Key Points
DLBCL can be detected in the blood by immunoglobulin high-throughput sequencing (Ig-HTS) with high specificity.
Although DLBCL can be detected in leukocytes or plasma by Ig-HTS, plasma has greater sensitivity and more accurately reflects disease.
Abstract
Recent studies have shown limited utility of routine surveillance imaging for diffuse large B-cell lymphoma (DLBCL) patients achieving remission. Detection of molecular disease by immunoglobulin high-throughput sequencing (Ig-HTS) from peripheral blood provides an alternate strategy for surveillance. We prospectively evaluated the utility of Ig-HTS within 311 blood and 105 tumor samples from 75 patients with DLBCL, comparing Ig-HTS from the cellular (circulating leukocytes) and acellular (plasma cell-free DNA) compartments of peripheral blood to clinical outcomes and 18fluoro-deoxyglucose positron emission tomography combined with computed tomography (PET/CT; n = 173). Clonotypic immunoglobulin rearrangements were detected in 83% of patients with adequate tumor samples to enable subsequent monitoring in peripheral blood. Molecular disease measured from plasma, compared with circulating leukocytes, was more abundant and better correlated with radiographic disease burden. Before treatment, molecular disease was detected in the plasma of 82% of patients compared with 71% in circulating cells (P = .68). However, molecular disease was detected significantly more frequently in the plasma at time of relapse (100% vs 30%; P = .001). Detection of molecular disease in the plasma often preceded PET/CT detection of relapse in patients initially achieving remission. During surveillance time points before relapse, plasma Ig-HTS demonstrated improved specificity (100% vs 56%, P < .0001) and similar sensitivity (31% vs 55%, P = .4) compared with PET/CT. Given its high specificity, Ig-HTS from plasma has potential clinical utility for surveillance after complete remission.
Introduction
Diffuse large B-cell lymphoma (DLBCL), the most common subtype of non-Hodgkin lymphoma (NHL), remains clinically heterogeneous. After therapy with first-line regimens, a significant number of DLBCL patients do not achieve a complete response (CR), and an additional subset of patients ultimately relapse after CR.1,2 Assessment of therapeutic response and monitoring for relapse are therefore important components of clinical care, and current guidelines recommend evaluation every 3 to 6 months for 5 years.3 Although some guidelines suggest x-ray computed tomography (CT) for 2 years after the completion of therapy,3 recent studies have shown limited utility of routine surveillance imaging,4,5 and newer guidelines therefore discourage this approach.6 Separately, despite significant utility of functional imaging for posttreatment evaluation,7 response assessment using interim 18fluoro-deoxyglucose positron emission tomography combined with CT (PET/CT) during immunochemotherapy can produce false-positive results and is less well established for DLBCL than Hodgkin lymphoma.8,9 More sensitive and specific methods for longitudinal disease monitoring are therefore needed.
DNA containing tumor-specific sequences can be detected in either the cellular (ie, circulating tumor cells [CTCs]) or cell-free (ie, circulating tumor DNA [ctDNA]) fraction of the blood in numerous cancers, including DLBCL.10-15 The junction between the immunoglobulin variable, (diversity), and joining genes provides a unique DNA clonotype that is shared by all malignant B cells and can be sensitively detected by next-generation sequencing. High-throughput sequencing of immunoglobulin genes (Ig-HTS) from circulating leukocytes has been used as a biomarker for disease in diverse B-cell malignancies,16-19 including an initial report in DLBCL.16 However, Ig-HTS from plasma (ctDNA) and circulating leukocytes (CTCs) has not been systematically compared with other clinical measures of disease and outcome. Using 311 blood samples across 75 DLBCL patients, we explore the clinical utility of Ig-HTS for detection of molecular disease from plasma ctDNA and peripheral blood CTCs for disease monitoring compared with PET/CT and ultimate clinical outcomes.
Methods
Patient selection
Patients were prospectively enrolled as part of either of 2 groups: cohort 1 patients were seen at Stanford University with newly diagnosed or recurrent DLBCL per World Health Organization 2008 criteria between 2010 and 2014. All treatments and radiographic examinations were performed as part of standard clinical care, using combination immunochemotherapy with curative intent. Treatment selection was at the discretion of the treating physician and in accordance with institutional standards and national guidelines.20 A subset of patients with historically poor risk factors for treatment with rituximab, cyclophosphamide, doxorubicin, vincristine, and prednisone (R-CHOP), including MYC translocation,21,22 double-hit translocations,23 high Ki-67,24 and HIV-associated NHLs,25 were treated with alternative regimens such as dose-adjusted etoposide, prednisone, vincristine, cyclophosphamide, doxorubicin, and rituximab (DA-EPOCH-R). Cohort 2 patients were seen as part of the Australasian Leukaemia and Lymphoma Group (ALLG) NHL-21 clinical trial (ACTRN12609001077257). As part of the NHL-21 trial, PET/CT scans and blood specimens were taken at diagnosis before treatment and an interim time point after 4 cycles of R-CHOP given every 14 days. Patients with residual DLBCL after 4 cycles of R-CHOP as identified by interim PET/CT received early treatment intensification and autologous stem cell transplantation. Of 140 patients enrolled on ALLG-NHL-21, 8 had sufficient tumor and pre-/posttreatment plasma samples and were therefore included in our study. Cell of origin was determined by molecular profiling26 and/or immunohistochemistry27 where available. Fluorescence in situ hybridization for BCL2, BCL6, and MYC translocations was performed using break-apart probes. This study conformed to the Declaration of Helsinki, with written informed consent provided by all participants, and was approved by all participating hospital/research institute Human Research Ethics/Institutional Review Board Committees. Patient characteristics are shown in Table 1 and are further detailed in supplemental Table 1 available on the Blood Web site.
