Abstract
Inhibitors of B-cell receptor (BCR) and pre-BCR signaling were successfully introduced into patient care for various subtypes of mature B-cell lymphoma (eg, ibrutinib, idelalisib). Acute lymphoblastic leukemia (ALL) typically originates from pre-B cells that critically depend on survival signals emanating from a functional pre-BCR. However, whether patients with ALL benefit from treatment with (pre-) BCR inhibitors has not been explored. Recent data suggest that the pre-BCR functions as tumor suppressor in the majority of cases of human ALL. However, a distinct subset of human ALL is selectively sensitive to pre-BCR antagonists.
Introduction
Acute lymphoblastic leukemia (ALL) represents the most common cancer in children, accounting for 26% of malignancies diagnosed in ages 0 to 14. An estimated 2670 children and 410 adolescents were diagnosed with ALL in the United States in 2014.1 Outcomes for patients with pre-B ALL have substantially improved over the past decades, reaching overall survival rates of 90% for children2 and 45% for adults.3 Owing to its frequent occurrence in children, ALL remains 1 of the leading causes of person-years of life lost in the United States (362 000 person-years of life lost in 2010).1 In addition, ∼20% of patients experience a bone marrow relapse after initially successful treatment and more than 60% of these patients will die of their disease.
Cellular origins define oncogenic signaling requirements of ALL cells
With the goal to decrease the frequency of ALL relapse and reduce side effects of cytotoxic therapy, recent efforts have introduced targeted therapies that focus on specific vulnerabilities of ALL cells. The basic premise for these studies has been that oncogenes in ALL will promote growth factor independence by delivering survival and proliferation signals that are normally provided by a favorable environment or as the outcome of positive selection. ALL typically originates from pro- and pre-B cells during early B-cell development—ie, cell types that critically depend on survival signals that emanate from an active cytokine receptor (eg, interleukin-7 receptor [IL7R] and/or an active pre–B-cell receptor [BCR]).
Recent studies revealed that a defined subset of ALL (termed Ph-like) is indeed driven by and particularly dependent on oncogenic cytokine receptor signaling (eg, through lesions of IL7R, CRLF2, JAK1, JAK2, JAK3, EPOR, PDGFR).4-9 Although genetically heterogeneous, this group of ALL has the oncogenic activation of signal transducer and activator of transcription 5 (STAT5)6,7 and a characteristic, STAT5-dependent, gene expression pattern in common.8 ALL driven by oncogenic cytokine receptor signaling accounts for ∼10% of ALL cases in children and ∼25% in young adults and is associated with poor clinical outcome.10,11 Importantly, ALL cells in this subgroup were found to be sensitive to lesion-specific tyrosine kinase inhibitors, including dasatinib, ruxolitinib, and crizotinib.10,11 Although the concept of lesion-specific tyrosine kinase inhibitor treatment of ALL driven by oncogenic cytokine receptor signaling is strongly supported by experimental and clinical evidence, targeted therapy of pre-BCR signaling has not been explored in human ALL.
Pre-BCR checkpoint control during normal B-cell development
During early B-cell development, developing pre-B cells are selected for productive rearrangement of VHDJH gene segments, which enables expression of a functional pre-BCR on the cell surface. Early pre-B cells are programmed to die unless they are rescued by “tonic” survival signals from a productively assembled pre-BCR.12 In mice, bone marrow progenitor cells produce approximately 10 million pre-B cells daily,13 the vast majority of which is eliminated at the pre-BCR checkpoint.14 Hence, in mouse mutants unable to express a pre-BCR, B-cell development is blocked at the pro–B-cell stage of differentiation.15,16 The differentiation block at the pre-BCR checkpoint, however, can be rescued by transgenic introduction of productively rearranged VHDJH gene segments, encoding a functional immunoglobulin μ heavy chain (µHC).17,18 Two models have been proposed for the initiation of pre-BCR signaling: Earlier work identified Galectin-1 expressed on the surface of bone marrow stroma cells as a potential ligand of the pre-BCR.19 However, a more recent study demonstrated autonomous, ligand-independent activation of the pre-BCR.20 This is based on the interaction between the surrogate light chain of the pre-BCR with a conserved asparagine (N)-linked glycosylation site at position 46 (N46) in the first conserved domain of the µHC.
