Key Points
GATA2 deficiency-associated bone marrow disorder can present with features that overlap with idiopathic aplastic anemia.
GATA2 marrows have severely decreased hematogones, monocytes, NK cells, and B cells; variable dysplasia; and clonal cytogenetic abnormalities.
Abstract
Germ-line GATA2 gene mutations, leading to haploinsufficiency, have been identified in patients with familial myelodysplastic syndrome/acute myeloid leukemia, monocytopenia and mycobacterial infections, Emberger syndrome, and dendritic cell, monocyte, B-, and NK-cell deficiency. GATA2 mutations have also been reported in a minority of patients with congenital neutropenia and aplastic anemia (AA). The bone marrow (BM) from patients with GATA2 deficiency is typically hypocellular, with varying degrees of dysplasia. Distinguishing GATA2 patients from those with AA is critical for selecting appropriate therapy. We compared the BM flow cytometric, morphologic, and cytogenetic features of 28 GATA2 patients with those of 32 patients being evaluated for idiopathic AA. The marrow of GATA2 patients had severely reduced monocytes, B cells, and NK cells; absent hematogones; and inverted CD4:CD8 ratios. Atypical megakaryocytes and abnormal cytogenetics were more common in GATA2 marrows. CD34+ cells were comparably reduced in GATA2 and AA. Using these criteria, we prospectively identified 4 of 32 patients with suspected AA who had features suspicious for GATA2 mutations, later confirmed by DNA sequencing. Our results show that routine BM flow cytometry, morphology, and cytogenetics in patients who present with cytopenia(s) can identify patients for whom GATA2 sequencing is indicated.
Introduction
Inherited and sporadic germ-line mutations of GATA2 leading to haploinsufficiency were first described in a series of patients with overlapping syndromes of GATA2 deficiency including monocytopenia with mycobacterial infections (monoMAC),1,2 immunodeficiency (dendritic cell, monocyte, B- and NK-cell lymphoid deficiency [DCML]),3,4 primary lymphedema and myelodysplastic syndrome (MDS; Emberger syndrome),5 and familial MDS/acute myeloid leukemia (MDS/AML).6 GATA2 mutations have also been identified in a subset of patients presenting with chronic neutropenia,7 pediatric bone marrow failure,8 and young adults with aplastic anemia (AA),9 highlighting the clinical heterogeneity and variable hematologic phenotypes associated with a single genetic defect.
GATA2 is a zinc-finger transcription factor required for maintenance and proliferation of hematopoietic progenitor cells during gestation and after birth. Absence of GATA2 in mice leads to severe anemia incompatible with life.10-12 GATA2 haploinsufficiency results in defective hematopoietic stem cell homeostasis.13 GATA2 also plays a critical role in early erythroid development,14,15 thrombopoiesis,16 myeloid/monocytic/dendritic cell maturation,17,18 and vascular/lymphatic development.19,20 Germ-line GATA2 mutations in patients with GATA2 deficiency have been documented in coding and noncoding regions.2,4-6 The corresponding GATA2 protein changes can be broadly classified as missense (dysfunctional protein), null (absent protein), regulatory (reduced wild-type protein), and uniallelic (wild-type protein expression from only 1 allele).21 Evidence suggests that GATA2 haploinsufficiency is the underlying mechanism responsible for bone marrow (BM) failure, immunodeficiency, MDS/AML, and lymphedema. Detailed genotypic and phenotypic analysis has shown no significant association between genotype and many of the clinical manifestations with the exception of lymphedema and severe infections, which are seen preferentially in patients with null mutations.21,22
Although a small subset of GATA2 patients present with de novo AML, many develop MDS with a high risk of evolution to AML or chronic myelomonocytic leukemia (CMML). MDS is heralded by cytopenias involving red cells, neutrophils, and/or platelets and can be the initial presentation or occur later in the disease process after a period of immunodeficiency associated with severe reduction in monocytes, B cells, and NK cells.21 Unlike de novo MDS, GATA2-related MDS occurs in younger patients with unique morphologic features including bone marrow hypocellularity, multilineage dysplasia, most pronounced in the megakaryocytic lineage, and increased reticulin fibrosis. By flow cytometry, abnormal granulocytic maturation, monocytopenia, and NK-cell and B-cell lymphopenia can be detected in the BM.23
A clinical history of infection, peripheral blood cytopenias, and BM hypocellularity are features common to both GATA2 deficiency and acquired AA. Of note, a subset of GATA2-deficient patients had been previously diagnosed with AA based on cytopenias and hypocellularity seen on BM biopsies.24 The frequency of GATA2 mutations in otherwise de novo hypocellular MDS is unknown, but the recent identification of GATA2 mutations in AA9 suggests that GATA2 deficiency may present with pancytopenia without clinically evident immunodeficiency.
Consequently, the identification of patients with AA who might benefit from GATA2 mutation testing is important for therapeutic management. GATA2-deficient patients are at risk for life-threatening infections and solid malignancies, as well as the increased risk of AML. Additionally, current management protocols for the treatment of acquired AA include immunosuppressive regimens, which may not be optimal for a constitutional BM failure disease. If GATA2 mutations are identified, it is important to screen family members who may be potential donors, as BM transplantation is the only definitive therapy for GATA2 deficiency.25
We analyzed BM flow cytometric, morphologic, and cytogenetic features of a cohort of patients with GATA2 mutations and cytopenias. We compared these features with those of untreated patients with pancytopenia and suspected or confirmed AA and identified 4 new patients with BM features suspicious for a GATA2 mutation who were subsequently confirmed to be GATA2 deficient by DNA sequencing. We also report BM and flow cytometric findings of 4 individuals with documented GATA2 mutations and normal hematopoietic indices. Finally, we summarize the hematologic spectrum associated with GATA2 gene mutations.
Patients, materials, and methods
Patients
The first cohort included 32 individuals with germ-line GATA2 mutations, enrolled in protocols approved by the institutional review boards at our institution, in accordance with the Declaration of Helsinki. Twenty-eight of the 32 individuals had clinical symptoms of GATA2 deficiency, and 4 were asymptomatic family members. The 28 symptomatic patients were chosen on the basis of their cytopenias and the availability of comprehensive BM morphologic and flow cytometric evaluation. A detailed summary of their clinical, laboratory, BM pathologic, and genetic features is summarized in Table 1 and has been recently reviewed.21 The second cohort included 32 untreated patients with pancytopenia and confirmed or suspected diagnosis of idiopathic AA referred to our institution for therapy. In the AA cohort, leukocyte telomere content was normal for age, and chromosome breakage analysis, performed to rule out Fanconi anemia in all patients <40 years old, was negative. Additionally, no patients in the AA cohort had evidence of a paroxysmal nocturnal hemoglobinuria clone. A detailed summary of their laboratory, BM pathologic, and cytogenetic features is summarized in Table 2. The control group comprised 6 healthy volunteers.
Clinical, laboratory, BM pathologic, and genetic features of 28 patients with GATA2 deficiency (modified from Spinner et al21 )
| Patient . | Age (years) . | Gender . | Mutation . | Peripheral blood (normal range × 109/L) . | Bone marrow . | Infections . | |||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Genomic . | Protein . | WBC (4.23-9.07) . | ANC (1.56-6.13) . | ALC (1.18-3.74) . | AMC (0.30-0.82) . | Hb (13.7-17.5 g/dL) . | Plt (161-347) . | B-cells (0.07- 0.7) . | T-cells (0.67-2.95) . | NK-cells (0.08- 1.15) . | Pathology . | Cytogenetics . | Viral . | NTM . | Fungal . | Other . | |||
| 2.II.3 | 39 | M | c.1192C>T | p.R398W missense | 4.06 | 3.65 | 0.34 | 0.06 | 8.2 | 26 | 0 | 0.34 | 0.1 | MDS-RAEB-2 | +8, t(1;7)(q10; p10) | HPV | MAC | H capsulatum, C glabrata | Serratia S epidermidis |
| 4.II.1 | 52 | F | c.1017+572C>T | reduced expression | 3.51 | 2.43 | 0.83 | 0.01 | 12.5 | 190 | 0.002 | 0.83 | 0.002 | MDS-RCMD | N/A | HPVCMV | MAC | H capsulatum | Serratia |
| 5.II.1 | 44 | F | c.1061C>T | p.T354M missense | 5.69 | 4.15 | 0.51 | 0.06 | 9.8 | 122 | 0.004 | 0.51 | 0.002 | MDS-RCMD | −6, ring | HPV | MAC M abscessus M fortuitum | None | MRSA P aeruginosa |
| 8.I.1 | 33 | M | c.243_244delAinsGC | p.G81fs null allele | 6.12 | 5.26 | 0.8 | 0 | 11 | 65 | 0.004 | 0.79 | 0.01 | MDS-RCMD | Normal | HPV | MAC | Aspergillus, Penicillium | P aeruginosa |
| 9.III.1 | 22 | M | c.1192C>T | p.R398W missense | 0.79 | 0.31 | 0.38 | 0 | 7 | 26 | 0.006 | 0.37 | 0.005 | MDS-RCMD | Normal | HPV | M Fortuitum | None | None |
| 11.II.1 | 40 | F | c.1017+572C>T | Reduced expression | 4.34 | 2.89 | 1.44 | 0.1 | 13.7 | 342 | 0.062 | 1.34 | 0.043 | MDS-RCUD | Normal | HPV VZV | None | Candida | S aureus |
| 12.I.1 | 27 | M | c.1083_1094del12 | p.R361del4 in-frame deletion | 3.47 | 2.56 | 0.35 | 0.08 | 8.3 | 41 | 0.001 | 0.35 | 0 | MDS-RCMD | −7, +21 | HPVHSVHCV | M kansasii | Candida | E coli P acnes VRE |
| 13.I.1 | 60 | F | c.1-200_871 +527del2033ins7 | p.M1del290 null allele | 3.87 | 2.22 | 1.49 | 0.001 | 13.6 | 348 | 0.021 | 1.44 | 0.022 | MDS-RCUD | Normal | HPV | None | None | None |