Patient characteristics
| . | Entire cohort (n = 75) . | Ig reporter identified (n = 57) . | No Ig reporter identified (n = 18) . | P . |
|---|---|---|---|---|
| Average age (years) | 58.3 | 57.5 | 60.8 | .3 |
| Male, n (%) | 44 (58.7) | 34 (59.6) | 10 (55.6) | .79 |
| Female, n (%) | 31 (41.3) | 23 (40.4) | 8 (44.4) | |
| Diagnosis | .23 | |||
| DLBCL, NOS | 54 | 39 | 15 | |
| DLBCL, transformed | 13 | 10 | 3 | |
| PTLD | 8 | 8 | 0 | |
| Stage, n (%) | .0181 | |||
| 1 | 7 (9.3) | 7 (12.3) | 0 (0.0) | |
| 2 | 11 (14.7) | 10 (17.5) | 1 (5.6) | |
| 3 | 8 (10.7) | 3 (5.3) | 5 (27.8) | |
| 4 | 49 (65.3) | 37 (64.9) | 12 (66.7) | |
| IPI, n (%) | .86 | |||
| 0 to 1 | 13 (18.8) | 9 (17.6) | 4 (23.5) | |
| 2 | 23 (33.3) | 17 (33.3) | 6 (35.3) | |
| 3 | 21 (30.4) | 15 (29.4) | 6 (35.3) | |
| 4 to 5 | 12 (17.4) | 10 (19.6) | 2 (11.8) | |
| LDH elevated, n (%) | 34 (48.6) | 29 (53.7) | 5 (31.3) | .11 |
| Cell of origin | ||||
| GCB | 38 (50.7) | 27 (47.4) | 11 (61.1) | .75 |
| Non-GCB | 17 (22.7) | 13 (22.8) | 4 (22.2) | |
| Not available/unclassifiable | 20 (26.7) | 17 (29.8) | 3 (16.7) | |
| Translocations | ||||
| BCL2 | 9/49 (18.4) | 6/35 (17.1) | 3/14 (21.4) | 1 |
| BCL6 | 6/41 (14.6) | 5/29 (17.2) | 1/12 (8.3) | .65 |
| MYC | 8/48 (16.7) | 6/35 (17.1) | 2/13 (15.4) | 1 |
| Treatment | ||||
| DA-EPOCH-R | 22 | |||
| R-CHOP like | 39 | |||
| Modified Magrath | 4 | |||
| Other | 9 | |||
| No treatment | 1 |
| . | Entire cohort (n = 75) . | Ig reporter identified (n = 57) . | No Ig reporter identified (n = 18) . | P . |
|---|---|---|---|---|
| Average age (years) | 58.3 | 57.5 | 60.8 | .3 |
| Male, n (%) | 44 (58.7) | 34 (59.6) | 10 (55.6) | .79 |
| Female, n (%) | 31 (41.3) | 23 (40.4) | 8 (44.4) | |
| Diagnosis | .23 | |||
| DLBCL, NOS | 54 | 39 | 15 | |
| DLBCL, transformed | 13 | 10 | 3 | |
| PTLD | 8 | 8 | 0 | |
| Stage, n (%) | .0181 | |||
| 1 | 7 (9.3) | 7 (12.3) | 0 (0.0) | |
| 2 | 11 (14.7) | 10 (17.5) | 1 (5.6) | |
| 3 | 8 (10.7) | 3 (5.3) | 5 (27.8) | |
| 4 | 49 (65.3) | 37 (64.9) | 12 (66.7) | |
| IPI, n (%) | .86 | |||
| 0 to 1 | 13 (18.8) | 9 (17.6) | 4 (23.5) | |
| 2 | 23 (33.3) | 17 (33.3) | 6 (35.3) | |
| 3 | 21 (30.4) | 15 (29.4) | 6 (35.3) | |
| 4 to 5 | 12 (17.4) | 10 (19.6) | 2 (11.8) | |
| LDH elevated, n (%) | 34 (48.6) | 29 (53.7) | 5 (31.3) | .11 |
| Cell of origin | ||||
| GCB | 38 (50.7) | 27 (47.4) | 11 (61.1) | .75 |
| Non-GCB | 17 (22.7) | 13 (22.8) | 4 (22.2) | |
| Not available/unclassifiable | 20 (26.7) | 17 (29.8) | 3 (16.7) | |
| Translocations | ||||
| BCL2 | 9/49 (18.4) | 6/35 (17.1) | 3/14 (21.4) | 1 |
| BCL6 | 6/41 (14.6) | 5/29 (17.2) | 1/12 (8.3) | .65 |
| MYC | 8/48 (16.7) | 6/35 (17.1) | 2/13 (15.4) | 1 |
| Treatment | ||||
| DA-EPOCH-R | 22 | |||
| R-CHOP like | 39 | |||
| Modified Magrath | 4 | |||
| Other | 9 | |||
| No treatment | 1 |
Characteristics of all patients enrolled in this study. NOS, not otherwise specified; PTLD, posttransplant lymphoproliferative disorder.
Sample collection, processing, DNA purification, and quantification
Tumor samples for clonotype determination comprised archived formalin-fixed, paraffin-embedded (FFPE) tissue blocks, archived DNA, or freshly isolated and immediately frozen tumor biopsy specimens. Peripheral blood samples were collected in K2EDTA Vacutainer tubes (BD) and processed within 4 hours of collection. Tumor DNA was isolated using the AllPrep DNA/RNA FFPE Kit (Qiagen), DNA from peripheral blood mononuclear cells with the DNeasy Blood & Tissue Kit (Qiagen), and cell-free DNA using the QIAamp Circulating Nucleic Acid Kit (Qiagen). Detailed sample collection and processing methods are available in the supplemental Methods.
Ig-HTS
Clonotypic immunoglobulin status and molecular disease were assessed using the LymphoSIGHT platform as previously described,17 with minor modifications. Briefly, genomic tumor DNA was amplified using locus-specific primers to allow for the amplification of all known alleles of the germ-line IGH and IGK sequences. The products were sequenced, yielding the identities and frequencies of the different clonotypes. Clonotype frequencies within a sample were determined by calculating the number of sequencing reads for each clonotype divided by the total number of passed sequencing reads in the sample. Lymphoma immunoglobulin reporters were defined using a frequency threshold exceeding 5% in tumor biopsies.