Tonic survival signals from the pre-BCR are transduced by the immunoglobulin α (Igα) and Igβ signaling chains.21,22 Hence, B-cell development in mice lacking functional Igα21 and Igβ222 expression is arrested at the pre-BCR checkpoint. Conditional ablation of tonic BCR signaling results in rapid B-cell depletion,23,24 highlighting the critical importance of tonic pre-BCR and BCR signaling at the pre-BCR checkpoint and later stages of B-cell development. Interestingly, loss of tonic pre-BCR signaling can be rescued by activation of phosphatidylinositol 3-kinase (PI3K)-AKT signaling,25 identifying PI3K-AKT as a central survival pathway downstream of the (pre-) BCR. Tonic pre-BCR signaling involves constitutive activity of the proximal pre–BCR-associated SRC family kinases Lyn, Fyn, and Blk26 as well as the SYK and ZAP70 tyrosine kinases,27 which then activate PI3K.28,29 In addition to its well-established role in mature B-cell survival,23 recent work highlighted the role of PI3K signaling and in particular the PI3K p110δ (PIK3CD) isoform for pre-BCR signaling and pre–B-cell survival during early B-cell development.30
In addition to activation of a downstream signaling cascade, tonic pre-BCR signaling induces important transcriptional changes that mediate positive selection and survival at the pre-BCR checkpoint. De novo expression of the pre-BCR induces strong expression of BCL6, which promotes survival by BCL6-mediated repression of CDKN2A and TP53.31 In the absence of BCL6, positive selection at the pre-BCR checkpoint is compromised, and even pre-B cells that express a functional pre-BCR die of CDKN2A- and p53-mediated apoptosis.32 Interestingly, BCL6 has the same function in human ALL cells and mediates survival and drug resistance.33 At the pre-BCR checkpoint and in human ALL cells, BCL6 is opposed by BACH2, a transcription factor that mediates negative pre–B-cell selection and reverses BCL6-mediated survival signals and drug resistance.34,35
Targeting the BCR signaling pathway in mature B-cell lymphoma
Unlike BCR signaling in mature B-cell lymphoma, targeting of pre-BCR signaling has not been explored as a treatment option for patients with ALL. The discovery that most subtypes of B-cell lymphoma critically depend on BCR signaling36,37 has led to the development of new targeting strategies that focus on BCR signaling at the level of SRC kinases (Lyn, Fyn, and Blk), SYK/ZAP70, Bruton tyrosine kinase (BTK), and PI3Kδ.38-42
The majority of mature B-cell lymphoma subtypes depend on tonic or chronic active BCR signaling (∼85%); however, classical Hodgkin disease,43 angioimmunoblastic lymphoma,44 AIDS-related B-cell lymphoma,45 and primary mediastinal B-cell lymphoma 46,47 together account for ∼15% of human B-cell lymphoma that lack BCR function. In many cases, tumor clones show evidence of negative selection of a functional BCR.43,44 The classification of B-cell lymphoma based on (1) chronic active, (2) tonic, and (3) lack of BCR signaling informed the development of new treatment strategies and has led to the successful introduction of BTK (chronic active)48-50 and PI3Kδ (tonic BCR signaling)51 inhibitors into patient care. Small molecule inhibition of BTK (eg, ibrutinib) and PI3Kδ (idelalisib) in activated B-cell–like diffuse large B-cell lymphoma, chronic lymphocytic leukemia, and mantle cell lymphoma has achieved major clinical successes in the treatment of these diseases.48-51 It should be noted, however, that clinical responses to inhibition of BTK (eg, ibrutinib) and PI3Kδ (idelalisib) in patients are not exclusively dependent on the role of BTK and PI3Kδ in BCR signaling. Because BTK and PI3K signaling is also active in chemokine and integrin receptor signaling, inhibition of these pathways may contribute to the effects observed in patients.
