstrated enhanced sensitivity of the UTX mutant cell line PF-382, as compared with the PEER cell line and the EZH2 mutant cell line TALL-1 (Figure 5A-D). Moreover, TALL-1 cells were strongly resistant to DZNep treatment even at 100 μM, presumably because of very low H3K27me3 levels (Figure 5C-D). As expected, DZNep treatment of PF-382 and PEER resulted in decreased levels of H3K27me334 as determined by western blot (Figure 5E). In parallel, we confirmed that MOHITO cells showed enhanced sensitivity toward DZNep treatment upon Utx knockdown as compared with control cells (Figure 5F). Notably, although control and Utx knockdown MOHITO cells showed comparable baseline levels of H3K27me3, DZNep treatment induced a stronger decrease in H3K27me3 expression in the Utx knockdown samples as compared with the empty vector controls (Figure 5G). Hence, UTX-deficient T-ALL shows collateral sensitivity to H3K27me3 inhibition.
Utx-driven cell lines are more sensitive for H3K27me3 inhibition. (A) gDNA sequencing chromatograms representing 2 missense mutations in the EZH2 gene detected in the T-ALL cell line TALL-1. (B) DNA sequencing chromatogram representing a nonsense mutation in the UTX gene detected in the human T-ALL cell line PF-382. (C) Western blot analysis of EZH2, H3K27me3, and tubulin in the human T-ALL cell lines PF-382, TALL-1, and PEER. EZH2 protein and H3K27me3 levels are quantified by Image J and normalized to tubulin levels. (D) Luminescence-based viability assay after 24 hours of DZNep administration in 3 different T-ALL cell lines (TALL-1: black; PEER: red; and PF-382: green) using a range of DZNep concentrations from 0 nM to 100 μM. The experiment was done using 6 replicates, and repeated twice independently. The viability score for each concentration was significantly different between the 3 T-ALL cell lines (*Kruskal-Wallis test, P < .0001). (E) Western blot analysis of H3K27me3 and tubulin 24 hours after DZNep administration (1000 nM and 500 nM) in 2 different T-ALL cell lines (PEER: red and PF-382: green). H3K27me3 levels are quantified by Image J and normalized to tubulin levels. DZNep-treated samples are compared with expression levels in untreated samples. (F) Luminescence-based viability assay after 24 hours of DZNep administration in Utx knockdown and control MOHITO samples (vector: black and Utx sh#1: green). The experiment was done in fourfold (unpaired Student t test: *P < .0001). (G) Western blot analysis of H3K27me3 and tubulin 24 hours after DZNep administration (1000 nM and 500 nM) in Utx knockdown and control MOHITO samples (vector: black and Utx sh#1: green). H3K27me3 levels are quantified by Image J and normalized to tubulin levels. DZNep-treated samples are compared with expression levels in untreated samples.
Utx-driven cell lines are more sensitive for H3K27me3 inhibition. (A) gDNA sequencing chromatograms representing 2 missense mutations in the EZH2 gene detected in the T-ALL cell line TALL-1. (B) DNA sequencing chromatogram representing a nonsense mutation in the UTX gene detected in the human T-ALL cell line PF-382. (C) Western blot analysis of EZH2, H3K27me3, and tubulin in the human T-ALL cell lines PF-382, TALL-1, and PEER. EZH2 protein and H3K27me3 levels are quantified by Image J and normalized to tubulin levels. (D) Luminescence-based viability assay after 24 hours of DZNep administration in 3 different T-ALL cell lines (TALL-1: black; PEER: red; and PF-382: green) using a range of DZNep concentrations from 0 nM to 100 μM. The experiment was done using 6 replicates, and repeated twice independently. The viability score for each concentration was significantly different between the 3 T-ALL cell lines (*Kruskal-Wallis test, P < .0001). (E) Western blot analysis of H3K27me3 and tubulin 24 hours after DZNep administration (1000 nM and 500 nM) in 2 different T-ALL cell lines (PEER: red and PF-382: green). H3K27me3 levels are quantified by Image J and normalized to tubulin levels. DZNep-treated samples are compared with expression levels in untreated samples. (F) Luminescence-based viability assay after 24 hours of DZNep administration in Utx knockdown and control MOHITO samples (vector: black and Utx sh#1: green). The experiment was done in fourfold (unpaired Student t test: *P < .0001). (G) Western blot analysis of H3K27me3 and tubulin 24 hours after DZNep administration (1000 nM and 500 nM) in Utx knockdown and control MOHITO samples (vector: black and Utx sh#1: green). H3K27me3 levels are quantified by Image J and normalized to tubulin levels. DZNep-treated samples are compared with expression levels in untreated samples.