Molecular disease measurement by Ig-HTS
The lymphoma-derived sequences identified in biopsies were used as a target to assess the presence of molecular disease in peripheral blood samples. For molecular disease quantitation, we generated multiple sequencing reads for each rearranged B cell in the reaction. To determine the absolute measure of the total lymphoma-derived molecules present in the sample, we added a known quantity of reference IgH sequence and counted the associated sequencing reads. The known quantity of reference IgH sequence was derived from a pool of plasmids containing 8 unique IgH clonotypes. The resulting factor (number of molecules per sequence read) was then applied to the lymphoma-associated clonal rearrangement reads to obtain an absolute measure of the total lymphoma-derived molecules in the reaction. Finally, we calculated the total leukocytes in the reaction by measuring the total DNA by picogreen and RT-PCR using genomic markers. The final molecular disease measurement was calculated for each sample both as a fraction of clonal immunoglobulin molecules per input DNA (defined as the number of clonal immunoglobulin molecules per 1 × 106 input genome equivalents of DNA) and as a concentration of clonal immunoglobulin molecules per volume (defined as the number of immunoglobulin reporter molecules divided by the input volume of the blood specimen). Molecular disease was considered positive for any nonzero result.
Peripheral blood samples were defined as occurring in any of 4 time points: pretreatment, during treatment, surveillance, and progressive disease. Pretreatment time points were defined as blood drawn before any treatment. During treatment time points were defined as blood drawn after the start of therapy until 21 days after the completion of therapy. Surveillance time points were defined as blood drawn after a documented complete response and after completion of treatment. Progressive disease time points were defined as blood drawn at the time of documented progression of disease by imaging or pathology but before the start of subsequent therapy.
PET/CT and metabolic tumor volume
PET/CT scans from patients in cohort 1 were reviewed by a single independent reviewer (D.M.K.). PET/CT scans from patients in cohort 2 before therapy and after 4 cycles of R-CHOP were reviewed per ALLG-NHL-21 protocol. For clinical care of patients, responses in cohort 1 were determined using International Harmonization Project criteria for malignant lymphoma.28 Responses in cohort 2 were determined using the 5-point Deauville criteria with a score >3 defining positivity.29 For calculations of sensitivity and specificity of surveillance PET/CT imaging, International Harmonization Project criteria were used in both cohorts.
Metabolic tumor volume (MTV) was defined as the volume of lymphoma visualized on PET/CT scans with a standardized uptake value greater than or equal to an absolute threshold of 4.0. Computer-aided analysis of PET images for MTV calculations was performed using MIM Maestro software suite, version 5.1 (MIM Software, Cleveland, OH) and using METAVOL30 (University of California, Los Angeles, CA), with exclusion of urinary, myocardial, and brain 18FDG uptake. For correlations between MTV and Ig-HTS on patients where such assessments were not available on exactly the same date, Ig-HTS value from the closest date was used as long as these were within 28 days. For generation of maximum intensity projection PET images, DICOM (Digital Imaging and Communications in Medicine) images were processed with OsiriX software.31
Statistical analysis
Comparisons between categorical variables were performed by Fisher’s exact test, ordinal variables including Ann Arbor stage and International Prognostic Index (IPI) by χ2 test, and patient age by Student t test. Pearson correlation was used to compare Ig-HTS to MTV. The analytical sensitivity of the LymphoSIGHT assay was estimated at detection of 1 clonal molecule in a background of 1 × 106 nonclonal molecules. As the average input into the Ig-HTS reaction for peripheral blood mononuclear cells was 1.03 × 106 genomes, the nominal sensitivity for this assay is 1 in 1 × 106. For detection of plasma molecular disease (ie, ctDNA), the average input into the Ig-HTS reaction was 2.18 × 103 genomes; therefore, the nominal sensitivity of Ig-HTS on plasma is ∼1 in 2.18 × 103. Sensitivity and specificity of Ig-HTS and PET/CT were calculated in comparison with eventual clinical outcome as the gold standard in patients achieving complete remission, over a median follow-up time of 34.2 months. Median follow-up from diagnosis for the entire cohort was 22.6 months (range, 0.7-285.2 months).
Results
Patients and sample collection
A total of 75 patients were accrued with characteristics shown in Table 1. The majority (76%) of patients presented with Ann Arbor stage III to IV disease but encompassed a range of IPI risk categories. Thirteen patients had an antecedent low-grade lymphoma. At the time of first evaluation, 68 patients had a new diagnosis of DLBCL and 7 patients had relapsed disease. A total of 105 tumor samples, 311 blood samples (39 pretreatment, 123 during treatment, 102 during surveillance, and 47 at time of progressive disease [PD]), and 173 PET/CTs were evaluated (supplemental Table 1).
Identification of clonotypic immunoglobulin rearrangements in pathologic specimens
Before disease monitoring, clonal immunoglobulin rearrangements were identified from a tumor biopsy. Immunoglobulin clonotype reporters (immunoglobulin reporters) included heavy-chain VDJ rearrangement (IGH-VDJ), heavy-chain DJ rearrangement (IGH-DJ), and κ light-chain VJ rearrangement (IGK). Clonotypic immunoglobulin reporters were identified in 83% (57 of 69) of patients with adequate DNA; 6 patients had insufficient DNA isolated from their tumor specimen (<1.5 ng). Of these 57 patients, 86% (49 of 57) had identification of clonal reporters on the first tumor specimen (Figure 1A). Clinical features of patients with immunoglobulin reporters identified were generally similar to those without reporters identified, with the exception of stage (Table 1). No significant relationship was found to cell of origin classification or to relevant cytogenetic lesions in BCL2, BCL6, or MYC, although these analyses were limited by small sample size. The success rate of clonotypic immunoglobulin reporter identification was confirmed in a subsequent validation cohort of 21 additional consecutive patients seen at Stanford University. The rate of clonotypic immunoglobulin reporter identification in this real-world cohort (71%) was lower but not significantly different from our initial cohort (P = .23; data not shown).