Tumor suppressor role of pre-BCR signaling in cytokine receptor/STAT5-dependent subsets of ALL
Like mature B-cell lymphoma, pre-B ALL originates from B-cell precursors that critically depend on survival signals emanating from a functional (pre-) BCR. In apparent contradiction to this notion, a series of previous studies suggested a role of pre-BCR signaling as a tumor suppressor in ALL cells. For instance, in BCR-ABL1–driven (Ph+) ALL, the BCR-ABL1 kinase activity mimics and supplants activity of the pre-BCR.52-55 Although pre-BCR signaling in ALL cells is frequently inactivated through multiple defects, including deletion of the surrogate light chain segment VPREB1,56 reconstitution of pre-BCR expression54 or its signaling chain Igα55 causes cell death. In agreement with a tumor suppressor role of the pre-BCR, several studies identified defective expression of the pre-BCR signaling molecules BLNK (SLP-65)57 and BTK58 in human ALL and demonstrated in genetic mouse models that Blnk and Btk cooperate in preventing malignant transformation of pre–B cells59,60 (Table 1). Importantly, pre-BCR signaling via BLNK negatively regulates STAT5 activity, which represents a central mediator of oncogenic cytokine receptor signaling in ALL cells.61 Thereby, BLNK binds to and inactivates JAK3 upstream of STAT5.61 Besides pre-BCR and BLNK, transcription factors (eg, PAX5, EBF1) that drive expression of BLNK60 and other components of the pre-BCR signaling pathway also result in suppression of cytokine receptor/STAT5 signaling in mouse models of ALL (Table 1).7 Besides PAX5,62 IKZF1 is a strong transcriptional activator of pre-BCR signaling.63 Although genomic lesions of BLNK (∼2% of ALL cases) are relatively rare, deletion of transcription factors that promote pre-BCR expression and activity are frequent in ALL. Deletions of PAX5 occur in up to 25% of ALL cases64 and IKZF1 deletions, resulting in expression of a dominant-negative protein, are found in >80% of cases of Ph+ ALL.65 Studying 830 ALL cases from 4 clinical trials, in the majority of cases, ALL cells lacked pre-BCR signaling (pre-BCR−),54,66 which represents a functional equivalent to classical Hodgkin lymphoma and mediastinal B-cell lymphoma among mature B-cell malignancies. Pre-BCR− tumors account for ∼85% of ALL cases: virtually all cases of BCR-ABL1, Ph-like (CRLF2, JAK, EPOR, PDGFR) and ETV6-RUNX1, and the vast majority of MLL-rearranged and hyperdiploid ALL cases lack expression of a functional pre-BCR. Among the Children’s Oncology Group P9906 and AALL0232 cohorts, in 101 of 486 ALL cases (21%) abnormal cytokine receptor signaling was observed (Table 1). Oncogenic hyperactivation of cytokine receptor signaling was the consequence of CRLF2 overexpression or rearrangement (n = 59; 12%), FLT3 mutation (n = 12; 2.5%), IL7R mutation (n = 9; 2%), or rearrangement of other cytokine receptors including PDGFRB (n = 4; 1%) and EPOR (n = 1; 0.2%). In other cases, oncogenic cytokine receptor signaling was caused by JAK1, JAK2, or JAK3 mutation or rearrangement (n = 35; 7%), ABL1 gene rearrangement (n = 5; 1%), or SH2B3 mutation or deletion (n = 9; 2%). In 28 cases, multiple lesions were detected. ALL clones that are driven by oncogenic cytokine receptor signaling typically express constitutively active STAT5 (Table 1). Consistent with pre-BCR–mediated attenuation of cytokine receptor/STAT5 signaling,7,60,67 tumor clones are selected for defective expression of the pre-BCR in cytokine receptor/STAT5-dependent subsets of ALL.