| 13.II.1 | 33 | M | c.1-200_871 +527del2033ins7 | p.M1del290 null allele | 1.14 | 0.95 | 0.08 | 0.03 | 9.7 | 22 | 0 | 0.08 | 0.001 | MDS-RAEB-1 | −7 | HPVHSV | MAC | H capsulatum, N udagawae | S epidermidis P aeruginosa Klebsiella |
| 15.I.1 | 14 | F | c.1186C>T | p.R396W missense | 2.67 | 1.5 | 1 | 0.01 | 11.9 | 174 | 0.005 | 0.97 | 0.024 | MDS-RCMD | +8, +mar | HSV | MAC | None | E histolytica |
| 17.I.1 | 32 | M | c.1061C>T | p.T354M missense | 1.44 | 0.69 | 0.64 | 0 | 13.5 | 61 | 0.003 | 0.49 | 0.145 | MDS-RCMD | +8 | HPV | MAC | T rubrum | None |
| 20.I.3 | 15 | M | c.769_778dup | p.Y260fsX24 null allele | 3.12 | 2.86 | 0.19 | 0.01 | 8.5 | 88 | 0 | 0.19 | 0 | MDS-RCMD | −7 | HPV MCV | MAC | None | Bartonella C difficile |
| 21.II.1 | 33 | M | c.1192C>T | p.R398W missense | 8.2 | 7.3 | 0.9 | 0 | 7.7 | 21 | 0.002 | 0.9 | 0.001 | MDS-RCMD | -Y | HPV MCV | MAC | None | None |
| 22.I.1 | 26 | F | c.941_951dup | p.A318fsX12 null allele | 0.48 | 0.23 | 0.25 | 0 | 9.4 | 17 | N/A | N/A | N/A | MDS-RCMD | −6 | HPV | M kansasii | None | None |
| 24.I.1 | 45 | F | c.1018-1G>A | p.del340-381 in-frame deletion | 1.34 | 1.14 | 0.07 | 0.03 | 10.5 | 138 | 0 | 0.07 | 0 | MDS-RCMD | der(22) t(1;22)(q12;p23) | HPVVZV | MAC | None | None |
| 31.II.1 | 32 | M | c.1187G>A | p.R396Q missense | 1.87 | 1.6 | 0.27 | 0 | 10.5 | 47 | 0.004 | 0.26 | 0 | MDS-RCMD | Normal | HPV | Granulo-matous lymphadenitis | Aspergillus | None |
| 31.II.2 | 29 | M | c.1187G>A | p.R396Q missense | 1.8 | 0.48 | 1.3 | 0.01 | 9.5 | 113 | 0.065 | 1.23 | 0.004 | MDS-RCMD | N/A | None | None | None | Polymicrobial infections |
| 33.III.1 | 17 | F | c.1099insG | p.D367GfsX15 truncated protein | 4.53 | 3.76 | 0.72 | 0 | 11.8 | 97 | 0.001 | 0.71 | 0.004 | MDS-RCMD | Normal | None | MAC | None | Brucella, C difficile |
| 34.II.2 | 17 | M | c.1116_1130del15bp | p.C373del5 in-frame deletion | 1.73 | 0.96 | 0.7 | 0.04 | 11.3 | 116 | 0.007 | 0.69 | 0.005 | MDS-RCUD | −7 | None | None | None | None |
| 35.III.3 | 33 | M | c.1061C>T | p.T354M missense | 3.17 | 2.19 | 0.91 | 0.02 | 14.3 | 106 | 0.007 | 0.9 | 0.003 | MDS-RCMD | +8 | None | Granulo-matous lymphadenitis | None | Polymicrobial infections |
| 38.I.1 | 25 | F | c.417dupT | p.V140CfsX44 null allele | 2.34 | 1.4 | 0.91 | 0 | 13.7 | 163 | 0.011 | 0.84 | 0.053 | MDS-RCUD | Normal | HPV | None | None | MRSA |
| 39.I.1 | 25 | F | c.988C>T | p.R330X null allele | 2.96 | 2.53 | 0.31 | 0.02 | 9.6 | 263 | 0.006 | 0.3 | 0 | MDS-RCMD | +8 | HPV HSVEBV | None | None | None |
| 39.I.2 | 25 | F | c.988C>T | p.R330X null allele | 1.64 | 1.3 | 0.25 | 0.02 | 8.6 | 232 | 0.001 | 0.25 | 0.005 | MDS-RCMD | +8 | HPV HSV EBV | None | None | Salmonella Neisseria |
| 41.I.1 | 44 | F | c.1009C>T | p.R337X null allele | 6.43 | 5.04 | 1.2 | 0.01 | 10.2 | 724 | 0.002 | 1.18 | 0.01 | MDS-RCMD | Normal | HPVHSV | None | Exophiala | Polymicrobial infections |
| 42.I.1 | 24 | M | c.1186C>T | p.R396W missense | 2.47 | 1.35 | 0.34 | 0.01 | 12.4 | 86 | 0.001 | 0.34 | 0 | MDS-RCMD | Normal | HPVEBV | MAC, M szulgai | None | Bacterial infections |
| 50 | 31 | M | c.1082G>A | p.R361H missense | 0.75 | 0.44 | 0.26 | 0.01 | 8.1 | 118 | 0.017 | 0.24 | 0.001 | MDS-RAEB1 | Normal | None | None | None | None |
| 51 | 19 | F | c.1017+572C>T | reduced expression | 1.44 | 0.43 | 0.97 | 0 | 10.4 | 152 | 0.042 | 0.89 | 0.042 | MDS-RCMD | +1, +8, der(1;7)(q10;p10) | None | None | None | None |
| 52 | 28 | F | Uniallelic | reduced expression | 2 | 0.43 | 1.57 | 0 | 10.5 | 153 | 0.028 | 1.52 | 0.022 | MDS-RCMD | +8 | None | None | None | None |
| Patient . | Age (years) . | Gender . | Mutation . | Peripheral blood (normal range × 109/L) . | Bone marrow . | Infections . | |||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Genomic . | Protein . | WBC (4.23-9.07) . | ANC (1.56-6.13) . | ALC (1.18-3.74) . | AMC (0.30-0.82) . | Hb (13.7-17.5 g/dL) . | Plt (161-347) . | B-cells (0.07- 0.7) . | T-cells (0.67-2.95) . | NK-cells (0.08- 1.15) . | Pathology . | Cytogenetics . | Viral . | NTM . | Fungal . | Other . | |||
| 2.II.3 | 39 | M | c.1192C>T | p.R398W missense | 4.06 | 3.65 | 0.34 | 0.06 | 8.2 | 26 | 0 | 0.34 | 0.1 | MDS-RAEB-2 | +8, t(1;7)(q10; p10) | HPV | MAC | H capsulatum, C glabrata | Serratia S epidermidis |
| 4.II.1 | 52 | F | c.1017+572C>T | reduced expression | 3.51 | 2.43 | 0.83 | 0.01 | 12.5 | 190 | 0.002 | 0.83 | 0.002 | MDS-RCMD | N/A | HPVCMV | MAC | H capsulatum | Serratia |
| 5.II.1 | 44 | F | c.1061C>T | p.T354M missense | 5.69 | 4.15 | 0.51 | 0.06 | 9.8 | 122 | 0.004 | 0.51 | 0.002 | MDS-RCMD | −6, ring | HPV | MAC M abscessus M fortuitum | None | MRSA P aeruginosa |
| 8.I.1 | 33 | M | c.243_244delAinsGC | p.G81fs null allele | 6.12 | 5.26 | 0.8 | 0 | 11 | 65 | 0.004 | 0.79 | 0.01 | MDS-RCMD | Normal | HPV | MAC | Aspergillus, Penicillium | P aeruginosa |
| 9.III.1 | 22 | M | c.1192C>T | p.R398W missense | 0.79 | 0.31 | 0.38 | 0 | 7 | 26 | 0.006 | 0.37 | 0.005 | MDS-RCMD | Normal | HPV | M Fortuitum | None | None |
| 11.II.1 | 40 | F | c.1017+572C>T | Reduced expression | 4.34 | 2.89 | 1.44 | 0.1 | 13.7 | 342 | 0.062 | 1.34 | 0.043 | MDS-RCUD | Normal | HPV VZV | None | Candida | S aureus |
| 12.I.1 | 27 | M | c.1083_1094del12 | p.R361del4 in-frame deletion | 3.47 | 2.56 | 0.35 | 0.08 | 8.3 | 41 | 0.001 | 0.35 | 0 | MDS-RCMD | −7, +21 | HPVHSVHCV | M kansasii | Candida | E coli P acnes VRE |
| 13.I.1 | 60 | F | c.1-200_871 +527del2033ins7 | p.M1del290 null allele | 3.87 | 2.22 | 1.49 | 0.001 | 13.6 | 348 | 0.021 | 1.44 | 0.022 | MDS-RCUD | Normal | HPV | None | None | None |
| 13.II.1 | 33 | M | c.1-200_871 +527del2033ins7 | p.M1del290 null allele | 1.14 | 0.95 | 0.08 | 0.03 | 9.7 | 22 | 0 | 0.08 | 0.001 | MDS-RAEB-1 | −7 | HPVHSV | MAC | H capsulatum, N udagawae | S epidermidis P aeruginosa Klebsiella |
| 15.I.1 | 14 | F | c.1186C>T | p.R396W missense | 2.67 | 1.5 | 1 | 0.01 | 11.9 | 174 | 0.005 | 0.97 | 0.024 | MDS-RCMD | +8, +mar | HSV | MAC | None | E histolytica |
| 17.I.1 | 32 | M | c.1061C>T | p.T354M missense | 1.44 | 0.69 | 0.64 | 0 | 13.5 | 61 | 0.003 | 0.49 | 0.145 | MDS-RCMD | +8 | HPV | MAC | T rubrum | None |
| 20.I.3 | 15 | M | c.769_778dup | p.Y260fsX24 null allele | 3.12 | 2.86 | 0.19 | 0.01 | 8.5 | 88 | 0 | 0.19 | 0 | MDS-RCMD | −7 | HPV MCV | MAC | None | Bartonella C difficile |
| 21.II.1 | 33 | M | c.1192C>T | p.R398W missense | 8.2 | 7.3 | 0.9 | 0 | 7.7 | 21 | 0.002 | 0.9 | 0.001 | MDS-RCMD | -Y | HPV MCV | MAC | None | None |
| 22.I.1 | 26 | F | c.941_951dup | p.A318fsX12 null allele | 0.48 | 0.23 | 0.25 | 0 | 9.4 | 17 | N/A | N/A | N/A | MDS-RCMD | −6 | HPV | M kansasii | None | None |
| 24.I.1 | 45 | F | c.1018-1G>A | p.del340-381 in-frame deletion | 1.34 | 1.14 | 0.07 | 0.03 | 10.5 | 138 | 0 | 0.07 | 0 | MDS-RCMD | der(22) t(1;22)(q12;p23) | HPVVZV | MAC | None | None |
| 31.II.1 | 32 | M | c.1187G>A | p.R396Q missense | 1.87 | 1.6 | 0.27 | 0 | 10.5 | 47 | 0.004 | 0.26 | 0 | MDS-RCMD | Normal | HPV | Granulo-matous lymphadenitis | Aspergillus | None |
| 31.II.2 | 29 | M | c.1187G>A | p.R396Q missense | 1.8 | 0.48 | 1.3 | 0.01 | 9.5 | 113 | 0.065 | 1.23 | 0.004 | MDS-RCMD | N/A | None | None | None | Polymicrobial infections |
| 33.III.1 | 17 | F | c.1099insG | p.D367GfsX15 truncated protein | 4.53 | 3.76 | 0.72 | 0 | 11.8 | 97 | 0.001 | 0.71 | 0.004 | MDS-RCMD | Normal | None | MAC | None | Brucella, C difficile |
| 34.II.2 | 17 | M | c.1116_1130del15bp | p.C373del5 in-frame deletion | 1.73 | 0.96 | 0.7 | 0.04 | 11.3 | 116 | 0.007 | 0.69 | 0.005 | MDS-RCUD | −7 | None | None | None | None |
| 35.III.3 | 33 | M | c.1061C>T | p.T354M missense | 3.17 | 2.19 | 0.91 | 0.02 | 14.3 | 106 | 0.007 | 0.9 | 0.003 | MDS-RCMD | +8 | None | Granulo-matous lymphadenitis | None | Polymicrobial infections |
| 38.I.1 | 25 | F | c.417dupT | p.V140CfsX44 null allele | 2.34 | 1.4 | 0.91 | 0 | 13.7 | 163 | 0.011 | 0.84 | 0.053 | MDS-RCUD | Normal | HPV | None | None | MRSA |
| 39.I.1 | 25 | F | c.988C>T | p.R330X null allele | 2.96 | 2.53 | 0.31 | 0.02 | 9.6 | 263 | 0.006 | 0.3 | 0 | MDS-RCMD | +8 | HPV HSVEBV | None | None | None |
| 39.I.2 | 25 | F | c.988C>T | p.R330X null allele | 1.64 | 1.3 | 0.25 | 0.02 | 8.6 | 232 | 0.001 | 0.25 | 0.005 | MDS-RCMD | +8 | HPV HSV EBV | None | None | Salmonella Neisseria |
| 41.I.1 | 44 | F | c.1009C>T | p.R337X null allele | 6.43 | 5.04 | 1.2 | 0.01 | 10.2 | 724 | 0.002 | 1.18 | 0.01 | MDS-RCMD | Normal | HPVHSV | None | Exophiala | Polymicrobial infections |
| 42.I.1 | 24 | M | c.1186C>T | p.R396W missense | 2.47 | 1.35 | 0.34 | 0.01 | 12.4 | 86 | 0.001 | 0.34 | 0 | MDS-RCMD | Normal | HPVEBV | MAC, M szulgai | None | Bacterial infections |
| 50 | 31 | M | c.1082G>A | p.R361H missense | 0.75 | 0.44 | 0.26 | 0.01 | 8.1 | 118 | 0.017 | 0.24 | 0.001 | MDS-RAEB1 | Normal | None | None | None | None |
| 51 | 19 | F | c.1017+572C>T | reduced expression | 1.44 | 0.43 | 0.97 | 0 | 10.4 | 152 | 0.042 | 0.89 | 0.042 | MDS-RCMD | +1, +8, der(1;7)(q10;p10) | None | None | None | None |
| 52 | 28 | F | Uniallelic | reduced expression | 2 | 0.43 | 1.57 | 0 | 10.5 | 153 | 0.028 | 1.52 | 0.022 | MDS-RCMD | +8 | None | None | None | None |
ALC, absolute lymphocyte count; AMC, absolute monocyte count; ANC, absolute neutrophil count; EBV, Epstein-Barr virus; Hb, hemoglobin; HCV, hepatitis C virus; HPV, human papilloma virus; HSV, herpes simplex virus; MAC, M avium complex; MCV, molluscum contagiosum virus; MDS, myelodysplastic syndrome; MRSA, methicillin-resistant S aureus; NTM, non-tuberculous Mycobacterium; Plt, platelet count; RAEB, refractory anemia with excess blasts; RCMD, refractory cytopenia with multilineage dysplasia; RCUD, refractory cytopenia with unilineage dysplasia; VZV, varicella zoster virus; WBC, white blood cell.