Calibration and performance of Ig-HTS. (A) The overall success rate of tumor clonotype identification is shown per patient (left bar) and further broken down when considering number of biopsy specimens (right two bars). (B) When considering all samples assayed, fresh/frozen tumor biopsy specimens had a significantly higher rate of tumor clonotype detection compared with formalin-fixed paraffin-embedded (FFPE) specimens (Fisher exact, P = .007). (C) The success rate of tumor clonotype identification from biopsy specimens was associated with the available yield of DNA input for the reaction. An increasing success rate can be seen with increased input mass. Samples with less than minimum required DNA input (<1.5 ng) were not assayed and are therefore excluded from all panels.
Calibration and performance of Ig-HTS. (A) The overall success rate of tumor clonotype identification is shown per patient (left bar) and further broken down when considering number of biopsy specimens (right two bars). (B) When considering all samples assayed, fresh/frozen tumor biopsy specimens had a significantly higher rate of tumor clonotype detection compared with formalin-fixed paraffin-embedded (FFPE) specimens (Fisher exact, P = .007). (C) The success rate of tumor clonotype identification from biopsy specimens was associated with the available yield of DNA input for the reaction. An increasing success rate can be seen with increased input mass. Samples with less than minimum required DNA input (<1.5 ng) were not assayed and are therefore excluded from all panels.
Success of immunoglobulin reporter identification was significantly higher in fresh/frozen tissue compared with FFPE (P = .007; Figure 1B) and with higher levels of available input DNA (Figure 1C). IGH-VDJ reporters were identified more frequently than IGH-DJ and IGK reporters (39 of 57, 23 of 57, and 25 of 57 patients, respectively), and ≥2 reporters were identified in 53% (30 of 57) patients.
Detection of molecular disease by Ig-HTS as ctDNA vs CTCs
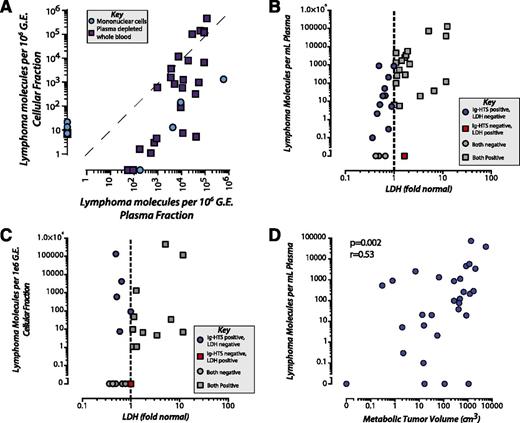
In patients with tumor-derived immunoglobulin reporters successfully identified, we assessed the ability of Ig-HTS to then detect molecular disease from peripheral blood samples. Circulating DNA can be found in either of 2 fractions: circulating cells (ie, CTCs) or cell-free DNA in plasma (ie, ctDNA). Surprisingly, we found that molecular disease burden was quantitatively and qualitatively higher in plasma as ctDNA compared with circulating cells as CTCs. In individual blood collections where molecular disease was detectable within both CTCs and ctDNA, we found ∼150-fold (median) higher allele fraction in the plasma as ctDNA compared with the corresponding cellular sample (Figure 2A). Because the plasma fraction contains substantially lower amounts of DNA compared with the cellular fraction for a given volume of blood, we normalized for input. Nevertheless, we found an ∼2.3-fold higher absolute level of molecular disease per milliliter of blood in plasma than in circulating cells in patients with DLBCL, where disease was detectable in both fractions (supplemental Figure 1).
Molecular disease in plasma vs leukocytes. (A) By analyzing cellular and plasma fractions isolated from 82 samples acquired from 25 patients, we found that clonotype was more abundant per million genome equivalent (G.E.) in the plasma fraction. This included 2 types of cellular fractions: peripheral blood mononuclear cells (blue circles) and plasma-depleted whole blood (purple squares). For 11 samples from 7 different patients, tumor clonotype was detected solely within the plasma fraction and not the cellular fraction, including 2 samples taken at the time of CNS relapse. A total of 7 samples from 4 patients had the tumor clonotype detected only in the cellular fraction and not in the plasma. (B) Thirty-four paired samples with both plasma Ig-HTS and LDH and (C) 26 paired samples with both cellular Ig-HTS and LDH at time points of overt disease (diagnosis or progression/relapse). Plasma Ig-HTS was significantly more sensitive than LDH (88% vs 59%, P = .01). Cellular Ig-HTS was not more sensitive than LDH (62% vs 59%, P = 1.0). (D) Detection of molecular disease in the plasma fraction of blood was significantly correlated with MTV as measured by PET/CT scans (n = 32) at paired time points across 27 patients (P = .002, Pearson r = 0.53). No significant correlation was seen between MTV and cellular Ig-HTS. Plasma samples were taken within 28 days of PET/CT scan (median, 1.5 days).
Molecular disease in plasma vs leukocytes. (A) By analyzing cellular and plasma fractions isolated from 82 samples acquired from 25 patients, we found that clonotype was more abundant per million genome equivalent (G.E.) in the plasma fraction. This included 2 types of cellular fractions: peripheral blood mononuclear cells (blue circles) and plasma-depleted whole blood (purple squares). For 11 samples from 7 different patients, tumor clonotype was detected solely within the plasma fraction and not the cellular fraction, including 2 samples taken at the time of CNS relapse. A total of 7 samples from 4 patients had the tumor clonotype detected only in the cellular fraction and not in the plasma. (B) Thirty-four paired samples with both plasma Ig-HTS and LDH and (C) 26 paired samples with both cellular Ig-HTS and LDH at time points of overt disease (diagnosis or progression/relapse). Plasma Ig-HTS was significantly more sensitive than LDH (88% vs 59%, P = .01). Cellular Ig-HTS was not more sensitive than LDH (62% vs 59%, P = 1.0). (D) Detection of molecular disease in the plasma fraction of blood was significantly correlated with MTV as measured by PET/CT scans (n = 32) at paired time points across 27 patients (P = .002, Pearson r = 0.53). No significant correlation was seen between MTV and cellular Ig-HTS. Plasma samples were taken within 28 days of PET/CT scan (median, 1.5 days).