Characteristics of pre-BCR+ and pre-BCR− ALL subsets
| Gene . | Function . | Type of defect . | Functional outcome . | Phenotype in disease model . | Frequency in human ALL (%) . | Reference . | ||
|---|---|---|---|---|---|---|---|---|
| Total . | Pre-BCR+ . | Pre-BCR− . | ||||||
| CRLF2 | Cytokine receptor | Overexpression | Cytokine receptor signaling ↑ | Propensity to ALL | 12* | 0 | 15 | 9-11 |
| Rearrangement | ||||||||
| FLT3 | Cytokine receptor | Point mutation | Cytokine receptor signaling ↑ | Propensity to leukemia | 2.5 | 0 | 3 | 9-11 |
| IL7R | Cytokine receptor | Point mutation | Cytokine receptor signaling ↑ | Propensity to ALL | 2 | 0 | 2 | 9-11 |
| PDGFRB | Cytokine receptor | Rearrangement | Cytokine receptor signaling ↑ | Propensity to leukemia | 1 | 0 | 1 | 9-11 |
| EPOR | Cytokine receptor | Rearrangement | Cytokine receptor signaling ↑ | <1 | 0 | <1 | 9-11 | |
| JAK1, JAK2, JAK3 | Tyrosine kinase | Point mutation | Cytokine receptor signaling ↑ | Propensity to ALL | 7* | 0 | 9 | 9-11 |
| Rearrangement | ||||||||
| ABL1 | Tyrosine kinase | BCR-ABL1 | Cytokine receptor signaling ↑ | ALL | 3*-27† | 0 | 4-35 | 9-11 |
| NUP214, ETV6, RCSD1 | Propensity to leukemia | 1 | 0 | 2 | 9-11 | |||
| SH2B3 | Linker | Deletion | Cytokine receptor signaling ↑ | Pro–B-cell block | 2 | 0 | 2 | 9-11 |
| IGHM | Ig heavy chain | Nonproductive V(D)J | No functional pre-BCR | B-cell autoimmunity | 83 | 0 | >80 | 54, 66 |
| VPREB1 | Surrogate light chain | Focal deletion | No functional pre-BCR | Pro–B-cell block | 15 | 0 | 20 | 56 |
| CD79A | (Pre-) BCR signaling chain | Transcriptional silencing | No functional pre-BCR | Pro–B-cell block | 80 | 0 | >80 | 54, 55 |
| CD79B | (Pre-) BCR signaling chain | Transcriptional silencing | No functional pre-BCR | Pro–B-cell block | 80 | 0 | >80 | 54, 55 |
| SYK | (Pre-) BCR tyrosine kinase | Aberrant splicing, mutation | Pre-BCR signaling ↓ | Pro–B-cell block | <10 | 0 | <10 | 54 |
| BLNK | (Pre-) BCR linker | Aberrant splicing | Pre–B-cell differentiation ↓ | Pre–B-cell block | 40 | 0 | ∼50 | 53, 57 |
| Focal deletion | Cytokine receptor signaling ↑ | Propensity to ALL | 2 | 2 | 64 | |||
| BTK | (Pre-) BCR tyrosine kinase | Aberrant splicing | Pre–B-cell differentiation ↓ | Pre–B-cell block | 30 | 0 | ∼25 | 58-60 |
| Propensity to ALL | ||||||||
| PAX5 | Transcription factor | Deletion, point mutation | Cytokine receptor signaling ↑ | Propensity to ALL | ∼25 | 10 | ∼25 | 64 |
| BACH2 | Transcription factor | Deletion 6q15 | Pre-BCR signaling ↓ | Propensity to ALL | 20 | 0 | ∼25 | 34 |
| IKZF1 | Transcription factor | Deletion, point mutation | Pre-BCR signaling ↓ | Propensity to ALL | 20-84‡ | 0 | ∼25 | 64, 65 |
| IKZF3 | Transcription factor | Deletion | Pre-BCR signaling ↓ | Propensity to ALL | 2 | 0 | 2 | 64, 65 |
| EBF1 | Transcription factor | Deletion, point mutation | Pre-BCR signaling ↓ | Propensity to ALL | 6 | 0 | 8 | 64, 65 |
| PBX1 | Transcription factor | TCF3-PBX1 rearrangement | Pre-BCR signaling ↑ | Propensity to ALL | 3*-5† | 40 | 0 | 66, 68 |
| 1q23 duplication | 1 | 5 | 0 | 66 | ||||
| PRDM1 | Transcription factor | Deletion 6q21 | Pre-BCR signaling ↑ | Plasma cell defect | 1 | 5 | 0 | 66 |