Discussion
A role for UTX as tumor suppressor was initially postulated in several human tumors including multiple myeloma, esophageal, and renal cancer.15 Notably, UTX deletions and mutations have also been identified in patients with Kabuki syndrome, a rare congenital anomaly syndrome.35 Interestingly, 6 Kabuki patients have been reported who developed different types of cancer, thereby suggesting an increased susceptibility to cancer for Kabuki patients carrying abnormalities in UTX.36 In this study, we identified gender-restricted somatic loss-of-function mutations targeting the histone demethylase UTX in male T-ALL patients and demonstrated its tumor suppressor function.
Random inactivation of one copy of the X-chromosome in female cells is the mechanism that ensures equal dosage of chromosome X genes between male and female cells. However, some genes can escape chromosome X-inactivation and show bi-allelic expression in normal female tissues. Previous studies demonstrated that UTX can escape chromosome X-inactivation in normal female tissues,23,37,38 and accordingly, shows higher expression in female embryonic stem cells and female tissues derived from brain, liver, neurons, and sexual organs in comparison with their male counterpart.36-38 The fact that UTX escapes chromosome X-inactivation in normal cells has important implications for gender-related tumor susceptibility. Indeed, given the previously established role for UTX as a tumor suppressor in human cancer, inactivation of only one single UTX copy in males will contribute to tumor development. In contrast, female cells are protected against such single copy loss of UTX because they still express UTX from the second allele (supplemental Figure 1A).
Most notably, T-ALL is characterized by a skewed gender distribution with a male to female ratio of 3:1. In contrast to other X-linked tumor suppressor genes in T-ALL (PHF639 and RPL1040 ), UTX escapes chromosome X-inactivation in female T-ALL blasts, suggesting that females are protected against single copy loss of one UTX allele. Therefore, UTX is the first X-linked tumor suppressor gene that might partially explain the skewed gender distribution in T-ALL toward males on a genetic level.
The PRC2 complex mediates H3K27 methylation, and therefore, counteracts UTX activity. Trimethylation of lysine 27 on histone 3 is a chromatin state that is usually associated with transcriptional gene repression. Depending on the tissue-type and cellular context, the PRC2 complex can function as an oncogene (eg, in B-cell lymphoma41 ) or tumor suppressor gene (eg, in T-ALL9,11,12 ). In addition, Ezh2 loss was sufficient to induce murine T-ALL development, further supporting a tumor suppressive activity for EZH2 in the context of T-ALL.12
Given our finding that the H3K27me3 modulator UTX is targeted by loss-of-function alterations in primary T-ALL, we explored it’s tumor suppressor activity using in vitro and in vivo perturbation model systems. Firstly, Utx knockdown provided murine leukemia cells with an oncogenic advantage after interleukin depletion using the IL2-IL7 dependent MOHITO culture system.18 Secondly, the loss of Utx resulted in significant acceleration of leukemia onset in a NOTCH1-induced T-ALL mouse model.20 In addition, differences in tumor immunophenotype between Utx-driven mouse leukemias and control tumors suggests that the tumor suppressor activity of Utx could depend on the maturation arrest or the cell of origin during murine T-cell transformation. For the first time, these functional in vitro and in vivo data firmly establish Utx as a bona fide tumor suppressor involved in murine T-cell transformation.
Transcriptional profiling of the in vivo transplant model revealed that the loss of Utx drives a unique gene expression signature in NOTCH1-driven mouse leukemias. Interestingly, in a previous study, we could demonstrate that shRNA mediated knockdown of Nf1, one of the down regulated genes after Utx knockdown, was also able to induce a strong acceleration in T-ALL onset in the NOTCH1 T-ALL mouse model.42 Hence, Nf1 loss seems to mimic the oncogenic effect of Utx loss in the same in vivo leukemic model system.
Notably, an important part of the gene expression profiles of the Utx knockdown samples most likely reflects the immunophenotypic differences observed between control and Utx-deficient tumors. Nevertheless, ChIP-seq analysis confirmed that some of the transcriptional differences are associated with accumulation of H3K27me3 at the promoter regions, and therefore, most probably reflect direct consequences of loss of Utx during malignant T-cell transformation.
Epigenetic drugs are currently developed and evaluated in different cancer subtypes. Since we identified UTX as a bona fide tumor suppressor gene in the pathogenesis of T-ALL, we hypothesized that the loss of UTX would render leukemia cells more vulnerable to treatment with particular chromatin modifying drugs. Because loss of UTX is associated with enhanced H3K27me3 levels, we explored the effect of DZNep, an epigenetic compound that specifically targets the H3K27me3 mark. Previous studies reported a strong apoptotic effect after DZNep administration in cancer cells, whereas normal cells were not affected.34 Importantly, in our study, we showed in both murine and human in vitro T-ALL model systems that loss of UTX/Utx renders the leukemic population more sensitive to DZNep treatment. In contrast, the EZH2 mutant T-ALL cell line TALL-1 seems resistant to DZNep treatment. Importantly, such proof of principle data should be further validated in a preclinical setting in which H3K27me3 modifying agents are formally tested in well-characterized genetic animal models of T-ALL.
In conclusion, our results provide new insights into the pathogenesis and gender distribution in T-ALL. Furthermore, we identified UTX as an X-linked tumor suppressor gene and showed that Utx can act as a bona fide tumor suppressor in T-ALL. All together, our study reveals how maintaining and pharmacologically restoring the precise balance of H3K27 methylation can restrain T-cell leukemia.
The online version of this article contains a data supplement.
There is an Inside Blood Commentary on this article in this issue.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank the members of both the Wendel laboratory and the Speleman laboratory for experimental support and discussion during this research project, Aline Eggermont for excellent technical assistance, the Geoffrey Beene Translational Oncology Core Facility at MSKCC for the sequencing support, the Flow Cytometry Facility at MSKCC for cell sorting assistance, the MSK animal facility and Research Animal Resource Center for assistance with mouse experiments, and the Laboratory of Comparative Pathology for histologic analysis.
This study was supported by the Fund for Scientific Research (FWO) Flanders (postdoctoral grants to P.V.V., P.R., and T.T., grants to J.V.d.M., B.P., and P.V. [Senior Clinical Investigators of FWO-Flanders], Odysseus grant to P.V.V. and T.T., project grants G.0198.08, G.0564.13N, G.0550.13N, and G.0869.10N to F.S., grants G065614, 3GA00113N, and G.0C47.13N to P.V.V., and grants G0B2913N and 3G002711 to T.T.), the Flemish League Against Cancer (PhD grant to J.V.d.M. and postdoctoral grant to F.M.), the Agency for Innovation by Science and Technology (PhD grant to K.D.), the GOA-UGent (grant #12051203 to F.S.), Belgian Foundation Against Cancer, the Cancer Plan from the Federal Public Service of Health (F.S.), the Children Cancer Fund Ghent (F.S.), the Belgian Program of Interuniversity Poles of Attraction IUAP, the Belgian Foundation Against Cancer (project grants 2010-187 to T.L. and 365O9110 to F.S.), the NCI R01-CA142798-01 and U01CA105492-08 (H-G.W.), the Leukemia Research Foundation (H-G.W.), LLS SCOR (#7006-13), LLS TRP (#6141-14), and the Burroughs Wellcome Foundation (A.M.M.).
Authorship
Contribution: J.V.d.M. performed analysis of genomic data in T-ALL patient samples and performed in vitro and in vivo experiments; V.S., K.M., K.D., F.M., P.R., T.P., and B.V. assisted with in vitro and in vivo experiments; F.F., M.R., and A.M.M. performed and supervised ChIP-sequencing experiments and data analysis; B.M. supervised array CGH and gene expression analysis; P.V., E.D., T.L., B.D.M., B.P., N.V.R., and Y.B. collected primary T-ALL patient samples; T.T. supervised immunophenotypic analysis of mouse leukemias; and P.V.V., H-G.W., F.S., and J.V.d.M. designed the experiments and wrote the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Pieter Van Vlierberghe, Center for Medical Genetics Ghent, Ghent University Hospital, Medical Research Building, 2nd Floor, Room 120.032, De Pintelaan 185, 9000 Ghent, Belgium; e-mail: pieter.vanvlierberghe@ugent.be.
References
Author notes
H.-G.W. and P.V.V. contributed equally to this study.



This feature is available to Subscribers Only
Sign In or Create an Account Close Modal