At the time of diagnosis, molecular disease was detected in 82% (18 of 22) of patients in the plasma and 71% (10 of 14) of patients in CTCs (P = .68). However, at the time of relapse/progression, detection from plasma as ctDNA significantly outperformed detection from circulating cells. All patients (11 of 11, 100%) had detectable disease at the time of relapse/progression in the plasma compared with only 30% (3 of 10) of patients detected in circulating cells (P = .001). At the sample level, 81% (38/48) of plasma samples were detectable at the time of diagnosis or progression compared with 53% (20 of 38) of cellular samples (P = .011). Additionally, in patients with paired plasma and cellular samples available throughout the duration of treatment, we observed a greater dynamic range and sensitivity of detection in the plasma compared with circulating cells (supplemental Figure 2).
Detection of molecular disease in plasma in relation to lactate dehydrogenase and MTV
The blood biomarker most widely used for DLBCL is serum lactate dehydrogenase (LDH), with higher LDH levels correlating with disease burden. Additionally, elevated LDH levels are independently predictive of adverse clinical outcomes.32,33 To compare the performance of Ig-HTS detection of disease from plasma or circulating cells, we assessed blood draws from patients with active disease (time of initial diagnosis or relapse) as measured by PET/CT. At these paired time points, LDH had a sensitivity of 59% (20 of 34) compared with 88% (30 of 34) by plasma Ig-HTS (P = .01; Figure 2B). Molecular disease measured from circulating cells had similar sensitivity to LDH at the same time points where data were available (62%; 16 of 26; P = 1.0; Figure 2C). Examples of the increased sensitivity from plasma Ig-HTS compared with LDH can be seen in paired samples from 4 patients through the course of treatment (supplemental Figure 3).
Additionally, Ig-HTS measurements from plasma reflected the overall tumor burden. Plasma Ig-HTS was significantly correlated with MTV measured from PET/CT, a quantitative measurement of disease burden correlated with adverse outcomes34 (P = .002; Figure 2D; supplemental Figure 4). Ig-HTS measurement of CTCs did not correlate with MTV (P = .12).
Clinical disease monitoring by Ig-HTS during therapy
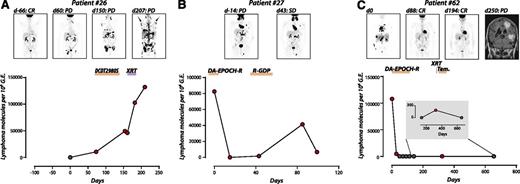
Examples of the utility of Ig-HTS from plasma for monitoring disease status during therapy are shown in Figure 3, with detectable molecular disease shown in red. In cases with PD (Figure 3A), stable disease (Figure 3B), and CR followed by progression (Figure 3C), quantitative measurements of molecular disease in the plasma reflect the changes in disease burden shown by PET/CT. Notably, during treatment of isolated central nervous system (CNS) relapse in patient 62, molecular disease was detectable at low levels in the plasma but not in the cellular fraction of the same sample (Figure 3C; day 323, 184 molecules per million genome equivalents; cellular data in supplemental Figure 2).
Ig-HTS monitoring of therapeutic response. Time courses of plasma molecular disease during therapy; red, positive molecular disease; black, negative molecular disease. (A) Patient 26 achieved a CR after primary therapy with DA-EPOCH-R for stage IVE DLBCL, but had progression based on a day 60 PET/CT. Plasma molecular disease at this time point was positive. In the ensuing disease progression during several lines of therapy, disease burden can be seen to increase by both plasma molecular disease and PET/CT. (B) Patient 27 was treated for relapsed stage IVAE DLBCL with DA-EPOCH-R with stable disease shown by PET/CT on day 43. Plasma molecular disease decreased, but remained detectable on days 15 and 43. PD was detected by CT on day 77 (data not shown), corresponding with an increased disease burden by plasma molecular disease that again subsides, but remains detectable, following treatment with R-GDP. (C) Patient 62 was diagnosed with stage IVBE DLBCL including leukemic disease, and disease burden can be seen to decline as the patient achieves a CR. An isolated CNS relapse detected by magnetic resonance imaging on day 250 is also detected by Ig-HTS (day 323).
Ig-HTS monitoring of therapeutic response. Time courses of plasma molecular disease during therapy; red, positive molecular disease; black, negative molecular disease. (A) Patient 26 achieved a CR after primary therapy with DA-EPOCH-R for stage IVE DLBCL, but had progression based on a day 60 PET/CT. Plasma molecular disease at this time point was positive. In the ensuing disease progression during several lines of therapy, disease burden can be seen to increase by both plasma molecular disease and PET/CT. (B) Patient 27 was treated for relapsed stage IVAE DLBCL with DA-EPOCH-R with stable disease shown by PET/CT on day 43. Plasma molecular disease decreased, but remained detectable on days 15 and 43. PD was detected by CT on day 77 (data not shown), corresponding with an increased disease burden by plasma molecular disease that again subsides, but remains detectable, following treatment with R-GDP. (C) Patient 62 was diagnosed with stage IVBE DLBCL including leukemic disease, and disease burden can be seen to decline as the patient achieves a CR. An isolated CNS relapse detected by magnetic resonance imaging on day 250 is also detected by Ig-HTS (day 323).
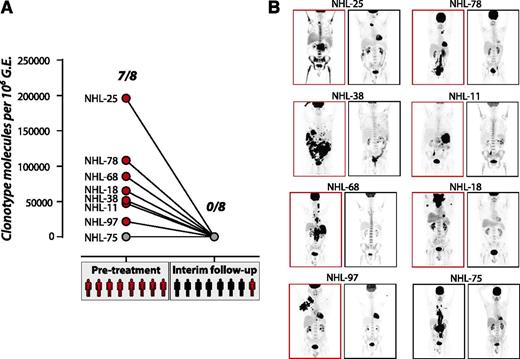
We further investigated the performance of plasma Ig-HTS for therapeutic monitoring from patients enrolled in the ALLG-NHL-21 trial, uniformly treated with R-CHOP therapy. Eight patients had both tumor specimens with sufficient DNA for clonotype identification and the required plasma samples before and after 4 cycles of R-CHOP (Figure 4). Despite only ∼1 mL plasma being available for this analysis, 87.5% (7 of 8) of samples had detectable disease at diagnosis. Seven of the 8 patients achieved a CR by PET/CT after 4 cycles; of these, 7 of 7 had no disease detectable by plasma, agreeing with PET/CT. However, in 1 patient who achieved only a partial response after 4 cycles of R-CHOP, molecular disease was not detected in the plasma at this interim time point. An additional 2 of the 7 patients who achieved a CR by PET/CT after 4 cycles ultimately relapsed; both of these patients had undetectable molecular disease after 4 cycles of therapy.