| Gene . | Function . | Type of defect . | Functional outcome . | Phenotype in disease model . | Frequency in human ALL (%) . | Reference . | ||
|---|---|---|---|---|---|---|---|---|
| Total . | Pre-BCR+ . | Pre-BCR− . | ||||||
| CRLF2 | Cytokine receptor | Overexpression | Cytokine receptor signaling ↑ | Propensity to ALL | 12* | 0 | 15 | 9-11 |
| Rearrangement | ||||||||
| FLT3 | Cytokine receptor | Point mutation | Cytokine receptor signaling ↑ | Propensity to leukemia | 2.5 | 0 | 3 | 9-11 |
| IL7R | Cytokine receptor | Point mutation | Cytokine receptor signaling ↑ | Propensity to ALL | 2 | 0 | 2 | 9-11 |
| PDGFRB | Cytokine receptor | Rearrangement | Cytokine receptor signaling ↑ | Propensity to leukemia | 1 | 0 | 1 | 9-11 |
| EPOR | Cytokine receptor | Rearrangement | Cytokine receptor signaling ↑ | <1 | 0 | <1 | 9-11 | |
| JAK1, JAK2, JAK3 | Tyrosine kinase | Point mutation | Cytokine receptor signaling ↑ | Propensity to ALL | 7* | 0 | 9 | 9-11 |
| Rearrangement | ||||||||
| ABL1 | Tyrosine kinase | BCR-ABL1 | Cytokine receptor signaling ↑ | ALL | 3*-27† | 0 | 4-35 | 9-11 |
| NUP214, ETV6, RCSD1 | Propensity to leukemia | 1 | 0 | 2 | 9-11 | |||
| SH2B3 | Linker | Deletion | Cytokine receptor signaling ↑ | Pro–B-cell block | 2 | 0 | 2 | 9-11 |
| IGHM | Ig heavy chain | Nonproductive V(D)J | No functional pre-BCR | B-cell autoimmunity | 83 | 0 | >80 | 54, 66 |
| VPREB1 | Surrogate light chain | Focal deletion | No functional pre-BCR | Pro–B-cell block | 15 | 0 | 20 | 56 |
| CD79A | (Pre-) BCR signaling chain | Transcriptional silencing | No functional pre-BCR | Pro–B-cell block | 80 | 0 | >80 | 54, 55 |
| CD79B | (Pre-) BCR signaling chain | Transcriptional silencing | No functional pre-BCR | Pro–B-cell block | 80 | 0 | >80 | 54, 55 |
| SYK | (Pre-) BCR tyrosine kinase | Aberrant splicing, mutation | Pre-BCR signaling ↓ | Pro–B-cell block | <10 | 0 | <10 | 54 |
| BLNK | (Pre-) BCR linker | Aberrant splicing | Pre–B-cell differentiation ↓ | Pre–B-cell block | 40 | 0 | ∼50 | 53, 57 |
| Focal deletion | Cytokine receptor signaling ↑ | Propensity to ALL | 2 | 2 | 64 | |||
| BTK | (Pre-) BCR tyrosine kinase | Aberrant splicing | Pre–B-cell differentiation ↓ | Pre–B-cell block | 30 | 0 | ∼25 | 58-60 |
| Propensity to ALL | ||||||||
| PAX5 | Transcription factor | Deletion, point mutation | Cytokine receptor signaling ↑ | Propensity to ALL | ∼25 | 10 | ∼25 | 64 |
| BACH2 | Transcription factor | Deletion 6q15 | Pre-BCR signaling ↓ | Propensity to ALL | 20 | 0 | ∼25 | 34 |
| IKZF1 | Transcription factor | Deletion, point mutation | Pre-BCR signaling ↓ | Propensity to ALL | 20-84‡ | 0 | ∼25 | 64, 65 |
| IKZF3 | Transcription factor | Deletion | Pre-BCR signaling ↓ | Propensity to ALL | 2 | 0 | 2 | 64, 65 |
| EBF1 | Transcription factor | Deletion, point mutation | Pre-BCR signaling ↓ | Propensity to ALL | 6 | 0 | 8 | 64, 65 |
| PBX1 | Transcription factor | TCF3-PBX1 rearrangement | Pre-BCR signaling ↑ | Propensity to ALL | 3*-5† | 40 | 0 | 66, 68 |
| 1q23 duplication | 1 | 5 | 0 | 66 | ||||
| PRDM1 | Transcription factor | Deletion 6q21 | Pre-BCR signaling ↑ | Plasma cell defect | 1 | 5 | 0 | 66 |
| Signaling . | Function . | Abnormality . | Functional outcome . | Phenotype in disease model . | Frequency in human ALL (%) . | Reference . | ||
|---|---|---|---|---|---|---|---|---|