Clinical, laboratory, BM pathologic, and cytogenetic features of 28 patients with AA
| Patient . | Age (years) . | Gender . | Peripheral blood (normal range × 109/L) . | Bone marrow . | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| WBC (4.23-9.07) . | ANC (1.56-6.13) . | ALC (1.18-3.74) . | AMC (0.30-0.82) . | Hb (13.7-17.5 g/dL) . | Plt (161-347) . | B-cells (0.07-0.71) . | T-cells (0.67-2.95) . | NK-cells (0.08-1.15) . | Pathology . | Cytogenetics . | |||
| 1 | 22 | F | 4.16 | 1.28 | 2.62 | 0.15 | 8.4 | 28 | 0.18 | 2.31 | 0.12 | Hypocellular with trilineage hypoplasia | Normal |
| 2 | 53 | F | 2.77 | 0.1 | 2.67 | 0 | 9.9 | 88 | 0.78 | 1.81 | 0.07 | Hypocellular with trilineage hypoplasia | Normal |
| 3 | 73 | F | 1.68 | 0.58 | 1.01 | 0.07 | 9.3 | 10 | 0.12 | 0.76 | 0.13 | Hypocellular with trilineage hypoplasia | Normal |
| 4 | 28 | F | 1.74 | 0.83 | 0.76 | 0.12 | 9.5 | 84 | 0.03 | 0.67 | 0.05 | Hypocellular with trilineage hypoplasia | Normal |
| 5 | 26 | M | 1.76 | 0.66 | 0.73 | 0.35 | 10.1 | 22 | 0.07 | 0.25 | 0.40 | Hypocellular with trilineage hypoplasia | Normal |
| 6 | 44 | F | 2.23 | 0.95 | 1.11 | 0.14 | 9.7 | 99 | 0.04 | 0.92 | 0.13 | Hypocellular with trilineage hypoplasia | Normal |
| 7 | 21 | F | 3.82 | 1.49 | 1.97 | 0.31 | 12 | 59 | 0.18 | 1.65 | 0.13 | Hypocellular with trilineage hypoplasia | Normal |
| 8 | 28 | F | 2.27 | 0.87 | 0.96 | 0.29 | 11.4 | 68 | 0.08 | 0.79 | 0.08 | Hypocellular with trilineage hypoplasia | Normal |
| 9 | 20 | M | 4.03 | 0.55 | 3.4 | 0.08 | 8.3 | 8 | 0.38 | 2.87 | 0.14 | Hypocellular with trilineage hypoplasia | Normal |
| 10 | 33 | F | 3.33 | 1.87 | 1 | 0.32 | 10.6 | 66 | 0.10 | 0.76 | 0.13 | Hypocellular with trilineage hypoplasia | Normal |
| 11 | 15 | F | 3.49 | 1.25 | 1.97 | 0.24 | 8.8 | 23 | 0.28 | 1.58 | 0.10 | Hypocellular with trilineage hypoplasia | Normal |
| 12 | 49 | F | 5.84 | 2.2 | 3.5 | 0.15 | 6.6 | 4 | 0.56 | 2.80 | 0.13 | Hypocellular with trilineage hypoplasia | Normal |
| 13 | 39 | F | 1.96 | 0.51 | 1.34 | 0.07 | 10.6 | 26 | 0.24 | 1.01 | 0.08 | Hypocellular with trilineage hypoplasia | Normal |
| 14 | 12 | M | 1.58 | 0.27 | 1.29 | 0.02 | 9.1 | 8 | 0.12 | 0.97 | 0.17 | Hypocellular with trilineage hypoplasia | Normal |
| 15 | 49 | F | 3.33 | 1.53 | 1.67 | 0.12 | 8.8 | 11 | 0.10 | 1.47 | 0.10 | Hypocellular with trilineage hypoplasia | Normal |
| 16 | 29 | F | 2.65 | 1.47 | 0.98 | 0.17 | 10.2 | 99 | 0.07 | 0.74 | 0.15 | Hypocellular with trilineage hypoplasia | Normal |
| 17 | 59 | M | 1.54 | 0.32 | 1.16 | 0.04 | 9.6 | 48 | 0.15 | 0.95 | 0.06 | Hypocellular with trilineage hypoplasia | Normal |
| 18 | 56 | F | 1.29 | 0.39 | 0.65 | 0.25 | 8.8 | 52 | 0.04 | 0.59 | 0.01 | Hypocellular with trilineage hypoplasia | Normal |
| 19 | 16 | M | 1.23 | 0.21 | 0.98 | 0.03 | 12.3 | 29 | 0.08 | 0.84 | 0.05 | Hypocellular with trilineage hypoplasia | Normal |
| 20 | 45 | M | 1.09 | 0.07 | 0.96 | 0.05 | 6.4 | 2 | 0.22 | 0.72 | 0.01 | Hypocellular with trilineage hypoplasia | Normal |
| 21 | 48 | F | 1.69 | 0.07 | 1.55 | 0.06 | 9.6 | 10 | 0.12 | 1.34 | 0.09 | Hypocellular with trilineage hypoplasia | Normal |
| 22 | 31 | F | 2.46 | 0.7 | 1.55 | 0.16 | 8.7 | 21 | 0.10 | 1.37 | 0.07 | Hypocellular with trilineage hypoplasia | Normal |
| 23 | 33 | M | 1.81 | 0.04 | 1.72 | 0.01 | 12.2 | 12 | 0.19 | 1.43 | 0.09 | Hypocellular with trilineage hypoplasia | Normal |
| 24 | 63 | M | 1.22 | 0.2 | 0.94 | 0.08 | 9.5 | 17 | 0.29 | 0.51 | 0.07 | Hypocellular with trilineage hypoplasia | Normal |
| 25 | 21 | M | 11.22 | 6.96 | 3.57 | 0.58 | 8.1 | 11 | 0.92 | 2.55 | 0.07 | Hypocellular with trilineage hypoplasia | Normal |
| 26 | 21 | M | 4.07 | 0.44 | 3.59 | 0.04 | 7.7 | 17 | 0.41 | 3.10 | 0.05 | Hypocellular with trilineage hypoplasia | Normal |
| 27 | 29 | M | 2.47 | 0.71 | 1.59 | 0.09 | 8.8 | 57 | 0.09 | 1.38 | 0.11 | Hypocellular with trilineage hypoplasia | Normal |
| 28 | 27 | F | 2.20 | 0.51 | 1.64 | 0.06 | 7.3 | 10 | 0.15 | 1.41 | 0.07 | Hypocellular with trilineage hypoplasia | Normal |
| Patient . | Age (years) . | Gender . | Peripheral blood (normal range × 109/L) . | Bone marrow . | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| WBC (4.23-9.07) . | ANC (1.56-6.13) . | ALC (1.18-3.74) . | AMC (0.30-0.82) . | Hb (13.7-17.5 g/dL) . | Plt (161-347) . | B-cells (0.07-0.71) . | T-cells (0.67-2.95) . | NK-cells (0.08-1.15) . | Pathology . | Cytogenetics . | |||
| 1 | 22 | F | 4.16 | 1.28 | 2.62 | 0.15 | 8.4 | 28 | 0.18 | 2.31 | 0.12 | Hypocellular with trilineage hypoplasia | Normal |
| 2 | 53 | F | 2.77 | 0.1 | 2.67 | 0 | 9.9 | 88 | 0.78 | 1.81 | 0.07 | Hypocellular with trilineage hypoplasia | Normal |
| 3 | 73 | F | 1.68 | 0.58 | 1.01 | 0.07 | 9.3 | 10 | 0.12 | 0.76 | 0.13 | Hypocellular with trilineage hypoplasia | Normal |
| 4 | 28 | F | 1.74 | 0.83 | 0.76 | 0.12 | 9.5 | 84 | 0.03 | 0.67 | 0.05 | Hypocellular with trilineage hypoplasia | Normal |
| 5 | 26 | M | 1.76 | 0.66 | 0.73 | 0.35 | 10.1 | 22 | 0.07 | 0.25 | 0.40 | Hypocellular with trilineage hypoplasia | Normal |
| 6 | 44 | F | 2.23 | 0.95 | 1.11 | 0.14 | 9.7 | 99 | 0.04 | 0.92 | 0.13 | Hypocellular with trilineage hypoplasia | Normal |
| 7 | 21 | F | 3.82 | 1.49 | 1.97 | 0.31 | 12 | 59 | 0.18 | 1.65 | 0.13 | Hypocellular with trilineage hypoplasia | Normal |
| 8 | 28 | F | 2.27 | 0.87 | 0.96 | 0.29 | 11.4 | 68 | 0.08 | 0.79 | 0.08 | Hypocellular with trilineage hypoplasia | Normal |
| 9 | 20 | M | 4.03 | 0.55 | 3.4 | 0.08 | 8.3 | 8 | 0.38 | 2.87 | 0.14 | Hypocellular with trilineage hypoplasia | Normal |
| 10 | 33 | F | 3.33 | 1.87 | 1 | 0.32 | 10.6 | 66 | 0.10 | 0.76 | 0.13 | Hypocellular with trilineage hypoplasia | Normal |
| 11 | 15 | F | 3.49 | 1.25 | 1.97 | 0.24 | 8.8 | 23 | 0.28 | 1.58 | 0.10 | Hypocellular with trilineage hypoplasia | Normal |
| 12 | 49 | F | 5.84 | 2.2 | 3.5 | 0.15 | 6.6 | 4 | 0.56 | 2.80 | 0.13 | Hypocellular with trilineage hypoplasia | Normal |
| 13 | 39 | F | 1.96 | 0.51 | 1.34 | 0.07 | 10.6 | 26 | 0.24 | 1.01 | 0.08 | Hypocellular with trilineage hypoplasia | Normal |
| 14 | 12 | M | 1.58 | 0.27 | 1.29 | 0.02 | 9.1 | 8 | 0.12 | 0.97 | 0.17 | Hypocellular with trilineage hypoplasia | Normal |
| 15 | 49 | F | 3.33 | 1.53 | 1.67 | 0.12 | 8.8 | 11 | 0.10 | 1.47 | 0.10 | Hypocellular with trilineage hypoplasia | Normal |
| 16 | 29 | F | 2.65 | 1.47 | 0.98 | 0.17 | 10.2 | 99 | 0.07 | 0.74 | 0.15 | Hypocellular with trilineage hypoplasia | Normal |
| 17 | 59 | M | 1.54 | 0.32 | 1.16 | 0.04 | 9.6 | 48 | 0.15 | 0.95 | 0.06 | Hypocellular with trilineage hypoplasia | Normal |
| 18 | 56 | F | 1.29 | 0.39 | 0.65 | 0.25 | 8.8 | 52 | 0.04 | 0.59 | 0.01 | Hypocellular with trilineage hypoplasia | Normal |
| 19 | 16 | M | 1.23 | 0.21 | 0.98 | 0.03 | 12.3 | 29 | 0.08 | 0.84 | 0.05 | Hypocellular with trilineage hypoplasia | Normal |
| 20 | 45 | M | 1.09 | 0.07 | 0.96 | 0.05 | 6.4 | 2 | 0.22 | 0.72 | 0.01 | Hypocellular with trilineage hypoplasia | Normal |
| 21 | 48 | F | 1.69 | 0.07 | 1.55 | 0.06 | 9.6 | 10 | 0.12 | 1.34 | 0.09 | Hypocellular with trilineage hypoplasia | Normal |
| 22 | 31 | F | 2.46 | 0.7 | 1.55 | 0.16 | 8.7 | 21 | 0.10 | 1.37 | 0.07 | Hypocellular with trilineage hypoplasia | Normal |
| 23 | 33 | M | 1.81 | 0.04 | 1.72 | 0.01 | 12.2 | 12 | 0.19 | 1.43 | 0.09 | Hypocellular with trilineage hypoplasia | Normal |
| 24 | 63 | M | 1.22 | 0.2 | 0.94 | 0.08 | 9.5 | 17 | 0.29 | 0.51 | 0.07 | Hypocellular with trilineage hypoplasia | Normal |
| 25 | 21 | M | 11.22 | 6.96 | 3.57 | 0.58 | 8.1 | 11 | 0.92 | 2.55 | 0.07 | Hypocellular with trilineage hypoplasia | Normal |
| 26 | 21 | M | 4.07 | 0.44 | 3.59 | 0.04 | 7.7 | 17 | 0.41 | 3.10 | 0.05 | Hypocellular with trilineage hypoplasia | Normal |
| 27 | 29 | M | 2.47 | 0.71 | 1.59 | 0.09 | 8.8 | 57 | 0.09 | 1.38 | 0.11 | Hypocellular with trilineage hypoplasia | Normal |
| 28 | 27 | F | 2.20 | 0.51 | 1.64 | 0.06 | 7.3 | 10 | 0.15 | 1.41 | 0.07 | Hypocellular with trilineage hypoplasia | Normal |
Flow cytometric analysis
Flow cytometric analysis of BM aspirates was performed using a fluorescence-activated cell sorter (FACS) Canto II Analyzer (BD Biosciences) equipped with 3 lasers and 8 fluorescent detectors. The following antibodies were used: CD45, CD34, CD117, HLA DR, CD13, CD15 CD11b, CD11c, CD14, CD16, CD38, CD42b, CD36, CD64, CD33, CD123, CD61, CD71, CD5, CD19, CD10, CD20, CD23, CD10, kappa, lambda, CD3, CD7, CD2, CD4, CD8, CD57, CD56, CD27, and CD138. Briefly, cells were stained with the appropriate antibodies for 15 minutes. Red blood cells were lysed with BD FACS lysing solution, and cells were washed with phosphate-buffered saline containing 1% albumin. Cells were fixed in a 1% paraformaldehyde solution, and 1 × 105 events were acquired using FACSDiva (BD Biosciences). The list mode files were analyzed with FCS Express (DeNovo Software).
BM aspirates, biopsy, and immunohistochemistry
BM biopsies and aspirates were independently reviewed by 3 hematopathologists. Immunohistochemical and special stains (CD34, CD61, and reticulin) were performed on a Ventana Benchmark Ultra platform (Ventana Medical Systems). Reticulin fibrosis was graded using a previously published scheme.26
Cytogenetic analysis of BM
Cytogenetic studies were performed on 27 of the 32 individuals with GATA2 mutations (cohort 1) in the National Cancer Institute laboratory using standard methods. Briefly, metaphase chromosomes were obtained from 24- and 48-hour unstimulated cultures of buffy coat cells from fresh, heparinized BM aspirates. Chromosomes were G-banded using Wright’s stain. A minimum of 20 metaphase cells were fully analyzed at the 400 to 550 band level of resolution whenever present. Multiple digital images were captured and karyotyped using a CytoVision system (Leica Biosystems, San Jose, CA). Clones were defined and karyotypes were designated according to the International System for Cytogenetic Nomenclature 2013 (ISCN 2013).27
BM cytogenetic analyses were performed outside the National Institutes of Health using similar methods on the other 5 individuals with GATA2 mutations (cohort 1) and all 32 patients evaluated for AA (cohort 2). Results were reviewed by D.C.A., K.R.C., or K.A.G.
GATA2 mutation analysis
DNA and RNA were isolated from peripheral blood using Puregene DNA purification kit (Qiagen) or Rneasy (Qiagen). All exons and intronic regulatory sites of GATA2 were amplified as described previously.2 cDNA amplification of GATA2 was performed as described previously.28 Twenty-seven patients carried GATA2 mutations, whereas 1 patient had uni-allelic expression as previously described.21,28
Molecular analysis for T-cell receptor gene rearrangement
DNA was extracted from paraffin-embedded tissue sections, and a single multiplexed polymerase chain reaction was performed for the T-cell receptor gene locus, as previously described by Lawnicki et al.29
Statistics
Nonparametric tests were used to compare different groups (Mann-Whitney test), and statistical analysis was performed using GraphPad Prism (GraphPad Software).
Results
Comparison of peripheral blood indices between GATA2 and AA patients
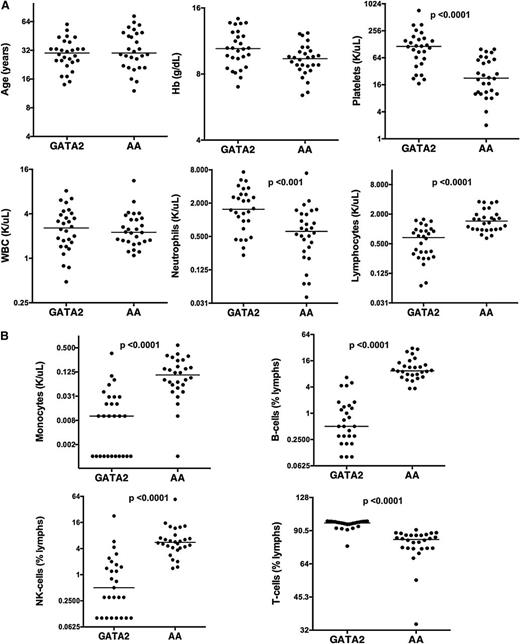
We compared 28 patients with GATA2 mutations/GATA2 deficiency and uni- or multilineage cytopenias (henceforth referred as GATA2) and 28 patients with untreated AA that were confirmed negative for GATA2 mutation by DNA sequencing analysis. The distribution of cytopenias in GATA2 patients at the time of BM biopsies was as follows: 11 patients had pancytopenia, 8 patients had anemia with thrombocytopenia, 6 patients had anemia, 1 patient had neutropenia with anemia, and 1 patient each had neutropenia and thrombocytopenia, respectively. Median ages of GATA2 (30 years; range, 14-60 years) and AA patients (30 years; range, 12-73 years) were comparable. GATA2 and AA had similar hemoglobin levels, but AA patients had significantly reduced platelet counts and severe thrombocytopenia in comparison with GATA2 (Table 3). Although overall peripheral blood white blood cell counts were comparable, GATA2 had relatively higher neutrophil counts and decreased lymphocytes compared with AA patients (Figure 1A).