Limits of disease detection during therapy. (A) Tumor clonotype was detected at varying levels within 1 mL of plasma from 7 of 8 patients before treatment. At an interim time point after 4 cycles of R-CHOP, disease was not detectable in any patient (red circles, detectable molecular disease; gray circles, undetectable molecular disease; red patients, detectable disease on PET/CT; black patients, no detectable disease on PET/CT). (B) PET/CT scans show the disease burden for 8 patients before treatment (left) and after 4 cycles of R-CHOP (right). NHL-75, with no tumor clonotype detected within the plasma at diagnosis, can be seen to have a large tumor burden. Although no patients had detectable disease at the interim time point, 1 case showed residual disease by PET scan. This patient relapsed 376 days later (NHL-11). Two additional cases showed no disease by PET/CT at the interim time point but went on to relapse 247 (NHL-75) and 267 days (NHL-68) later.
Limits of disease detection during therapy. (A) Tumor clonotype was detected at varying levels within 1 mL of plasma from 7 of 8 patients before treatment. At an interim time point after 4 cycles of R-CHOP, disease was not detectable in any patient (red circles, detectable molecular disease; gray circles, undetectable molecular disease; red patients, detectable disease on PET/CT; black patients, no detectable disease on PET/CT). (B) PET/CT scans show the disease burden for 8 patients before treatment (left) and after 4 cycles of R-CHOP (right). NHL-75, with no tumor clonotype detected within the plasma at diagnosis, can be seen to have a large tumor burden. Although no patients had detectable disease at the interim time point, 1 case showed residual disease by PET scan. This patient relapsed 376 days later (NHL-11). Two additional cases showed no disease by PET/CT at the interim time point but went on to relapse 247 (NHL-75) and 267 days (NHL-68) later.
Disease monitoring by Ig-HTS during surveillance
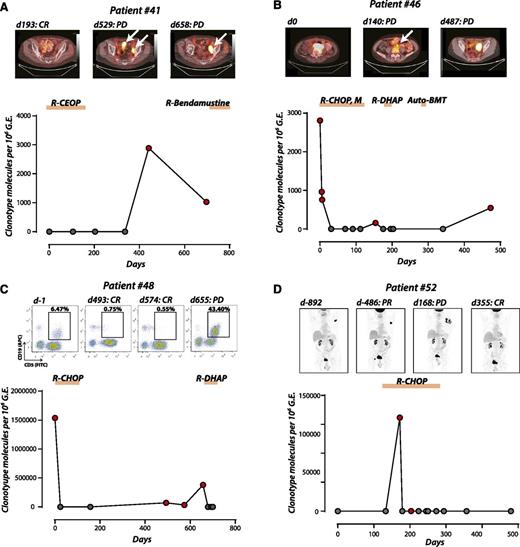
Having previously detected molecular disease in the plasma in 11 of 11 patients at the time of overt relapse, we sought to assess the performance of plasma Ig-HTS for detection of occult residual disease after CR. Although a more challenging task, detection of disease during surveillance after CR remains a major clinical need. Fifty-one assays from 25 patients were available during surveillance, of which 5 patients eventually relapsed. Three of these 5 patients (60%; patients 41, 46, and 48) had disease detectable in the plasma before their overt relapse (median lead time, 88 days; range, 14-162 days; Figure 5A-C). The remaining 2 relapsing patients had detectable disease in the plasma at the time of relapse. Similarly, plasma Ig-HTS identified a patient with antecedent follicular lymphoma at the time of transformation, with a striking increase in ctDNA (Figure 5D).
Ig-HTS surveillance for disease progression/relapse (A) Patient 41 was treated for stage 1A DLBCL with R-CEOP, achieved a CR, and was monitored thereafter. Plasma ctDNA was positive on day 441 within the plasma, before detection by PET/CT and diagnosis of PD (lead time = 88 days). (B) Patient 46, diagnosed with stage IIA DLBCL, was monitored during treatment encompassing 2 separate relapses. We observed a rapid decline in plasma disease to undetectable levels as this patient achieves a partial response. On days 42 and 84, residual disease was detected by PET/CT (data not shown) despite negative plasma Ig-HTS. A subsequent PET/CT detected progression (day 140) that was also detected by Ig-HTS. The patient achieved a CR by PET/CT after salvage chemotherapy on day 223, but ultimately relapsed as detected by plasma molecular disease on day 473, confirmed by PET/CT on day 487 (lead time = 14 days). (C) Patient 48 was diagnosed with stage IVBE DLBCL with leukemic phase and bone marrow involvement. After treatment with R-CHOP, the patient achieved a CR by flow cytometry on day 228 (data not shown), at which time disease was undetectable Ig-HTS. Plasma molecular disease was detected at a level of 71 860 molecules per million genome equivalents on day 493, despite no evidence of disease on peripheral blood flow cytometry on day 493 or PET/CT (data not shown). Plasma Ig-HTS was positive 162 days before the detection of PD by flow cytometry on day 655. (D) Patient 52 was initially expectantly observed with stage IIIA indolent follicular lymphoma, at which point plasma molecular disease was negative despite clear radiographic evidence. This patient then underwent clinical and histological transformation to DLBCL in a single site that coincided with a dramatic spike in plasma molecular disease. He was treated with R-CHOP chemotherapy for DLBCL, with subsequent CR on PET/CT and normalization of plasma molecular disease.
Ig-HTS surveillance for disease progression/relapse (A) Patient 41 was treated for stage 1A DLBCL with R-CEOP, achieved a CR, and was monitored thereafter. Plasma ctDNA was positive on day 441 within the plasma, before detection by PET/CT and diagnosis of PD (lead time = 88 days). (B) Patient 46, diagnosed with stage IIA DLBCL, was monitored during treatment encompassing 2 separate relapses. We observed a rapid decline in plasma disease to undetectable levels as this patient achieves a partial response. On days 42 and 84, residual disease was detected by PET/CT (data not shown) despite negative plasma Ig-HTS. A subsequent PET/CT detected progression (day 140) that was also detected by Ig-HTS. The patient achieved a CR by PET/CT after salvage chemotherapy on day 223, but ultimately relapsed as detected by plasma molecular disease on day 473, confirmed by PET/CT on day 487 (lead time = 14 days). (C) Patient 48 was diagnosed with stage IVBE DLBCL with leukemic phase and bone marrow involvement. After treatment with R-CHOP, the patient achieved a CR by flow cytometry on day 228 (data not shown), at which time disease was undetectable Ig-HTS. Plasma molecular disease was detected at a level of 71 860 molecules per million genome equivalents on day 493, despite no evidence of disease on peripheral blood flow cytometry on day 493 or PET/CT (data not shown). Plasma Ig-HTS was positive 162 days before the detection of PD by flow cytometry on day 655. (D) Patient 52 was initially expectantly observed with stage IIIA indolent follicular lymphoma, at which point plasma molecular disease was negative despite clear radiographic evidence. This patient then underwent clinical and histological transformation to DLBCL in a single site that coincided with a dramatic spike in plasma molecular disease. He was treated with R-CHOP chemotherapy for DLBCL, with subsequent CR on PET/CT and normalization of plasma molecular disease.
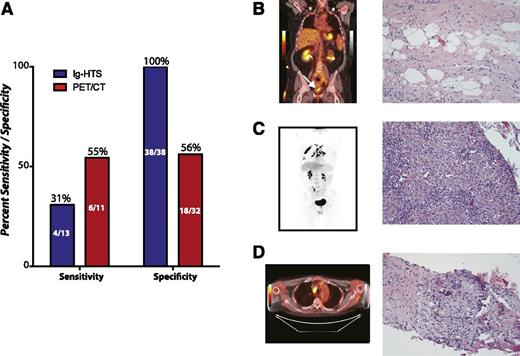
Overall, plasma Ig-HTS had no false positives, yielding a specificity of 100%. Surveillance PET/CT scans from the same 25 patients had a specificity of 56% (Fisher’s exact, P < .0001; Figure 6A; supplemental Table 2). The sensitivity of plasma Ig-HTS to predict a future relapse was only 31% at the individual sample level compared with 55% for PET/CT for the 5 relapsed patients (Fisher’s exact test, P = .4).
Superior specificity of Ig-HTS compared with PET/CT. (A) A comparison of the sensitivity and specificity of Ig-HTS and PET/CT in patients who had both tests performed during clinical surveillance, with the performance assessed by ultimate clinical outcome of the patient. PET/CT can be seen to have superior sensitivity to Ig-HTS, but the specificity of Ig-HTS was 100% and greater than that of PET/CT. (B-D) Examples of false-positive PET/CT scans with concurrent true-negative Ig-HTS. (B) A PET/CT of a patient in remission from stage IV DLBCL showed a positive signal at the site of previous disease (left, white arrow). Biopsy showed unremarkable fibro-adipose tissue. (C) A patient in remission from stage IV DLBCL with a positive PET/CT in the lungs (left) resulting from granulomatous inflammation (right). (D) A PET/CT of a patient in remission from stage IV DLBCL with signal in a para-aortic lymph node (left) that was shown to result from granulomatous lymphadenitis (right).
Superior specificity of Ig-HTS compared with PET/CT. (A) A comparison of the sensitivity and specificity of Ig-HTS and PET/CT in patients who had both tests performed during clinical surveillance, with the performance assessed by ultimate clinical outcome of the patient. PET/CT can be seen to have superior sensitivity to Ig-HTS, but the specificity of Ig-HTS was 100% and greater than that of PET/CT. (B-D) Examples of false-positive PET/CT scans with concurrent true-negative Ig-HTS. (B) A PET/CT of a patient in remission from stage IV DLBCL showed a positive signal at the site of previous disease (left, white arrow). Biopsy showed unremarkable fibro-adipose tissue. (C) A patient in remission from stage IV DLBCL with a positive PET/CT in the lungs (left) resulting from granulomatous inflammation (right). (D) A PET/CT of a patient in remission from stage IV DLBCL with signal in a para-aortic lymph node (left) that was shown to result from granulomatous lymphadenitis (right).
The high specificity of plasma Ig-HTS is further shown in 3 cases where follow-up imaging suggested probable residual disease that was subsequently proven to be falsely positive either by subsequent biopsy and/or serial clinical monitoring and long-term follow-up without interval therapy. This included cases of biopsy-proven fibro-adipose tissue (Figure 6B), granulomatous inflammation (Figure 6C), and granulomatous lymphadenitis (Figure 6D), during which time Ig-HTS monitoring of patient plasma remained negative.
Discussion
Accurate assessment of treatment response and posttreatment surveillance are important for clinical management of DLBCL patients. Although 18FDG PET/CT has become widely used for disease surveillance,4 its use in this setting has had mixed results.7,8,35 The specificity of PET/CT for malignancy is limited, as other conditions associated with increased glycolysis (eg, necrosis, infection, and inflammation) and the use of rituximab36 can confound interpretation. Moreover, the utility of PET/CT must be balanced against exposure to radiation and cost.37 Alternative methods are therefore needed.
The clonally rearranged immunoglobulin genes of mature B-cell malignancies offer specific somatic DNA sequences that can serve as reporters for tumor cells. We investigated the utility of Ig-HTS for monitoring disease from blood samples in consecutive patients with DLBCL compared with current clinical standards. This approach has been reported in a small cohort16 but has not yet been systematically compared with other circulating (ie, LDH) or radiographic (ie, PET/CT) biomarkers of disease. Before monitoring in the peripheral blood, clonotypic sequences must be identified using tumor biopsies; this step was successful in 83% of patients. Although success was higher with fresh/frozen samples compared with FFPE, either source can be used if sufficient DNA is available. As most patients have excisional lymph node biopsies stored in FFPE, this would likely be the source material in real world applications. This rate of successful clonotype identification in tumor biopsies is consistent with other reports38 ; we separately observed a similar frequency of success in a third independent cohort.
In patients with ≥1 identified immunoglobulin reporter, we detected molecular disease within peripheral blood at times of both diagnosis and relapse. Detection of circulating tumor cells has previously been demonstrated in hematologic malignancies, including lymphomas.17-19,39 Similarly, tumor-specific cell-free DNA in hematologic malignancies has been reported,12,40 but the utility of these 2 methods has not been compared in a large cohort. Assessing molecular disease in the plasma and cellular fractions from the same blood sample, we found quantitatively more tumor DNA in the plasma. Additionally, disease detection from plasma was more sensitive than from circulating cells or LDH at time points of overt disease, especially at relapse. Measurement of disease from ctDNA—but not from circulating tumor cells—was significantly correlated with tumor volume as measured by PET/CT scan. This suggests that ctDNA may be both more sensitive and more accurately reflect disease burden than measurement of circulating tumor cells.
The differences between circulating tumor cells and ctDNA described above likely reflect their inherently distinct biology. Although recent reports describe defects in homing pathways in germinal center B-cell like lymphomas (GCB-DLBCL) leading to circulating tumor cells,41 such cells are typically rare in the blood of DLBCL patients when assessed by conventional methods.42,43 Curiously, we detected circulating tumor cells even in patients with radiographically limited disease (stage I/II), suggesting that even at low clinical stage, DLBCL is a systemic disease. Nevertheless, we observed no correlation between the levels of circulating tumor cells and disease burden. In contrast, plasma ctDNA likely originates from tumor cell death by either apoptosis or necrosis, even when occurring in reservoirs of tumor cells that do not circulate.15 This is likely directly proportional to tumor burden and disease biology, including rates of cell proliferation and cell death.
We interrogated molecular disease levels in the plasma of consecutive patients during and after the course of their therapy. We found molecular disease levels reflected changes in tumor burden, including escalating levels with PD, low levels with stable disease, and declining or undetectable levels with CR. At the time of relapse, plasma ctDNA levels performed particularly well, invariably detecting disease at time of radiographic relapse. Furthermore, in select patients, plasma ctDNA levels detected radiographically occult disease before detection by PET/CT. However, this detection of occult disease was not universal, as each of 8 patients who eventually progressed had ≥1 negative ctDNA assay. Therefore, this assay is best used serially for disease monitoring rather than as a one-time test for presence of disease. Exemplifying this, all relapsing patients in the surveillance cohort had detectable disease before or at the time of relapse by plasma Ig-HTS.
This study was also the first to directly compare Ig-HTS to PET/CT imaging for detection and prediction of relapse in DLBCL. Compared with the gold standard of ultimate clinical outcome, surveillance imaging by PET/CT during time points of disease remission showed a higher sensitivity of 55% per scan compared with 31% per test for Ig-HTS (this difference was not statistically significant). Conversely, Ig-HTS showed a superior specificity of 100% compared with 56% for PET/CT, showing that detection of molecular disease by Ig-HTS does not suffer from the false-positive rate of PET/CT.
We conclude that Ig-HTS from plasma is a viable means for disease detection in DLBCL patients. We must note, however, that like any circulating biomarker, ctDNA detection by Ig-HTS cannot identify the site or extent of disease relapse. Therefore, this assay will likely be complementary to existing radiographic methods of surveillance. Although Ig-HTS cannot be used in patients whose tumor clonotype is unidentified, it is possible that this may be overcome by routinely obtaining higher quality tumor specimens. Ig-HTS performed well in detecting disease progression but showed a lower sensitivity for detecting disease associated with low tumor burden. More frequent monitoring may improve the sensitivity for low burden disease.
As monitoring with serial imaging in disease surveillance remains of limited benefit, surveillance with Ig-HTS has potential clinical utility in this setting. However, the specificity of this test warrants further prospective investigation, in combination with imaging modalities to lower false-positive rates or as a stand-alone assay for monitoring patients with DLBCL.
The online version of this article contains a data supplement.
There is an Inside Blood Commentary on this article in this issue.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank the patients and families who participated in this study for their generosity and support, as well as all the nurses, patient coordinators, and staff at Stanford University Medical Center and the ALLG who made this work possible.
This work was supported by a Stanford Cancer Institute’s Innovation Fund Award (to A.A.A.), Gabrielle’s Angel Foundation (to A.A.A.), and the Damon Runyon Cancer Research Foundation (to A.A.A.). M.R.G. is a Special Fellow of the Leukemia and Lymphoma Society. M.D. is supported by the National Institutes of Health, National Cancer Institute Director’s New Innovator Award Program grant 1-DP2-CA186569. A.A.A. and M.D. are Doris Duke Charitable Foundation Clinical Investigators.
Authorship
Contribution: M.R.G., D.M.K., and A.A.A. designed experiments, analyzed data, and wrote the manuscript; S.V.B. designed experiments and analyzed data; C.L.L., C.A.K., K.A.K., and M.F. performed experiments and analyzed data; F.S., K.T., C.G., C.K., S.K.,B.V., J.C., K.S.C., D.M., R.H.A., R.L., R.J.H., R.S.O., M.H., and M.K.G. participated in sample collection and annotation; M.D. designed experiments; and all authors revised the manuscript.
Conflict-of-interest disclosure: K.A.K. and M.F. are employees and holders of equity in Sequenta, Inc. The remaining authors declare no competing financial interests.
The current affiliation for M.R.G. is Eppley Institute for Research in Cancer and Allied Diseases, University of Nebraska Medical Center, Omaha, NE.
Correspondence: Ash A. Alizadeh, Division of Oncology, Department of Medicine, Stanford University School of Medicine, Lorry Lokey Building, SIM1 Room G2120B, 265 Campus Dr, Stanford, CA 94305; e-mail: arasha@stanford.edu.
References
Author notes
D.M.K. and M.R.G. contributed equally to this work.