| Total . | Pre-BCR+ . | Pre-BCR− . | ||||||
| STAT5 | Transcription factor | Hyperphosphorylation | Cytokine receptor signaling ↑ | Pro–B-cell survival ↓ | ∼40 | 0 | >50 | 9-11, 66 |
| BCL6 | Transcription factor | Overexpression | Pre-BCR signaling ↑ | Pre–B-cell survival ↓ | 13.5 | >95 | 0 | 31-33, 66 |
| Signaling . | Function . | Abnormality . | Functional outcome . | Phenotype in disease model . | Frequency in human ALL (%) . | Reference . | ||
|---|---|---|---|---|---|---|---|---|
| Total . | Pre-BCR+ . | Pre-BCR− . | ||||||
| STAT5 | Transcription factor | Hyperphosphorylation | Cytokine receptor signaling ↑ | Pro–B-cell survival ↓ | ∼40 | 0 | >50 | 9-11, 66 |
| BCL6 | Transcription factor | Overexpression | Pre-BCR signaling ↑ | Pre–B-cell survival ↓ | 13.5 | >95 | 0 | 31-33, 66 |
Pediatric ALL.
Adult ALL.
Ph+ ALL (BCR-ABL1).
Identification of a pre-BCR–dependent subset of human ALL
In ∼85% of human ALL cases, the dominant leukemic clones lack expression of a functional pre-BCR. However, we and others recently identified a distinct subset of human ALL that is selected for expression and activity of a functional pre-BCR.54,66,68 In about 13.5% of human ALL cases (112 of 830 cases studied),54,66 ALL cells exhibit tonic pre-BCR signaling (pre-BCR+), and were highly sensitive to inhibition of SYK, SRC, and BTK tyrosine kinases66,68 as well as PI3Kδ inhibition.66 In analogy to mature B-cell lymphoma, patient-derived pre-BCR+ ALL cells responded to treatment with ibrutinib and idelalisib in vitro. This group includes the ALL subset with TCF3-PBX1 rearrangement, which is selectively sensitive to ibrutinib.69 Treatment with the dual ABL1/BTK-SRC kinase inhibitor dasatinib induced leukemia regression and extended overall survival of nonobese diabetic/severe combined immunodeficiency mice that were transplanted with patient-derived pre-BCR+ ALL cells.64 In up to 50% of pre-BCR+ ALL cells, the leukemia cells carry an activating lesion of the homeodomain transcription factor PBX1, mostly through TCF3-PBX1 rearrangement66,68 and in some cases through duplication of a fragment at 1q23 encompassing PBX1. PBX1 is critical for normal B lymphopoiesis70 and chromatin immunoprecipitation sequencing and gene expression analyses revealed that PBX1 directly binds to and activates multiple components of the pre-BCR signaling pathway, including the surrogate light chain segments VPREB1 and IGLL1 (λ5), CD79A, CD79B, BLNK, and BTK.66 Given its critical role during early B-cell development and ability to activate pre-BCR expression, it is conceivable that gain of function lesions of PBX1 contribute to the development of pre-BCR+ ALL. Deletion of 6q21, encompassing PRDM1 (BLIMP1) represents a second recurrent genetic lesion in pre-BCR+ ALL cells (Table 1). Of note, lesions of PRDM1 are frequently observed in activated B-cell–like-diffuse large B-cell lymphoma,71 which are—like pre-BCR+ ALL—sensitive to treatment with ibrutinib and idelalisib. In human ALL, deletions of 6q21 are rare (∼1%); likewise, TCF3-PBX1 rearrangements only account for approximately 3% to 5% of all cases of human ALL (Table 1). For this reason, it is likely that other recurrent genetic lesions that define pre-BCR+ ALL (∼10% to 15%) still remain to be identified.