Summary of peripheral blood counts and immunophenotyping of GATA2 and AA patients
| Features . | GATA2 . | AA . | P . |
|---|---|---|---|
| Hemoglobin (median, g/L; normal range 137-175 g/L) | 105.00 | 94.00 | .03 |
| Platelets (median, x 109/L; normal range 161-347 109/L) | 115.00 | 23.00 | <.0001 |
| WBC (median, 109/L; normal range 4.23-9.07 109/L) | 2.57 | 2.25 | .7054 |
| Neutrophils (median, 109/L; normal range 1.56-6.13 109/L) | 1.55 | 0.62 | <.001 |
| Monocytes (median, 109/L; normal range 0.30-0.82 109/L) | 0.01 | 0.10 | <.001 |
| Lymphocytes (median, 109/L; normal range 1.18-3.74 109/L) | 0.67 | 1.45 | <.0001 |
| B cells (median, % lymphs; normal range 6-19%) (median, 109/L; normal range 0.07- 0.71 109/L) | 0.50 | 9.40 | <.0001 |
| 0.003 | 0.13 | <.0001 | |
| T cells (median, % lymphs; normal range 57-79%) (median, 109/L; normal range 0.67- 2.95 109/L) | 98.10 | 82.60 | <.0001 |
| 0.59 | 1.17 | <.0001 | |
| NK cells (median, % lymphs; normal range 7-31%) (median, 109/L; normal range 0.08- 1.15 109/L) | 0.50 | 5.60 | <.0001 |
| 0.003165 | 0.09122 | <.0001 |
| Features . | GATA2 . | AA . | P . |
|---|---|---|---|
| Hemoglobin (median, g/L; normal range 137-175 g/L) | 105.00 | 94.00 | .03 |
| Platelets (median, x 109/L; normal range 161-347 109/L) | 115.00 | 23.00 | <.0001 |
| WBC (median, 109/L; normal range 4.23-9.07 109/L) | 2.57 | 2.25 | .7054 |
| Neutrophils (median, 109/L; normal range 1.56-6.13 109/L) | 1.55 | 0.62 | <.001 |
| Monocytes (median, 109/L; normal range 0.30-0.82 109/L) | 0.01 | 0.10 | <.001 |
| Lymphocytes (median, 109/L; normal range 1.18-3.74 109/L) | 0.67 | 1.45 | <.0001 |
| B cells (median, % lymphs; normal range 6-19%) (median, 109/L; normal range 0.07- 0.71 109/L) | 0.50 | 9.40 | <.0001 |
| 0.003 | 0.13 | <.0001 | |
| T cells (median, % lymphs; normal range 57-79%) (median, 109/L; normal range 0.67- 2.95 109/L) | 98.10 | 82.60 | <.0001 |
| 0.59 | 1.17 | <.0001 | |
| NK cells (median, % lymphs; normal range 7-31%) (median, 109/L; normal range 0.08- 1.15 109/L) | 0.50 | 5.60 | <.0001 |
| 0.003165 | 0.09122 | <.0001 |
Peripheral blood indices of GATA2 and AA patients. (A) GATA2 and AA patients have comparable ages, hemoglobin levels, and WBC counts, but GATA2 patients have higher platelet, neutrophil count, and lower lymphocyte count compared with AA patients. (B) GATA2 patients have reduced peripheral blood monocytes, B cells, and NK cells, and relatively increased T cells compared with AA patients.
Peripheral blood indices of GATA2 and AA patients. (A) GATA2 and AA patients have comparable ages, hemoglobin levels, and WBC counts, but GATA2 patients have higher platelet, neutrophil count, and lower lymphocyte count compared with AA patients. (B) GATA2 patients have reduced peripheral blood monocytes, B cells, and NK cells, and relatively increased T cells compared with AA patients.
GATA2 had peripheral blood B-cell and NK-cell lymphopenia, monocytopenia and a concomitant relative increase in T cells (Table 3). Median values of these indices in AA patients were within the normal reference range and significantly different between the 2 cohorts. However, individually, the range of values showed overlap (Figure 1B).
BM flow cytometric analysis of mononuclear cells in GATA2 and AA patients
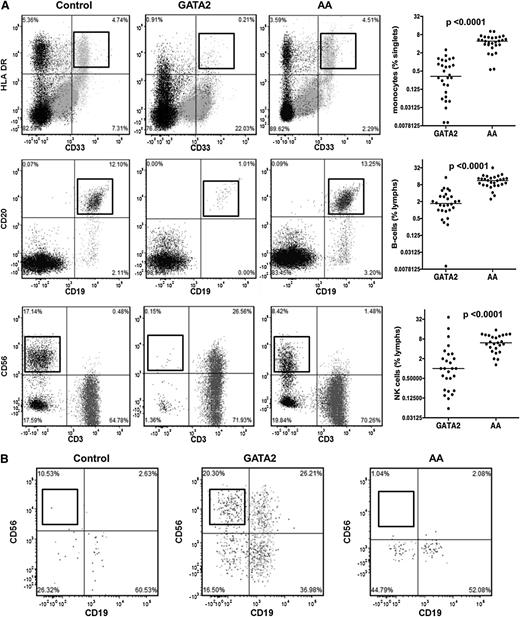
Flow cytometric analysis was performed on BM aspirates from 28 GATA2 patients and 28 AA patients who were negative for GATA2 mutations (Table 4). GATA2 patients had significantly reduced BM monocytes, B cells, and NK cells compared with AA patients and controls (Figure 2A).
Summary of flow cytometric features of BM aspirates from GATA2 and AA patients
| Bone marrow flow cytometry features . | GATA2 . | AA . | P . |
|---|---|---|---|
| Monocytes (median, % singlets) | 0.34 | 4.90 | <.0001 |
| CD34-positive cells (median, % singlets) | 0.42 | 0.39 | .2319 |
| B cells (median, % lymphs) | 1.71 | 11.26 | <.0001 |
| Hematogones (median, % lymphs) | 0.01 | 0.18 | <.0001 |
| NK cells (median, % lymphs) | 0.90 | 12.22 | <.0001 |
| T cells (median, % lymphs) | 96.70 | 77.40 | <.0001 |
| CD4:CD8 ratio (median) | 0.76 | 1.19 | <.01 |
| CD3+CD56+ T cells (median, % lymphs) | 18.50 | 3.10 | <.0001 |
| CD3+CD57+ T cells (median, % lymphs) | 20.55 | 6.51 | <.005 |
| Atypical CD19−CD56+ plasma cells | 10/18 | 0/28 |
| Bone marrow flow cytometry features . | GATA2 . | AA . | P . |
|---|---|---|---|
| Monocytes (median, % singlets) | 0.34 | 4.90 | <.0001 |
| CD34-positive cells (median, % singlets) | 0.42 | 0.39 | .2319 |
| B cells (median, % lymphs) | 1.71 | 11.26 | <.0001 |
| Hematogones (median, % lymphs) | 0.01 | 0.18 | <.0001 |
| NK cells (median, % lymphs) | 0.90 | 12.22 | <.0001 |
| T cells (median, % lymphs) | 96.70 | 77.40 | <.0001 |
| CD4:CD8 ratio (median) | 0.76 | 1.19 | <.01 |
| CD3+CD56+ T cells (median, % lymphs) | 18.50 | 3.10 | <.0001 |
| CD3+CD57+ T cells (median, % lymphs) | 20.55 | 6.51 | <.005 |
| Atypical CD19−CD56+ plasma cells | 10/18 | 0/28 |
Bone marrow flow cytometric analysis of monocytes and lymphoid subsets in GATA2 and AA patients. (A) GATA2 patients have reduced bone marrow monocytes (CD33+, HLA-DR+), mature B cells (CD19+, CD20+), and NK cells (sCD3−, CD56+) compared with AA patients. (B) A subset of GATA2 patients has atypical CD19−CD56+ plasma cells (gating performed on CD38+, CD138+ positive population), which are not identified in AA patients.
Bone marrow flow cytometric analysis of monocytes and lymphoid subsets in GATA2 and AA patients. (A) GATA2 patients have reduced bone marrow monocytes (CD33+, HLA-DR+), mature B cells (CD19+, CD20+), and NK cells (sCD3−, CD56+) compared with AA patients. (B) A subset of GATA2 patients has atypical CD19−CD56+ plasma cells (gating performed on CD38+, CD138+ positive population), which are not identified in AA patients.
Despite a significant B-cell lymphopenia, GATA2 patients had detectable plasma cells. Of the 18 GATA2 patients who had detailed flow cytometric analysis, 10 had atypical CD19−CD56+ plasma cell subsets (56%), consistent with previous reports.23 Plasma cells with an atypical immunophenotype were not identified in AA patients (Figure 2B).
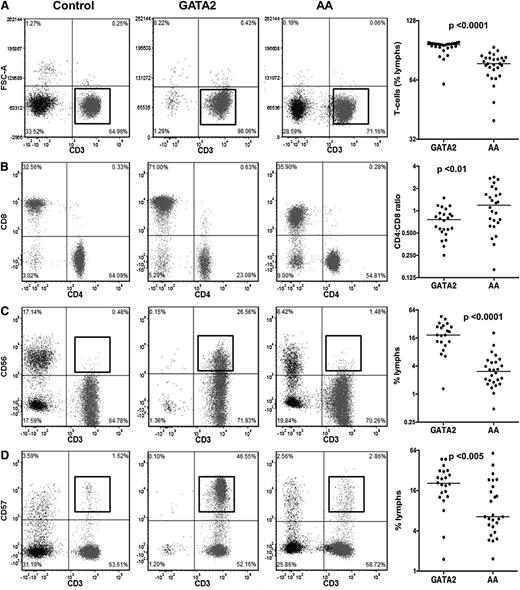
GATA2 patients had a relative increase in marrow T cells as a percentage of all lymphocytes compared with AA patients and controls (Figure 3A). A more detailed analysis of the BM T cells in these patients showed an inverted CD4:CD8 ratio (Figure 3B) and an expansion of CD3+CD56+ and CD3+CD57+ T-cell subsets, which was significantly higher than in controls and AA patients (Figure 3C-D).
Comparison of T-cell subsets between GATA2 and AA patients. (A) GATA2 patients have relatively increased bone marrow T cells, (B) inverted CD4:CD8 ratios, (C) increased CD3+CD56+, and (D) increased CD3+CD57+ T-cell subsets compared with AA patients.
Comparison of T-cell subsets between GATA2 and AA patients. (A) GATA2 patients have relatively increased bone marrow T cells, (B) inverted CD4:CD8 ratios, (C) increased CD3+CD56+, and (D) increased CD3+CD57+ T-cell subsets compared with AA patients.
We additionally identified atypical T-cell subsets in 4 of 28 GATA2 patients and 1 of 28 AA patients. T-cell receptor gene rearrangement studies were performed on marrow specimens of 10 of 28 GATA2 patients. Five GATA2 patients had abnormal rearrangement patterns (peaks of increased intensity in a polyclonal background), whereas the remainder had polyclonal rearrangement patterns. One patient with AA had a circulating clonal T-cell population with an immunophenotype consistent with T-large granular lymphocytes.
Analysis of precursor cells in GATA2 and AA patients
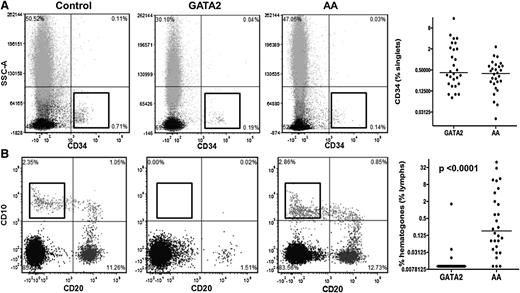
It has been reported that CD34+ cells are reduced in AA compared with hypocellular MDS and can aid in the differential diagnosis of these overlapping conditions.30 We found that, although both GATA2 and AA patients had reduced CD34+ cells with respect to controls, median values were comparable. However, the range of CD34+ cells was greater in GATA2 patients, due to 3 GATA2 patients with BM features indicative of myelodysplastic syndrome-refractory anemia with excess blasts (Figure 4A).
Comparison of bone marrow precursor cells between GATA2 and AA patients. (A) GATA2 and AA patients have comparable levels of CD34-positive cells. (B) GATA2 patients have absent hematogones compared with AA patients.
Comparison of bone marrow precursor cells between GATA2 and AA patients. (A) GATA2 and AA patients have comparable levels of CD34-positive cells. (B) GATA2 patients have absent hematogones compared with AA patients.
We then analyzed hematogone levels, which we defined as CD19+CD10+CD20− precursor B cells. GATA2 patients had nearly absent hematogones, consistent with previous reports.3 In contrast, AA patients had normal/mildly reduced levels of hematogones, expressed as a percentage of all lymphocytes (Figure 4B). BM flow cytometric findings are summarized in Table 4.
Morphologic analysis of BM biopsies from GATA2 and AA patients
BM biopsies from GATA2 patients were hypocellular-for-age, with median cellularity of 30% on core biopsies. Megakaryocytes were increased with atypical/dysplastic features in 23 of 28 biopsies, including micromegakaryocytes, nuclear hypolobation, and large megakaryocytes with separated and peripheralized nuclear lobes, as previously described.23 Erythroid dysplasia (nuclear budding, binucleation, prominent megaloblastoid change) was present in 11 of 28 and myeloid dysplasia (maturation asynchrony, hypogranularity) was present in 7 of 28 biopsies. BM reticulin fibrosis, graded as described previously,26 was uniformly increased (≥2+/4) in 13 of 28 biopsies (Table 5; Figure 5). Based on the 2008 World Health Organization classification, the following diagnostic categories were assigned: 22 patients, refractory cytopenia with multilineage dysplasia; 3 patients, refractory cytopenia with unilineage dysplasia; 2 patients, refractory anemia with excess blasts-1; 1 patient, refractory anemia with excess blasts-2.
Summary of morphologic features of BM biopsies from GATA2 and AA patients
| Features . | GATA2 . | AA . |
|---|---|---|
| No. of patients | 28 | 28 |
| Gender (M/F) | 13/15 | 11/17 |
| Age, years (median, range) | 30 (14-60) | 30 (12-73) |
| Median bone marrow cellularity (%) | 30 | 10 |
| Megakaryocytic dysplasia (%) | 23/28 (82) | 0/28 |
| Erythroid dysplasia (%) | 11/28 (39) | 0/28 |
| Myeloid dysplasia (%) | 7/28 (25) | 0/28 |
| Reticulin fibrosis (≥2/4) (%) | 13/28 (46) | 2/28 (focal) (7) |
| Abnormal cytogenetics (%) | 16/28 (58) | 0/28 |
| Features . | GATA2 . | AA . |
|---|---|---|
| No. of patients | 28 | 28 |
| Gender (M/F) | 13/15 | 11/17 |
| Age, years (median, range) | 30 (14-60) | 30 (12-73) |
| Median bone marrow cellularity (%) | 30 | 10 |
| Megakaryocytic dysplasia (%) | 23/28 (82) | 0/28 |
| Erythroid dysplasia (%) | 11/28 (39) | 0/28 |
| Myeloid dysplasia (%) | 7/28 (25) | 0/28 |
| Reticulin fibrosis (≥2/4) (%) | 13/28 (46) | 2/28 (focal) (7) |
| Abnormal cytogenetics (%) | 16/28 (58) | 0/28 |
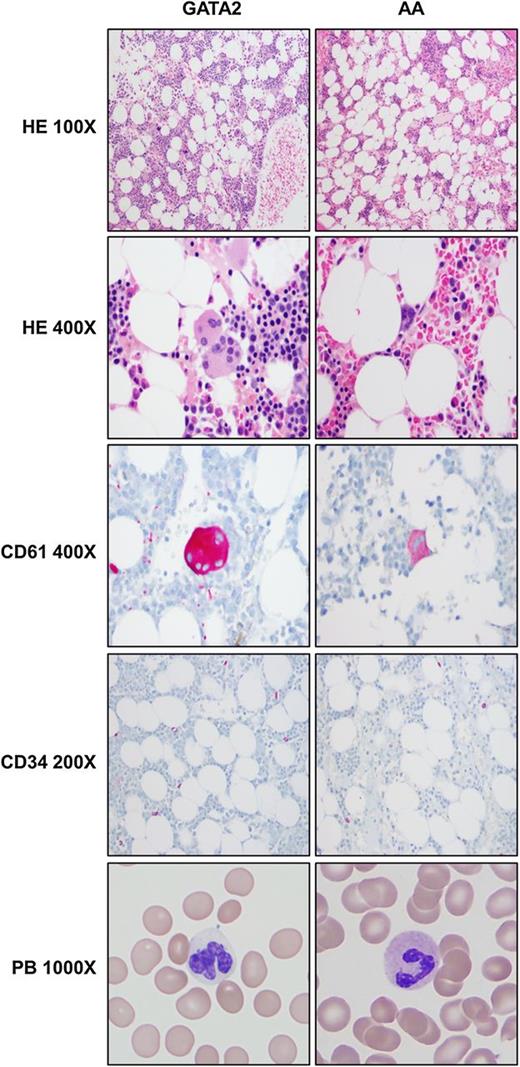
Comparison of peripheral blood and bone marrow morphologic and immunohistochemical features between GATA2 and AA patients. GATA2 patient marrows have greater overall cellularity, increased megakaryocytes with atypical features (highlighted by CD61), but overall similar numbers of CD34-positive cells compared with AA marrows. A subset of GATA2 patients has hypogranular circulating neutrophils.
Comparison of peripheral blood and bone marrow morphologic and immunohistochemical features between GATA2 and AA patients. GATA2 patient marrows have greater overall cellularity, increased megakaryocytes with atypical features (highlighted by CD61), but overall similar numbers of CD34-positive cells compared with AA marrows. A subset of GATA2 patients has hypogranular circulating neutrophils.
AA patients had severely hypocellular BM, with median cellularity of 10% on core biopsies and prominent lymphocytes, plasma cells, and mast cells in a stroma-rich background. Megakaryocytes were absent or rare and not overtly dysplastic. Erythroid and myeloid elements were markedly suppressed, and when present, showed progressive maturation with <10% of precursors displaying dyspoietic features in either erythroid or myeloid lineages. None of these 28 AA patients had morphologic or cytogenetic features diagnostic of hypocellular MDS based on World Health Organization 2008 criteria. Focal increase in reticulin fibrosis (grade 2+ of 4) was only seen in 2 of 28 AA biopsies; the biological significance of the increased focal fibrosis is uncertain. (Figure 5).
Cytogenetic analysis
Clonal cytogenetic abnormalities were identified in 16 (57%) of 28 GATA2 BMs; all but 3 have been previously reported.21 Recurring abnormalities included trisomy 8 alone in 5 patients, monosomy 7 alone in 3 patients, gain of 1q alone in 1 patient and in combination with 7q deletion and trisomy 8 in 2 patients, and loss of part or all of chromosome 6 in 2 patients. Trisomy 8 plus a marker chromosome, monosomy 7 with trisomy 21, and loss of the Y chromosome were found in 1 patient each. Ten of 28 GATA2 patients had normal cytogenetics, and the remaining 2 patients had inadequate studies (<20 normal metaphases). No cytogenetic abnormalities were detected in 28 AA patients that were negative for GATA2 mutations. (Table 5).
Flow cytometry findings in 4 new patients evaluated for AA with BM features suspicious for GATA2 deficiency
The characteristic BM features of GATA2 patients were used as guidelines to identify potential patients with GATA2 deficiency within the original 32 patients referred for confirmed or suspected AA. Four of 32 patients in cohort 2 were found to have features suspicious for GATA2 deficiency (Tables 4 and 5). Subsequent DNA sequencing confirmed the presence of GATA2 mutations in these 4 patients. The relevant clinical and BM morphologic features are described below, and BM flow cytometric features are summarized in Table 6.
Summary of clinical and bone marrow features of 4 new patients presenting with pancytopenia that were positive for GATA2 mutations
| Features . | P1, 43/F . | P2, 24/F . | P3, 28/F . | P4, 36/M . |
|---|---|---|---|---|
| Hemoglobin (g/dL, normal 13.7-17.5 g/dL) | 11.3 | 9.9 | 11.4 | 10.7 |
| Platelet count (K/μL, normal 161-347 K/μL) | 105.00 | 67.00 | 183.00 | 77.00 |
| Neutrophils (K/μL, normal 1.56-6.13 K/μL) | 1.75 | 1.73 | 1.25 | 1.24 |
| Monocytes (K/μL, normal 0.30-0.82 K/μL) | 0.04 | 0.06 | 0.01 | 0.00 |
| Lymphocytes (K/μL, normal 1.18-3.74 K/μL) | 1.01 | 0.36 | 1.23 | 1.05 |
| BM FC monocytes (% singlets) | 0.53 | 2.69 | 0.53 | 0.19 |
| BM FC CD34 (% singlets) | 0.45 | 0.24 | 0.45 | 0.32 |
| BM FC B cells (% lymphs) | 2.96 | 1.49 | 2.78 | 3.09 |
| BM FC hematogones (% lymphs) | 0.00 | 0.00 | 0.00 | 0.00 |
| BM FC NK cells (% lymphs) | 0.90 | 12.22 | 3.10 | 2.29 |
| BM FC T cells (% lymphs) | 94.80 | 84.90 | 93.10 | 93.90 |
| BM FC CD4:CD8 | 0.54 | 0.62 | 1.02 | 0.35 |
| BM FC CD3+CD56+ T cells (% lymphs) | 6.29 | 7.71 | 13.71 | 14.59 |
| BM FC CD3+CD57+ T cells (% lymphs) | 40.77 | 33.43 | 18.74 | 25.05 |
| BM FC CD19−CD56+ plasma cells | − | − | + | − |
| Atypical megakaryocytes | +++ | + | ++ | +++ |
| BM cytogenetics | +8 | +8 | Normal | +8 |
| GATA2 mutation | c.1114G>A | c.1123C>T | c.1017+572C>T | c.229+1G>A |
| A372T | L375F |
| Features . | P1, 43/F . | P2, 24/F . | P3, 28/F . | P4, 36/M . |
|---|---|---|---|---|
| Hemoglobin (g/dL, normal 13.7-17.5 g/dL) | 11.3 | 9.9 | 11.4 | 10.7 |
| Platelet count (K/μL, normal 161-347 K/μL) | 105.00 | 67.00 | 183.00 | 77.00 |
| Neutrophils (K/μL, normal 1.56-6.13 K/μL) | 1.75 | 1.73 | 1.25 | 1.24 |
| Monocytes (K/μL, normal 0.30-0.82 K/μL) | 0.04 | 0.06 | 0.01 | 0.00 |
| Lymphocytes (K/μL, normal 1.18-3.74 K/μL) | 1.01 | 0.36 | 1.23 | 1.05 |
| BM FC monocytes (% singlets) | 0.53 | 2.69 | 0.53 | 0.19 |
| BM FC CD34 (% singlets) | 0.45 | 0.24 | 0.45 | 0.32 |
| BM FC B cells (% lymphs) | 2.96 | 1.49 | 2.78 | 3.09 |
| BM FC hematogones (% lymphs) | 0.00 | 0.00 | 0.00 | 0.00 |
| BM FC NK cells (% lymphs) | 0.90 | 12.22 | 3.10 | 2.29 |
| BM FC T cells (% lymphs) | 94.80 | 84.90 | 93.10 | 93.90 |
| BM FC CD4:CD8 | 0.54 | 0.62 | 1.02 | 0.35 |
| BM FC CD3+CD56+ T cells (% lymphs) | 6.29 | 7.71 | 13.71 | 14.59 |
| BM FC CD3+CD57+ T cells (% lymphs) | 40.77 | 33.43 | 18.74 | 25.05 |
| BM FC CD19−CD56+ plasma cells | − | − | + | − |
| Atypical megakaryocytes | +++ | + | ++ | +++ |
| BM cytogenetics | +8 | +8 | Normal | +8 |
| GATA2 mutation | c.1114G>A | c.1123C>T | c.1017+572C>T | c.229+1G>A |
| A372T | L375F |
FC, flow cytometry.
Patient 1 is a 43-year-old woman who presented with intermittent pancytopenia and recurrent infections since 2004. A BM biopsy showed adequate hematopoiesis. She underwent extensive rheumatologic evaluation for recurrent fevers, panniculitis, granulomatous dermatitis, and hyperferritinemia, but no definitive diagnosis was rendered. In 2013, she was noted to have mild pancytopenia and hypocellular marrow with trisomy 8 and was referred for management. The BM was hypocellular with atypical megakaryocytes, hypogranular myeloid elements, normal erythroid maturation, and increased polyclonal plasma cells. Cytogenetic analysis was positive for trisomy 8. DNA sequencing revealed a GATA2 c.1114G>A mutation (A372T).
Patient 2 is a 24-year-old woman who had recurrent cytopenias since 2007. She had no history of infections or family history of hematologic malignancies. BM biopsy in 2010 showed adequate hematopoiesis, and cytogenetics was positive for trisomy 8. Immunosuppressive therapy was initiated in 2011. BM examination in 2012 showed mild hypocellularity and persistence of trisomy 8, and she was referred for management. The BM was hypocellular with atypical megakaryocytes, but no definitive morphologic evidence of dysplasia in myeloid and erythroid elements. Cytogenetic analysis was positive for trisomy 8. DNA sequencing revealed a GATA2 c.1123C>T mutation (L375F).
Patient 3 is a 28-year-old woman who presented with a history of pancytopenia and recurrent genital warts since 2009. The BM was hypocellular with atypical megakaryocytes and dyspoietic myeloid and erythroid elements. Cytogenetic analysis was normal. DNA sequencing revealed a GATA2 c.1017+572C>T mutation.
Patient 4 is a 38-year-old man with a history of recurrent warts and a 3-year history of progressive pancytopenia. A prior BM biopsy was normocellular with megakaryocytic atypia, mild myeloid, and erythroid atypia with trisomy 8. The BM was hypocellular with increased markedly atypical/dysplastic megakaryocytes, myeloid, and erythroid dyspoiesis. Cytogenetic analysis was positive for trisomy 8. DNA sequencing revealed a GATA2 c.229+1G>A mutation.
Family members with GATA2 mutations without hematologic manifestation
We also evaluated the bone marrow of 4 noncytopenic family members from 3 families who had documented GATA2 mutations, the first 3 of whom were previously described.21 Two individuals (family 4; 4.III.2, 4.III.3) had mutations in the intronic regulatory region of GATA2 (c.1017+572C>T); one developed lymphedema, whereas the other remains asymptomatic. The third individual (family 6; 6.I.1) had a 2-kb deletion spanning exons including the start codon (c.1017+512del28bp) and isolated lymphedema. The fourth individual (family 9; 9.II.2) has a GATA2 missense mutation (1192C>T, R398W) and remains asymptomatic. At the time of last follow-up, within the last year, all 4 individuals had adequate circulating monocytes, B cells, and NK cells. BMs were normocellular for age with rare hypolobated megakaryocytes but no morphologic features of myeloid or erythroid dysplasia. BM flow cytometry showed adequate monocytes, B cells, and NK cells, and cytogenetic analysis revealed normal karyotypes. The BM morphologic and flow cytometric findings are summarized in Table 7.
Summary of clinical and bone marrow features of 4 noncytopenic individuals with GATA2 mutations identified by screening family members of symptomatic GATA2 deficiency patients
| Features . | P1, 30/M 4.III.1* . | P2, 28/F 4.III.2* . | P3, 66/M 6.I.1* . | P4, 26/M 9.II.2 . |
|---|---|---|---|---|
| Symptoms | Lymphedema | None | Lymphedema | None |
| GATA2 mutation | c.1017+572C>T | c.1017+572C>T | c.1017+512 del28bp | c.1192C>T, p.R398W |
| Hemoglobin (g/dL, normal 13.7-17.5 g/dL) | 15.20 | 13.2 | 14.6 | 15.7 |
| Platelets (K/μL, normal 161-347 K/μL) | 199.00 | 170.00 | 230.00 | 212.00 |
| WBC (K/μL, normal 4.23-9.07 K/μL) | 7.44 | 5.83 | 4.38 | 7.15 |
| Neutrophils (K/μL, normal 1.56-6.13 K/μL) | 3.59 | 3.02 | 2.69 | 3.73 |
| Monocytes (K/μL, normal 0.30-0.82 K/μL) | 0.79 | 0.42 | 0.5 | 0.39 |
| Lymphs (K/μL, normal 1.18-3.74 K/μL) | 2.38 | 2.2 | 0.96 | 2.86 |
| BM monocytes (% singlets) | 3.56 | 5.8 | 5.49 | 5.02 |
| BM CD34 (% singlets) | 1.37 | 1.1 | 1.22 | 0.88 |
| BM B cells (% lymphs) | 10.85 | 20.08 | 9.21 | 12.89 |
| BM hematogones (% lymphs) | 7.77 | 5.38 | 0.00 | 3.66 |
| BM NK cells (% lymphs) | 3.47 | 8.06 | 13.52 | 12.70 |
| BM T cells (% lymphs) | 76.56 | 66.41 | 74.94 | 71.05 |
| BM CD4:CD8 | 0.75 | 0.78 | 1.28 | 1.02 |
| BM CD3+CD56+ T cells (% lymphs) | 3.91 | 1.50 | 9.80 | 6.49 |
| BM CD3+CD57+ T cells (% lymphs) | 10.48 | 3.96 | 20.20 | 8.79 |
| BM CD19−CD56+ plasma cells | − | + | + | − |
| BM cellularity | 50 | 55 | 35 | 50 |
| BM cytogenetics | Normal | Normal | Normal | Normal |
| Features . | P1, 30/M 4.III.1* . | P2, 28/F 4.III.2* . | P3, 66/M 6.I.1* . | P4, 26/M 9.II.2 . |
|---|---|---|---|---|
| Symptoms | Lymphedema | None | Lymphedema | None |
| GATA2 mutation | c.1017+572C>T | c.1017+572C>T | c.1017+512 del28bp | c.1192C>T, p.R398W |
| Hemoglobin (g/dL, normal 13.7-17.5 g/dL) | 15.20 | 13.2 | 14.6 | 15.7 |
| Platelets (K/μL, normal 161-347 K/μL) | 199.00 | 170.00 | 230.00 | 212.00 |
| WBC (K/μL, normal 4.23-9.07 K/μL) | 7.44 | 5.83 | 4.38 | 7.15 |
| Neutrophils (K/μL, normal 1.56-6.13 K/μL) | 3.59 | 3.02 | 2.69 | 3.73 |
| Monocytes (K/μL, normal 0.30-0.82 K/μL) | 0.79 | 0.42 | 0.5 | 0.39 |
| Lymphs (K/μL, normal 1.18-3.74 K/μL) | 2.38 | 2.2 | 0.96 | 2.86 |
| BM monocytes (% singlets) | 3.56 | 5.8 | 5.49 | 5.02 |
| BM CD34 (% singlets) | 1.37 | 1.1 | 1.22 | 0.88 |
| BM B cells (% lymphs) | 10.85 | 20.08 | 9.21 | 12.89 |
| BM hematogones (% lymphs) | 7.77 | 5.38 | 0.00 | 3.66 |
| BM NK cells (% lymphs) | 3.47 | 8.06 | 13.52 | 12.70 |
| BM T cells (% lymphs) | 76.56 | 66.41 | 74.94 | 71.05 |
| BM CD4:CD8 | 0.75 | 0.78 | 1.28 | 1.02 |
| BM CD3+CD56+ T cells (% lymphs) | 3.91 | 1.50 | 9.80 | 6.49 |
| BM CD3+CD57+ T cells (% lymphs) | 10.48 | 3.96 | 20.20 | 8.79 |
| BM CD19−CD56+ plasma cells | − | + | + | − |
| BM cellularity | 50 | 55 | 35 | 50 |
| BM cytogenetics | Normal | Normal | Normal | Normal |
From Spinner et al.21
Discussion
In addition to MonoMAC syndrome, Emberger syndrome, DCML deficiency, and familial MDS/AML, GATA2 mutations have also been documented in patients diagnosed with chronic neutropenia,7 aplastic anemia,9 and pediatric BM failure.8 These studies highlight the expanding spectrum of GATA2 deficiency-associated phenotypes. It is now acknowledged that GATA2 deficiency can present with pancytopenia and BM morphology that mimics aplastic anemia or de novo hypoplastic MDS.
Our analysis showed that all 28 symptomatic/proband GATA2 patients had characteristic severe peripheral blood monocytopenia and B-cell and NK-cell lymphopenia, in addition to varying degrees of anemia and/or thrombocytopenia and relatively preserved neutrophil counts in the majority of GATA2 patients. In contrast, although all 28 AA patients who were negative for GATA2 mutations had pancytopenia and significant neutropenia, they typically had only minimally reduced circulating monocytes, B cells, and NK cells by immunophenotyping. Although peripheral blood lymphoid immunophenotyping provides valuable additional data, it is often a specialized diagnostic assay and not performed routinely in the evaluation of patients with pancytopenia.
BM flow cytometry in GATA2 patients showed a concomitant reduction in monocytes, B cells, and NK cells, as expected, all significantly lower than in AA patients. The complete loss of B-cell precursors (CD19+CD10+CD20−) in GATA2 patients is consistent with results from a small cohort of DCML patients.3 The near total absence of hematogones with low, yet detectable mature B cells (CD19+CD10−CD20+) suggests that disrupted B-cell lymphopoiesis is an early event in disease progression and is supported by previous reports of reduced to absent BM hematogones in patients with de novo MDS.31 Previous studies have shown reduced hematogones in pediatric AA patients compared with age-matched controls.32 Although we did not control for age, our results show that hematogone levels are overall lower than normal but still detectable in AA patients in contrast to GATA2 patients.
Although GATA2 patients have decreased B cells, BM mature plasma cells were identified and often increased, including a subset with an abnormal CD19−CD56+ immunophenotype, seen in 10 of 28 (36%) patients, consistent with prior reports.23 CD56 has been reported as a marker of neoplastic plasma cells,33 but a recent study has shown that a minority of normal plasma cells have a CD19−CD56+ immunophenotype.34 Only 1 GATA2 patient with atypical plasma cells showed evidence of a clonal plasma cell expansion, consistent with smoldering plasma cell myeloma.23 BM and peripheral blood benign polyclonal plasmacytosis was first described in association with bacterial sepsis and AIDS.35 BM plasmacytosis has also been described in inflammation, infections, autoimmune disorders, and immunodeficiency states.36,37 The CD19−CD56+ plasma cells in GATA2 patients, especially in the setting of chronic infections, must not be interpreted as neoplastic without definitive evidence of clonality.
Enumeration of myeloid blasts is critical to the classification of MDS, and morphologic assessment remains the gold standard. However, this can be challenging in marrows with paucicellular aspirate smears. Analysis of CD34-positive cells by flow cytometry and immunohistochemistry of BM biopsies can provide additional useful information in these specimens, especially if aberrant patterns are identified. Our results show an overall reduction in CD34-positive cells in AA compared with controls, which is consistent with previous reports.30,38 Although CD34 immunostaining has been reported useful in differentiating hypocellular MDS and AA,39 we did not observe a significant difference in CD34-positive cells between GATA2 and AA patients (excluding the 3GATA2 patients with refractory anemia with excess blasts). Haploinsufficiency of GATA2 has been shown to reduce the hematopoietic primitive stem cell pool but not maturation capability.13 This could explain the baseline low levels of CD34-positive cells in GATA2 patients until myeloid transformation with the acquisition of additional somatic genetic alterations.40
GATA2 patients had relative peripheral blood and BM T-cell lymphocytosis. Increased bone marrow T cells with cytotoxic phenotypes, including CD3+CD56+ and CD3+CD57+ subsets and an inverted CD4:CD8 ratio is consistent with an acquired, activated T-cell repertoire described in GATA2 patients22 and also seen in age-related changes, immunodeficiency states, and chronic viral infections.41 The atypical T-cell populations we observed in 4 of 28 GATA2 patients were not clonal by T-cell receptor gene rearrangement studies and may represent restricted T-cell populations, secondary to chronic infections. Clonal or restricted CD8-positive T-cell expansions have been documented in AA patients, supporting an immune mechanism.42 We did not perform T-cell clonality analysis in our AA cohort routinely, except in 1 patient, who had a clonal T-large granular lymphocyte population. Additionally, patients with untreated AA have increased BM CD8-positive T cells and CD56-positive NK cells, but decreased NK-T cells.43,44 However, using routine lymphoid panels, we did not identify significant abnormalities in these lymphoid subsets in our AA patient cohort. Our results suggest that the presence of atypical T-cell populations in GATA2 patients should not be interpreted as neoplastic T-cell proliferations.
The identification of atypical megakaryocytes and increased reticulin fibrosis can help in distinguishing hypocellular MDS and AA.45 Our results show that immunostaining for megakaryocytes (with CD61 or factor VIIIR-Ag antibodies) and reticulin stains are valuable in the evaluation of a hypocellular BM. Although megakaryocytes with dysplastic features are increased in de novo MDS, both large and small megakaryocytes with separated and peripheralized nuclear lobes may provide a morphologic clue to an underlying GATA2 deficiency. In addition to early hematopoietic stem cells, GATA2 is expressed at high levels during thrombopoiesis and is required for megakaryocyte development,16,46 providing a possible explanation for the prominent dysmegakaryopoiesis in GATA2 patients. Recent studies have demonstrated that AML associated with the recurring inv(3)(q21q26.2) or t(3;3)(q21;26.2) rearrangements is driven by underexpression of GATA2 and overexpression of EVI1.47,48 These rearrangements are associated with AML occurring de novo or from an underlying MDS, which may be hypocellular and often has increased atypical megakaryocytes.49,50 Whether MDS/AML arising from these genetic aberrations represents a somatic variant of GATA2 deficiency merits further investigation.
The clinical diversity, phenotype variability, and lack of specific genotype-phenotype correlation with GATA2 mutations have been documented.21 Our description of 4 individuals with 3 different documented GATA2 mutations, no history of infections or peripheral cytopenias, and normal BM features supports this observation and suggests other factors cooperate with GATA2 mutation for development of immunodeficiency, BM failure, and neoplasia. Additionally, we did not detect a correlation between genotype and BM flow cytometry and morphologic features. Current data suggest that the hematologic spectrum of GATA2 deficiency is diverse (Table 8). The factors that trigger the onset of cytopenias, infections, or progression to MDS/AML are still being investigated. Dickinson et al22 have shown that serum Flt3L levels are elevated at the onset of cytopenias but are lower in more advanced disease. Although MDS/AML can occur without preceding cytopenias as described previously,6 the presence of additional acquired mutations suggests that multiple factors may determine disease evolution.40 The identification of individuals with GATA2 mutations and a normal phenotype in this study and others8 also underscores the need for screening of family members who are considered potential donors for GATA2 patients.
Hematologic features of phenotypes seen in GATA2 deficiency
| Phenotype . | Normal . | Immunodeficiency . | Bone marrow failure . | MDS/AML/CMML . |
|---|---|---|---|---|
| Infections | − | + | +/− | +/− |
| Peripheral blood | ||||
| No cytopenias | + | |||
| Monocytopenia | + | + | + | |
| B-, NK- lymphopenia | + | + | + | |
| Inverted CD4:CD8 ratio | + | + | + | |
| Anemia | +/− | +/− | ||
| Neutropenia | +/− | +/− | ||
| Thrombocytopenia | +/− | +/− | ||
| Increased blasts | +/− | |||
| Monocytosis | + (CMML, AML[monocytic]) | |||
| Bone marrow morphology | ||||
| Normocellular | + | |||
| Hypocellular | +/− | + | +/− | |
| Hypercellular | −/+ | |||
| No dysplasia | + | |||
| Atypical megakaryocytes | +/− | + | + | |
| Myeloid/erythroid dysplasia | + | |||
| Increased blasts | + (RAEB, AML, CMML) | |||
| Bone marrow flow cytometry | ||||
| No cytopenias | + | |||
| Monocytopenia | + | + | + | |
| B-, NK-lymphopenia | + | + | + | |
| Reduced hematogones | + | + | + | |
| Inverted CD4:CD8 ratio | + | + | + | |
| Abnormal myeloid maturation | + | |||
| CD19−CD56+ plasma cells | +/− | +/− | +/− | |
| Increased blasts | + (RAEB, AML, CMML) | |||
| Increased monos/precursors | + (CMML, AML[monocytic]) | |||
| Cytogenetics | ||||
| Normal | + | + | + | + |
| Abnormal | + | + |
| Phenotype . | Normal . | Immunodeficiency . | Bone marrow failure . | MDS/AML/CMML . |
|---|---|---|---|---|
| Infections | − | + | +/− | +/− |
| Peripheral blood | ||||
| No cytopenias | + | |||
| Monocytopenia | + | + | + | |
| B-, NK- lymphopenia | + | + | + | |
| Inverted CD4:CD8 ratio | + | + | + | |
| Anemia | +/− | +/− | ||
| Neutropenia | +/− | +/− | ||
| Thrombocytopenia | +/− | +/− | ||
| Increased blasts | +/− | |||
| Monocytosis | + (CMML, AML[monocytic]) | |||
| Bone marrow morphology | ||||
| Normocellular | + | |||
| Hypocellular | +/− | + | +/− | |
| Hypercellular | −/+ | |||
| No dysplasia | + | |||
| Atypical megakaryocytes | +/− | + | + | |
| Myeloid/erythroid dysplasia | + | |||
| Increased blasts | + (RAEB, AML, CMML) | |||
| Bone marrow flow cytometry | ||||
| No cytopenias | + | |||
| Monocytopenia | + | + | + | |
| B-, NK-lymphopenia | + | + | + | |
| Reduced hematogones | + | + | + | |
| Inverted CD4:CD8 ratio | + | + | + | |
| Abnormal myeloid maturation | + | |||
| CD19−CD56+ plasma cells | +/− | +/− | +/− | |
| Increased blasts | + (RAEB, AML, CMML) | |||
| Increased monos/precursors | + (CMML, AML[monocytic]) | |||
| Cytogenetics | ||||
| Normal | + | + | + | + |
| Abnormal | + | + |
AA and hypocellular MDS can show significant morphologic overlap, and the distinction between both entities can be challenging and subject to interobserver variability. None of our AA cases definitively fulfilled the morphologic or immunophenotypic criteria for the diagnosis of hypocellular MDS.51 The GATA2 deficiency marrows in this study showed morphologic features within the spectrum of AA and hypocellular MDS, underscoring the importance of considering GATA2 deficiency in the differential diagnosis of patients diagnosed with de novo hypocellular MDS.
In summary, we describe data acquired from routine BM analysis that helped identify 4 new GATA2-deficient patients from a cohort of patients being evaluated for cytopenias (including 2 patients without a significant infectious history) that were confirmed by GATA2 mutation analysis. We believe the frequency of GATA2 mutations in MDS/AML and in patients being evaluated for cytopenias is higher than currently reported. Although GATA2 testing is not routinely indicated in all patients presenting with cytopenias, a combination of routine diagnostic BM morphology, flow cytometry, immunohistochemistry, and cytogenetic analysis can help identify a subset of cytopenic patients who could benefit from GATA2 mutation analysis.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
This work was supported by the National Institutes of Health Division of Intramural Research, Clinical Center, National Institute of Allergy and Infectious Diseases, National Heart, Lung, and Blood Institute, and National Cancer Institute.
The content of this publication does not necessarily reflect the views or policies of the Department of Health and Human Services.
Authorship
Contribution: K.A.G. and K.R.C. analyzed the data, created the figures, and wrote the manuscript; A.P.H. performed genetic sequencing for all patients; D.M.T., N.S.Y., C.S.Z., J.C.-R., D.D.H., and S.M.H. provided clinical care and patient samples; K.R.C. and D.C.A. reviewed bone marrow biopsies and cytogenetics; S.D.R. provided peripheral blood immunophenotyping data; R.C.B., K.A.G., and K.R.C. performed flow cytometric analysis; and A.P.H., D.M.T., N.S.Y., D.D.H., R.C.B., and S.M.H. revised the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Katherine R. Calvo, Hematology Section, Department of Laboratory Medicine, National Institutes of Health, Clinical Center, 10 Center Dr, Building 10, Room 2C390, Bethesda, MD 20892-1508; e-mail: calvok@mail.nih.gov.