BCL6 as biomarker for pre–BCR-dependent cases of human ALL
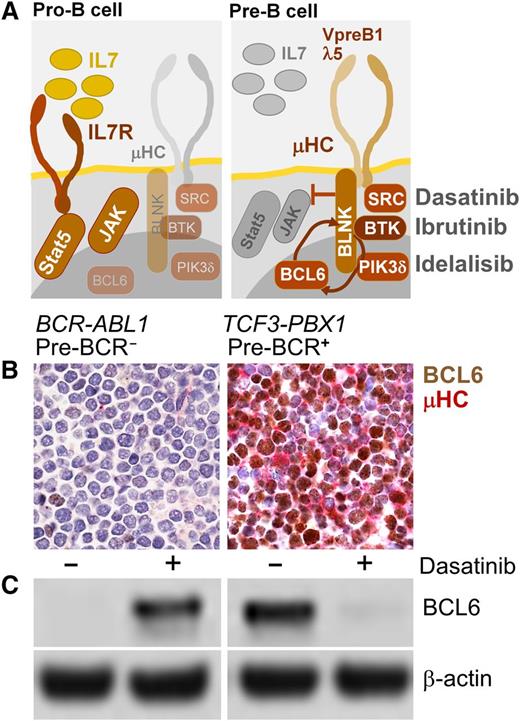
B-cell lymphoma cells express high levels of BCL6 in the context of tonic BCR signaling.72 Likewise, tonic pre-BCR signaling in ALL cells induces strong expression of BCL6.31 Consistent with induction of BCL6 by tonic pre-BCR signaling, all pre-BCR+ ALL cases studied showed constitutive and pre–BCR-dependent expression of BCL6, which was sensitive to small molecule inhibition of pre–BCR-signaling66 (Figure 1). Unlike pre-BCR+ ALL, cytokine receptor–dependent subsets of ALL express active STAT5 (Table 1), a negative regulator of BCL6. Illustrating the divergent biological behavior of pre-BCR+ ALL (eg, TCF3-PBX1) and cytokine receptor/STAT5-dependent ALL (eg, BCR-ABL1), the dual ABL1/SRC-BTK inhibitor dasatinib abolished BCL6 expression in pre-BCR+ ALL, but induced de novo expression of BCL6 in Ph+ ALL cells (Figure 1). Because functional assays to measure tonic pre-BCR signaling in ALL patient samples may not be practical for diagnostic purposes, immunostaining for BCL6 and immunoglobulin µHC expression may be a feasible alternative to rapidly identify patients that might benefit from treatment with inhibitors of tonic pre-BCR signaling (eg, SYK, SRC, BTK, PIK3δ). Besides BCL6/µHC double strainings, additional markers, including phospho-STAT5, are currently being tested with the goal to reliably identify patients with pre-BCR+ ALL. Our current cytogenetic and phenotypic definition of pre-BCR+ ALL is still incomplete and the identification of additional genetic and phenotypic characteristics will not only further elucidate the biology of pre-BCR+ ALL, but also help to identify patients that may benefit from treatment strategies targeting pre-BCR signaling.
BCL6 as biomarker to identify pre-BCR− and pre-BCR+ cases of human ALL. A schematic summarizing normal cytokine-dependent survival signaling in pro-B cells (pre-BCR−) and pre–BCR-dependent survival signaling in pre-B cells (A). SRC and BTK are targets of dasatinib, BTK is a target of ibrutinib, and PI3Kδ is the target if idelalisib (A), which are being used in clinical trials for patients with mature B-cell lymphoma. Functional assays to measure tonic pre-BCR signaling in ALL patient samples may not be practical for diagnostic purposes. Given that tonic pre-BCR signaling results in strong expression of BCL6,31 immunostainings for BCL6 and immunoglobulin µHC expression represents a feasible alternative to rapidly identify patients that might benefit from treatment with inhibitors of pre-BCR signaling (eg, ibrutinib, idelalisib). (B) Double stainings for BCL6 (brown; D8, Santa Cruz Biotech) and µHC (red; G20-127, BD Biosciences) were performed on paraffin-embedded sections from bone marrow clots. (B) Two patient-derived ALL cases were studied and counterstained with hematoxylin and eosin (100× magnification).66 One pre-BCR− ALL case (BCR-ABL1, left) and 1 pre-BCR+ ALL case (TCF3-PBX1, right) is shown. Panel C shows a western blot analysis of BCL6 expression (D8; Santa Cruz Biotech) in pre-BCR− (left) and pre-BCR+ (right) and ALL cells, with and without treatment with the dual ABL1/SRC-BTK inhibitor dasatinib (25 nmol/L for 24 hours), using β-actin (ab8227, Abcam) as loading control (C).
BCL6 as biomarker to identify pre-BCR− and pre-BCR+ cases of human ALL. A schematic summarizing normal cytokine-dependent survival signaling in pro-B cells (pre-BCR−) and pre–BCR-dependent survival signaling in pre-B cells (A). SRC and BTK are targets of dasatinib, BTK is a target of ibrutinib, and PI3Kδ is the target if idelalisib (A), which are being used in clinical trials for patients with mature B-cell lymphoma. Functional assays to measure tonic pre-BCR signaling in ALL patient samples may not be practical for diagnostic purposes. Given that tonic pre-BCR signaling results in strong expression of BCL6,31 immunostainings for BCL6 and immunoglobulin µHC expression represents a feasible alternative to rapidly identify patients that might benefit from treatment with inhibitors of pre-BCR signaling (eg, ibrutinib, idelalisib). (B) Double stainings for BCL6 (brown; D8, Santa Cruz Biotech) and µHC (red; G20-127, BD Biosciences) were performed on paraffin-embedded sections from bone marrow clots. (B) Two patient-derived ALL cases were studied and counterstained with hematoxylin and eosin (100× magnification).66 One pre-BCR− ALL case (BCR-ABL1, left) and 1 pre-BCR+ ALL case (TCF3-PBX1, right) is shown. Panel C shows a western blot analysis of BCL6 expression (D8; Santa Cruz Biotech) in pre-BCR− (left) and pre-BCR+ (right) and ALL cells, with and without treatment with the dual ABL1/SRC-BTK inhibitor dasatinib (25 nmol/L for 24 hours), using β-actin (ab8227, Abcam) as loading control (C).
Acknowledgments
The author thanks Drs Hassan Jumaa (Ulm, Germany), Ari Melnick (New York, NY), Art Weiss (San Francisco, CA), Rudi W. Hendriks (Rotterdam, Netherlands), and Louis M. Staudt (Bethesda) for critical discussions and encouragement.
This work is supported by grants from the National Institutes of Health/National Cancer Institute (R01CA137060, R01CA139032, R01CA157644, R01CA169458, and R01CA172558), the William Lawrence and Blanche Hughes Foundation, the California Institute for Regenerative Medicine (CIRM, TR2-01816), Leukaemia and Lymphoma Research, and Cancer Research UK. M.M. is a Scholar of the Leukemia and Lymphoma Society and a Senior Investigator of the Wellcome Trust.
Authorship
Contribution: M.M. developed the concept and wrote the manuscript.
Conflict-of-interest disclosure: The author declares no competing financial interests.
Correspondence: Markus Müschen, Department of Laboratory Medicine, University of California San Francisco, 513 Parnassus Ave, San Francisco CA 94143; e-mail: markus.muschen@ucsf.edu.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal