Key Points
IL-3, IL-5, and GM-CSF promote eosinophil survival by NF-κB–induced upregulation of Bcl-xL, which can be blocked by specific inhibitors.
Specific and constitutive deletion of the inhibitor of NF-κB (IκBα) in eosinophils in vivo reduced apoptosis during helminth infection.
Abstract
Eosinophils are associated with type 2 immune responses to allergens and helminths. They release various proinflammatory mediators and toxic proteins on activation and are therefore considered proinflammatory effector cells. Eosinophilia is promoted by the cytokines interleukin (IL)-3, IL-5, and granulocyte macrophage–colony-stimulating factor (GM-CSF) and can result from enhanced de novo production or reduced apoptosis. In this study, we show that only IL-5 induces differentiation of eosinophils from bone marrow precursors, whereas IL-5, GM-CSF, and to a lesser extent IL-3 promote survival of mature eosinophils. The receptors for these cytokines use the common β chain, which serves as the main signaling unit linked to signal transducer and activator of transcription 5, p38 mitogen-activated protein kinase, and nuclear factor (NF)-κB pathways. Inhibition of NF-κB induced apoptosis of in vitro cultured eosinophils. Selective deletion of IκBα in vivo resulted in enhanced expression of Bcl-xL and reduced apoptosis during helminth infection. Retroviral overexpression of Bcl-xL promoted survival, whereas pharmacologic inhibition of Bcl-xL in murine or human eosinophils induced rapid apoptosis. These results suggest that therapeutic strategies targeting Bcl-xL in eosinophils could improve health conditions in allergic inflammatory diseases.
Introduction
Eosinophils constitute about 1% to 4% of leukocytes in the peripheral blood of healthy individuals. They are multifaceted cells of the innate immune system and generally associated with type 2 immune responses against allergens and helminths whereby they mediate proinflammatory and protective functions, respectively. Moreover, eosinophils were reported to support survival of plasma cells in the bone marrow,1 promote differentiation of alternatively activated macrophages in fat tissue,2,3 induce immunoglobulin A secretion in the gut,4 and facilitate liver regeneration.5
Eosinophils develop in the bone marrow from eosinophil lineage–committed progenitors, which express the interleukin (IL)-5 receptor and the transcription factors Gata-1 and c/EBPα.6,7 Mice with a point mutation in the promoter region of Gata-1 lack the eosinophil lineage.8 Eosinophils upregulate expression of the chemokine receptor CCR3 and L-selectin (CD62L) before they leave the bone marrow as mature effector cells with a lifespan of ∼1 to 2 days in the spleen of normal mice.9,10 However, eosinophils have a longer lifespan in the intestine, thymus, and uterus, indicating that the local tissue milieu modulates the survival of eosinophils.11 We have previously shown that infection of mice with the gastrointestinal helminth Nippostrongylus brasiliensis causes massive eosinophilia in lung and peritoneum, which is not due to increased de novo production of eosinophils but is caused by a reduced rate of apoptosis.9
The cytokines IL-3, IL-5, and granulocyte macrophage–colony-stimulating factor (GM-CSF) can promote expansion of the eosinophil population, although it remains unclear whether this reflects primarily enhanced de novo generation of eosinophils or inhibition of apoptosis.12 IL-5 appears to be rather specific for eosinophils, whereas IL-3 and GM-CSF also induce maturation of other myeloid cells and all 3 cytokines can be expressed in eosinophils themselves.13,14 IL-5 is mainly produced by T helper 2 cells and type 2 innate lymphoid cells, as recently revealed by analyzing fluorescent IL-5 reporter mice.15 IL-5 transgenic mice show massive systemic eosinophilia.16 IL-5–depleting antibodies reduce eosinophilia in helminth-infected mice and in the blood and sputum of allergic asthma patients.17,18 However, IL-5–deficient and IL-5 receptor–deficient mice have almost normal numbers of eosinophils under steady-state conditions, indicating that IL-5 is not essential for eosinophil development.19,20 The receptors for IL-3, IL-5, and GM-CSF are composed of a ligand-specific α chain and the common β (βc) chain, which is the main signaling unit.21 Structural analysis revealed that these receptors form large complexes.22 The cytoplasmic tail of the IL-5 receptor α chain binds syntenin, which stabilizes oligomeric complexes of the IL-5 receptor and promotes the signaling output.23 Signal transduction from the βc chain involves several pathways that lead to activation of signal transducer and activator of transcription (STAT)5, p38 mitogen-activated protein kinase (MAPK), phosphatidylinositol 3-kinase (PI3K), and nuclear factor (NF)-κB. Lyn and Syk kinases have been shown to associate with the βc chain and they appear to be critical for IL-5– or GM-CSF–induced inhibition of apoptosis in human eosinophils.24
IL-5 can protect human eosinophils from spontaneous, tumor necrosis factor (TNF)-α–induced, or dexamethasone-induced apoptosis.25,26 Interestingly, glucocorticoids can enhance apoptosis at low IL-5 concentrations but promote survival when IL-5 levels are increased.27 The regulation of eosinophil survival is only partially understood and may be controlled by many different pathways. IL-5 signaling was shown to prevent activation of caspase-8 and caspase-3, whereas antiapoptotic Bcl-2 or Bcl-xL proteins were not detected in human eosinophils.25 However, it was also reported that IL-5 induced Bcl-2 messenger (m)RNA and protein expression in human eosinophils in vitro.28 Others demonstrated that Bcl-xL rather than Bcl-2 is induced by IL-5 or GM-CSF in ex vivo isolated human eosinophils and that antisense oligonucleotides for Bcl-xL reduced the survival of eosinophils in vitro.29
Here, we demonstrate that IL-5, but not IL-3 or GM-CSF, induced eosinophil development from bone marrow precursor cells. In contrast, apoptosis of mature eosinophils caused by growth factor withdrawal was inhibited by all 3 cytokines, albeit with different efficiency. Apoptosis could be promoted by inhibition of NF-κB but not by blocking p38-MAPK or STAT5. We created a new eosinophil-specific Cre-deleter mouse and demonstrated that selective deletion of IκBα in eosinophils increased the expression of Bcl-xL and promoted their survival in vivo. Bcl-xL retroviral transduction of eosinophil cultures prevented apoptosis in IL-5–deprived eosinophils, whereas small molecular inhibitors of Bcl-xL caused rapid apoptosis in the presence of IL-5. These findings indicate that eosinophil survival is largely regulated by βc chain–induced NF-κB activation and subsequent induction of Bcl-xL.
Materials and methods
Human eosinophils
Human eosinophils were prepared from leukoreduction system chambers obtained from the Department of Transfusion Medicine, University Hospital Erlangen. Chambers were flushed out with phosphate-buffered saline (PBS) containing heparin. The cell suspension was layered on Ficoll (1.077 g/mL; Biochrom, Berlin, Germany) and centrifuged at 1000g for 30 minutes at room temperature. The pellet containing erythrocytes and granulocytes was harvested and subjected to hypotonic lysis. Untouched eosinophils were isolated with a purity of >90% by magnetic cell sorting (Miltenyi Biotec, Bergisch Gladbach, Germany). Finally, eosinophils were adjusted to 106 cells per milliliter and cultured in the presence of 10 ng/mL of human IL-5 (R&D Systems, Minneapolis, MN). The experiments were approved by the Ethics Committee of the University Hospital Erlangen (approval no. 224_14B), and each participant gave written informed consent.
Mice
EpxCre mice were generated by bacterial artificial chromosome (BAC) recombineering. BAC clone 193P5 (encoding the eosinophil peroxidase gene epx) of the mouse genomic BAC library RPCI-23 was purchased from Invitrogen (Carlsbad, CA). An eYFP-Cre fusion protein cassette was inserted directly after the start codon of epx by homologous recombination in bacteria. After linearization with NotI, BAC DNA was purified by phenol/chloroform extraction and injected into oocytes. Cre activity is preserved in EpxCre mice, whereas the eYFP fluorescence is lost, which might be due to an altered tertiary structure of eYFP in this eYFP-Cre fusion protein. R-RFP mice31 were kindly provided by Hans-Jörg Fehling at the University of Ulm (Ulm, Germany), and R-eYFP mice32 were originally obtained from The Jackson Laboratory. Both mice encode fluorescent proteins behind a loxP-flanked Stop-cassette in the ubiquitously expressed Rosa26 locus. IκBαF/F mice have been described.33 They encode loxP-flanked IκBα gene. IL-5 transgenic mice (strain NJ.1638) express IL-5 in T cells under control of regulatory elements of the CD3δ gene.16 Mice were used between 6 and 12 weeks of age, and animal experiments were approved by the local government.
N brasiliensis infection
Third-stage larvae of N brasiliensis were washed in sterile 0.9% saline (37°C) and injected subcutaneously (500 organisms) into mice. Mice were provided water containing antibiotics (2 g/L of neomycin sulfate and 100 mg/L of polymyxin B sulfate; Sigma-Aldrich, St Louis, MO) for the first 5 days after infection. Mice were analyzed on day 9 after infection.
In vitro culture of eosinophils
Eosinophils were generated from bone marrow as described previously.34 In brief, bone marrow was flushed out from femur and tibia. Single-cell suspension was generated and erythrocytes were removed by hypotonic lysis. Bone marrow cells were placed in bone marrow medium (BM-medium) (RPMI 1640; Pan-Biotech, Aidenbach, Germany) containing 20% fetal bovine serum, 55 μM β-mercaptoethanol, 10 mM nonessential amino acids, 1 mM sodium pyruvate, 100 IU/mL of penicillin, 100 μg/mL of streptomycin, 2 mM glutamine (all from Life Technologies, Darmstadt, Germany), and 25 mM N-2-hydroxyethylpiperazine-N′-2-ethanesulfonic acid (HEPES) buffer. Cells were stimulated with 100 ng/mL of stem cell factor (SCF) and 100 ng/mL of fms-like tyrosine kinase 3 ligand (FLT3L) (both from PeproTech, Rocky Hill, NJ) for 4 days. From day 4 on, cells were kept in medium supplemented with 10 ng/mL of recombinant murine IL-5 (R&D Systems). At day 14, the cultures contained >90% bone marrow–derived eosinophils (BMDEs), indicated by high side scatter profile and expression of Siglec-F. In some experiments, BMDEs were stimulated with 10 ng/mL of GM-CSF or IL-3 (both from ImmunoTools, Friesoythe, Germany).
Protein inhibitors
BMDEs were seeded at 106 cells per milliliter in fresh BM-medium and incubated with 5 μM STAT5 inhibitor (CAS 285986-31-4; Merck Millipore, Nottingham, United Kingdom), 5 μM p38-MAPK inhibitor (SB 203580; Sigma-Aldrich), or 5 μM NF-κB inhibitor (Bay11-7085; Merck Millipore). After 2 hours of preincubation, cells were stimulated with 10 ng/mL of IL-5. Bcl-xL inhibitors Z-36, BHI-1, and HA14.1 (Enzo Life Sciences, Farmingdale, NY) were applied as a 50 μM solution in BM-medium containing 10 ng/mL of IL-5.
Flow cytometry
Single-cell suspensions were generated from lymph nodes, spleen, or PBS-perfused lung samples that had been cut into small pieces and mechanically dispersed using a 70-μm nylon strainer (BD Biosciences, San Jose, CA). Samples were washed once in fluorescence-activated cell sorter buffer (PBS, 2% fetal bovine serum, and 1 mg/mL of sodium azide), incubated with anti-CD16/CD32 blocking monoclonal antibody (clone 2.4G2) for 5 minutes at room temperature, and stained with diluted monoclonal antibody mixtures as described in supplemental Materials and Methods, available on the Blood Web site.
Samples were acquired on a FACSCanto II instrument (BD Biosciences) and analyzed by FlowJo software (Tree Star, Ashland, OR).
Retroviral transduction
Retroviral vector MIG-bcl-xL and empty vector MSCV-IRES-GFP were obtained from Addgene, Cambridge, MA. Retrovirus-containing supernatants were produced and collected from transfected Phoenix E cells using standard procedures. Bone marrow cells were stimulated for 2 days in BM-medium supplemented with SCF and FLT3L (both 100 ng/mL). Cells (3 × 106) were mixed with Phoenix cell supernatant containing retroviral particles supplemented with 4 μg/mL of polybrene. Spin transduction was performed by centrifugation at 1000g for 2 hours at 25°C. Cells were left in BM-medium containing SCF and FLT3L (both 100 ng/mL) for another 2 days. From day 4 on, SCF/FLT3L was replaced by 10 ng/mL of IL-5 as described earlier.
Western blot
Eosinophils were isolated from peritoneal lavage of IL-5 transgenic mice.16 The NE-PER Nuclear and Cytoplasmic Extraction Kit (Thermo Fisher Scientific, Waltham, MA) was used to generate cytosolic and nuclear protein fractions for western blot analysis. All buffers were supplemented with Halt Protease and Phosphatase Inhibitor Cocktail (Thermo Fisher Scientific). Protein fractions were separated on a 10% sodium dodecyl sulfate gel followed by transfer on a polyvinylidene fluoride membrane. Membranes were blocked with 3% bovine serum albumin for 1 hour at room temperature and incubated with the following primary antibodies: rabbit-anti-p65 (Santa Cruz Biotechnology), rabbit-anti-glyceraldehyde-3-phosphate dehydrogenase, and rabbit-anti-histone H2B (both from Cell Signaling Technology, Danvers, MA) overnight. After extensive washing, the membrane was incubated with the TrueBlot HRP–coupled donkey-anti-rabbit secondary antibody (Rockland Immunochemicals, Limerick, PA). Lastly, enhanced chemiluminescence substrate (PerkinElmer, Waltham, MA) was added, and signals were detected with a ChemoCam-Imager (Intas Science Imaging, Göttingen, Germany).
RT-PCR analysis
Bcl-2 and bcl-xL mRNA levels were analyzed in ex vivo isolated eosinophils (Siglec-F+RFP+SSChi) sort-purified with a purity of >95% from spleens of EpxCre/R-RFP/IκBF/F and EpxCre/R-RFP/IkBF/+ mice using the FACSAria instrument (BD Biosciences). Bcl-xL mRNA levels were further analyzed in BMDEs cultured for 16 hours without cytokines and then cultured for 3 hours in 10 ng/mL of IL-5 or GM-CSF in the presence or absence of 5 μM NF-κB inhibitor Bay11-7085. RNA was isolated using the RNeasy kit (QIAGEN, Hilden, Germany) and transcribed into complementary DNA with the SuperScript III Reverse Transcriptase and oligo-(dT)-primers (Invitrogen). The primer sequences are given in supplemental Materials and Methods. Analysis was performed on 7900HT (Applied Biosystems) and CFX Connect (Bio-Rad Laboratories) real-time polymerase chain reaction (RT-PCR) machines.
Results
IL-5, but not IL-3 or GM-CSF, promotes eosinophil development in vitro
IL-3, IL-5, and GM-CSF have been shown to increase the number of eosinophils, which could be due to enhanced de novo production from eosinophil-committed precursors or inhibition of apoptosis in mature eosinophils.35 To distinguish between these 2 possibilities, we first compared the efficiency and kinetics of eosinophil differentiation from murine bone marrow cells in response to recombinant IL-3, IL-5, and GM-CSF. Total bone marrow cells were first cultured for 2 days in medium containing FLT3L and SCF to increase the number of hematopoietic stem cells as described previously.34 Then, medium containing either IL-3, IL-5, or GM-CSF was used to culture and differentiate eosinophils as determined by flow cytometry on the following days. The frequency of eosinophils (SSChiSiglec-F+) was ∼1% at the onset of the culture and increased in the presence of IL-5 to over 80% by day 21, although the maximal number of eosinophils was already reached at day 12 and was maintained thereafter (Figure 1A-C). Unexpectedly, IL-3 and GM-CSF were not able to induce eosinophil development under these conditions (Figure 1A-C and supplemental Figure 1). Because both cytokines have been proposed to promote eosinophilia, we next analyzed their activity in prevention of spontaneous apoptosis caused by IL-5 deprivation. For this analysis, we generated BMDEs as described earlier and changed the culture conditions on day 12 to medium that contained no additional cytokines or medium supplemented with either IL-3, IL-5, or GM-CSF. Apoptotic eosinophils were determined over the next 4 days by flow cytometry after staining with anti-Siglec-F, annexin V, and propidium iodide. After 3 days in the absence of any added cytokines, <10% viable eosinophils were left. IL-5 and GM-CSF completely inhibited apoptosis, whereas IL-3 was less efficient (Figure 1D-E). On the basis of these experiments, we conclude that IL-3 and GM-CSF contribute to eosinophilia largely by inhibiting apoptosis rather than by promoting de novo differentiation.
Role of IL-5, IL-3, and GM-CSF in eosinophil development and survival. Bone marrow cells were initially stimulated with SCF/FLT3L for 4 days and further cultured in the presence of IL-5, IL-3, or GM-CSF. Cells were analyzed at indicated time points by flow cytometry. (A) Dot plots show the frequency of eosinophils (Siglec-F+SSChi) after 21 days in culture. (B) Eosinophil numbers are shown as mean + SD (n = 3). (C) Frequency of eosinophils (Siglec-F+SSChi) is shown as mean + SEM. Mature eosinophils were washed on day 12 after onset of culture and further cultured in the absence or presence of the indicated cytokines. Cells were harvested at the indicated time points and subjected to annexin V/propidium iodide (PI) staining for detection of apoptotic cells by flow cytometry. (D) Dot plots show representative stainings after 96 hours of culture. (E) Frequency of viable eosinophils (annexin V−PI−) shown as mean + SD (n = 3). SD, standard deviation; SEM, standard error of the mean.
Role of IL-5, IL-3, and GM-CSF in eosinophil development and survival. Bone marrow cells were initially stimulated with SCF/FLT3L for 4 days and further cultured in the presence of IL-5, IL-3, or GM-CSF. Cells were analyzed at indicated time points by flow cytometry. (A) Dot plots show the frequency of eosinophils (Siglec-F+SSChi) after 21 days in culture. (B) Eosinophil numbers are shown as mean + SD (n = 3). (C) Frequency of eosinophils (Siglec-F+SSChi) is shown as mean + SEM. Mature eosinophils were washed on day 12 after onset of culture and further cultured in the absence or presence of the indicated cytokines. Cells were harvested at the indicated time points and subjected to annexin V/propidium iodide (PI) staining for detection of apoptotic cells by flow cytometry. (D) Dot plots show representative stainings after 96 hours of culture. (E) Frequency of viable eosinophils (annexin V−PI−) shown as mean + SD (n = 3). SD, standard deviation; SEM, standard error of the mean.
IL-5–mediated survival operates via induction of the classical NF-κB pathway
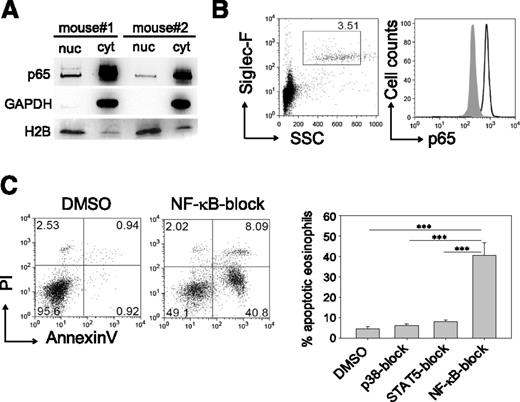
Next, we sought to identify the critical signaling pathways downstream of the βc chain, which is the signal-transducing unit of the IL-3, IL-5, and GM-CSF receptors. p65 (RelA), a component of the classical NF-κB transcription factor, was detected by western blot analysis in the cytoplasm and nucleus of eosinophils isolated from the peritoneal cavity of IL-5 transgenic mice (Figure 2A). Flow-cytometric analysis of peripheral blood from wild-type mice also revealed prominent expression of p65 in eosinophils (Figure 2B). To determine whether NF-κB signaling was required for IL-5–mediated protection against apoptosis, we added the NF-κB inhibitor Bay11-7085 at 5 μM to BMDE cultures on day 14 and observed a fourfold increased apoptosis rate compared to controls. In contrast, the block of p38 or STAT5 by the same concentration of specific inhibitors (SB203580 and CAS 285986-31-4, respectively) affected the proliferative response but did not result in apoptosis (Figure 2C and supplemental Figure 2). This finding is consistent with the observation that dominant-negative STAT5 inhibits proliferation but does not cause apoptosis of eosinophils.36 However, we found that partial apoptosis could be induced with high (20 μM) concentrations of the STAT5 inhibitor, indicating a minor contribution from the STAT5 pathway (supplemental Figure 2). Taken together, the antiapoptotic signal from the βc chain is largely mediated by NF-κB–regulated genes.
NF-κB inhibition impairs survival of mature eosinophils. (A) Western blot of p65 from cytoplasmic (cyt) and nuclear (nuc) protein fractions of peritoneal eosinophils isolated from 2 IL-5 transgenic mice.16 Glyceraldehyde-3-phosphate dehydrogenase (GAPDH) and histone H2B were used as cytoplasmic and nuclear marker proteins, respectively. (B) Blood eosinophils (Siglec-F+SSChigh) from wild-type BALB/c mice were stained intracellularly for p65 (black outline) and an isotype control antibody (gray shade). Plots are representative of 3 independently analyzed mice. (C) Eosinophils were pretreated with inhibitors for STAT5, p38, or NF-κB and then cultured in the presence of IL-5 for 72 hours. Dot plots show representative annexin V/propidium iodide (PI) staining of NF-κB–exposed eosinophils vs dimethylsulfoxide (DMSO) vehicle control. Bar graphs show mean + SEM of apoptotic eosinophils pooled from 3 independent experiments (n = 8-9; ***P < .0001).
NF-κB inhibition impairs survival of mature eosinophils. (A) Western blot of p65 from cytoplasmic (cyt) and nuclear (nuc) protein fractions of peritoneal eosinophils isolated from 2 IL-5 transgenic mice.16 Glyceraldehyde-3-phosphate dehydrogenase (GAPDH) and histone H2B were used as cytoplasmic and nuclear marker proteins, respectively. (B) Blood eosinophils (Siglec-F+SSChigh) from wild-type BALB/c mice were stained intracellularly for p65 (black outline) and an isotype control antibody (gray shade). Plots are representative of 3 independently analyzed mice. (C) Eosinophils were pretreated with inhibitors for STAT5, p38, or NF-κB and then cultured in the presence of IL-5 for 72 hours. Dot plots show representative annexin V/propidium iodide (PI) staining of NF-κB–exposed eosinophils vs dimethylsulfoxide (DMSO) vehicle control. Bar graphs show mean + SEM of apoptotic eosinophils pooled from 3 independent experiments (n = 8-9; ***P < .0001).
Selective deletion of IκBα in eosinophils promotes their survival in response to helminth infection
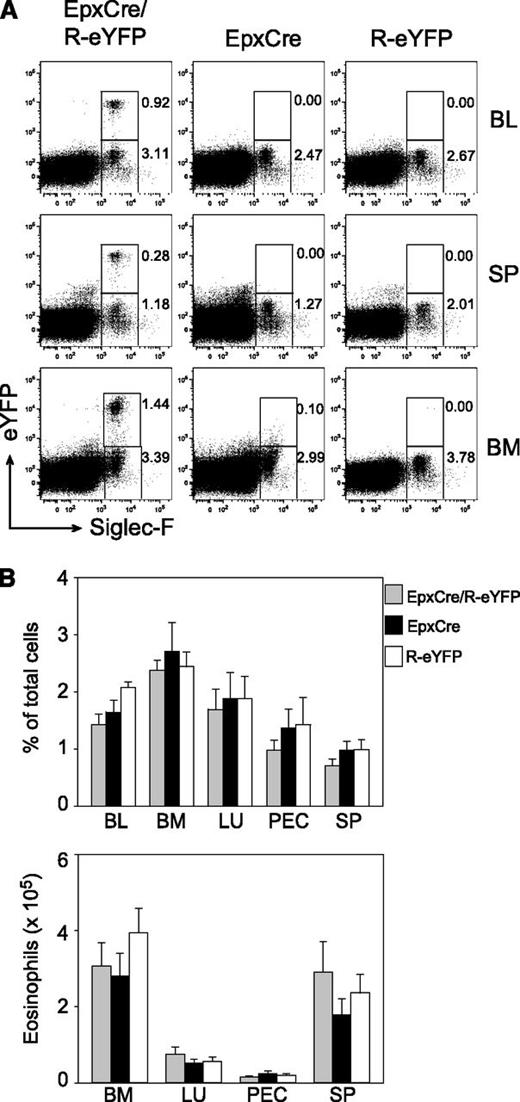
To further substantiate the critical role of NF-κB–regulated genes for eosinophil homeostasis in vivo, we created a new mouse model in which the inhibitor of NF-κB (IκBα) could be selectively deleted in eosinophils. Therefore, we generated BAC transgenic mice that express the Cre recombinase under control of regulatory elements of the eosinophil peroxidase gene epx (EpxCre mice). In contrast to basophils and mast cells in which high-level Cre expression has been shown to cause cell death due to Cre toxicity,37,38 we observed normal eosinophil frequencies and total numbers in blood, spleen, bone marrow, lung, and peritoneal cavity of EpxCre mice (Figure 3). By crossing EpxCre mice to R-eYFP mice, we found that Cre was expressed only in eosinophils, although only 20% to 30% of all eosinophils were marked with eYFP (Figure 3). This finding indicates that Cre recombination is not very efficient in eosinophils of Epx-Cre mice. However, YFP+ and YFP− eosinophils showed comparable nuclear morphology; identical surface expression of the eotaxin receptor CCR3, LPAM-1 (α4β7 integrin), and CD44; and the same mRNA expression levels for major basic protein and IL-13 (supplemental Figure 3). The reason for only partial Cre activity remains unclear but could be due to relatively low expression levels of Cre. Efficient Cre recombination has recently been demonstrated with another eosinophil-specific Cre mouse, which demonstrates that eosinophils are not resistant to Cre-mediated recombination per se.39 On occasion, we also observed ubiquitous Cre expression, indicating that transient expression in the germline can occur. Nevertheless, these mice appeared useful to study the role of NF-κB signaling in eosinophils in vivo.
EpxCre/R-eYFP mice selectively mark eosinophils. (A) Flow-cytometric analysis of Siglec-F+ eosinophils in blood, spleen, and bone marrow from naive EpxCre/R-eYFP, EpxCre, and R-eYFP mice. Numbers indicate frequencies of gated cells. (B) Frequencies (upper graph) and total number (lower graph) of eosinophils in blood, bone marrow, lung, peritoneum, and spleen from naive EpxCre/R-eYFP (gray bars), EpxCre (black bars), and R-eYFP (white bars) mice. Bar graphs show the mean + SEM (n = 9) from 3 independent experiments. BL, blood; BM, bone marrow; LU, lung; PEC, peritoneal cavity; SP, spleen.
EpxCre/R-eYFP mice selectively mark eosinophils. (A) Flow-cytometric analysis of Siglec-F+ eosinophils in blood, spleen, and bone marrow from naive EpxCre/R-eYFP, EpxCre, and R-eYFP mice. Numbers indicate frequencies of gated cells. (B) Frequencies (upper graph) and total number (lower graph) of eosinophils in blood, bone marrow, lung, peritoneum, and spleen from naive EpxCre/R-eYFP (gray bars), EpxCre (black bars), and R-eYFP (white bars) mice. Bar graphs show the mean + SEM (n = 9) from 3 independent experiments. BL, blood; BM, bone marrow; LU, lung; PEC, peritoneal cavity; SP, spleen.
We crossed EpxCre mice to IκBα-floxed mice to specifically delete IκBα in eosinophils. These mice were further crossed to R-RFP mice to mark the eosinophils with Cre activity. We observed the same frequency of RFP+ eosinophils in lung, spleen, peritoneum, small intestine, and bone marrow of EpxCre/R-RFP/IκBαF/F mice and EpxCre/R-RFP/IκBαF/+ controls, indicating that constitutive activation of the NF-κB pathway under steady-state conditions does not promote eosinophilia (Figure 4A). Next, we infected mice with the helminth N brasiliensis to determine whether NF-κB–regulated genes promote eosinophilia under inflammatory conditions. Here, we observed that the number of eosinophils with constitutively active NF-κB signaling increased in all examined tissues as compared to eosinophils from control mice (Figure 4A). In the spleen, we observed a trend to more total eosinophils and more RFP+ eosinophils in EpxCre/R-RFP/IκBαF/F mice as compared to EpxCre/R-RFP/IκBαF/+ mice (Figure 4B). Using downregulation of CD62L as an activation marker, we found that RFP+ and RFP− eosinophils were similarly activated in both strains of mice (Figure 4C).
Constitutively active NF-κB in eosinophils increases their survival during N brasiliensis infection. (A) Percentage of RFP+ cells among eosinophils (Siglec-F+SSChi) in naive (left) and day 9 N brasiliensis–infected (right) EpxCre/R-RFP/IκBF/+ (gray bars) and EpxCre/R-RFP/IκBF/F (black bars) mice. Bars show the mean + SEM from 6 to 9 mice per group from pooled experiments. *P < .05; **P < .01. (B) Number of total (left) and RFP+ (right) eosinophils in the spleen of EpxCre/R-RFP/IκBF/+ (gray bars) and EpxCre/R-RFP/IκBF/F (black bars) mice on day 10 after N brasiliensis infection. Bars show the mean + SEM from 3 to 6 mice per group from 3 pooled experiments. (C) Expression of CD62L on RFP+ and RFP− eosinophils from indicated mice. Dot plots are representative of 3 analyzed mice per group. (D) Dot plots show frequencies of late apoptotic eosinophils (Siglec-F+SSChiRFP+annexinV+PI+) in naive and day 9–infected EpxCre/R-RFP/IκBF/+ (gray bars) and EpxCre/R-RFP/IκBF/F (black bars) mice. Bar graph shows the mean + SEM of late apoptotic Siglec-FhiRFP+ eosinophils from 6 naive and infected mice per group. *P < .05. BAL, broncho-alveolar lavage; SI, small intestine.
Constitutively active NF-κB in eosinophils increases their survival during N brasiliensis infection. (A) Percentage of RFP+ cells among eosinophils (Siglec-F+SSChi) in naive (left) and day 9 N brasiliensis–infected (right) EpxCre/R-RFP/IκBF/+ (gray bars) and EpxCre/R-RFP/IκBF/F (black bars) mice. Bars show the mean + SEM from 6 to 9 mice per group from pooled experiments. *P < .05; **P < .01. (B) Number of total (left) and RFP+ (right) eosinophils in the spleen of EpxCre/R-RFP/IκBF/+ (gray bars) and EpxCre/R-RFP/IκBF/F (black bars) mice on day 10 after N brasiliensis infection. Bars show the mean + SEM from 3 to 6 mice per group from 3 pooled experiments. (C) Expression of CD62L on RFP+ and RFP− eosinophils from indicated mice. Dot plots are representative of 3 analyzed mice per group. (D) Dot plots show frequencies of late apoptotic eosinophils (Siglec-F+SSChiRFP+annexinV+PI+) in naive and day 9–infected EpxCre/R-RFP/IκBF/+ (gray bars) and EpxCre/R-RFP/IκBF/F (black bars) mice. Bar graph shows the mean + SEM of late apoptotic Siglec-FhiRFP+ eosinophils from 6 naive and infected mice per group. *P < .05. BAL, broncho-alveolar lavage; SI, small intestine.
To determine whether the increase of RFP+ eosinophils in infected EpxCre/R-RFP/IκBαF/F mice reflects reduced apoptosis, we determined the frequency of ex vivo isolated annexin V–positive eosinophils. Indeed, we observed significantly reduced apoptotic eosinophils in infected EpxCre/R-RFP/IκBαF/F mice as compared to eosinophils from control mice, whereas no differences were found for eosinophils isolated from naive mice (Figure 4D).
NF-κB induces expression of Bcl-xL, which is required and sufficient to protect eosinophils from apoptosis caused by growth factor deprivation
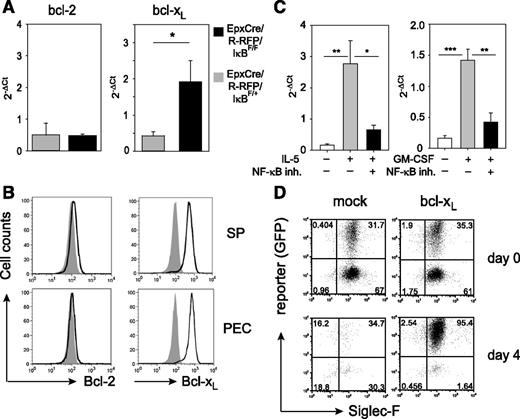
The reduced apoptosis in eosinophils from EpxCre/R-RFP/IκBαF/F mice indicated that NF-κB–regulated genes are critical for prevention of cell death. One antiapoptotic target gene of NF-κB is bcl-x, a member of the Bcl-2 family that encodes the proapoptotic Bcl-xS and the antiapoptotic Bcl-xL proteins. RT-PCR analysis revealed that eosinophils from N brasiliensis–infected EpxCre/R-RFP/IκBαF/F mice expressed higher levels of bcl-xL mRNA as compared with control mice, whereas expression levels of bcl-2 mRNA were similar (Figure 5A). Consistent with a previous report from human eosinophils,29 murine eosinophils isolated from IL-5 transgenic mice expressed Bcl-xL, but not Bcl-2, on the protein level as determined by intracellular flow-cytometric analysis (Figure 5B). Furthermore, inhibition of NF-κB blocked the IL-5– or GM-CSF–induced upregulation of bcl-xL mRNA in BMDEs, which demonstrates the requirement of NF-κB signaling for cytokine-induced Bcl-xL expression (Figure 5C).
Bcl-xL promotes survival of eosinophils with constitutively active NF-κB. (A) RNA was isolated from sort-purified eosinophils of EpxCre/R-RFP/IκBF/+ (gray bars) and EpxCre/R-RFP/IκBF/F (black bars) mice. Expression of bcl-2 and bcl-xL was determined by RT-qPCR. Porphobilinogen deaminase was used as housekeeping control. Bar graphs show the mean + SEM of 4 mice per group. *P < .05. (B) Eosinophils (Siglec-F+SSChi) from spleen and peritoneum of IL-5 transgenic mice were stained intracellularly for expression of Bcl-2 and Bcl-xL (black outline) and compared to isotype control staining (gray shade). (C) RT-qPCR for bcl-xL in BMDEs stimulated for 3 hours with IL-5 (left graph) or GM-CSF (right graph) in the presence or absence of the NF-κB inhibitor (inh.) Bay11-7085. Bar graphs show the mean + SEM of 6 separate samples per group from 2 independent experiments. *P < .05; **P < .01; ***P < .001. (D) BMDEs from wild-type mice were retrovirally transduced with either mock or bcl-xL–expressing retroviruses. Dot plots show frequencies of transduced eosinophils (reporter+Siglec-F+) before (day 0) and after (day 4) IL-5 withdrawal. Similar results were obtained in 2 other independent experiments. 2−ΔCt, the difference of Ct values obtained by RT-PCR analysis between the gene of interest and the house-keeping gene; GFP, green fluorescent protein; qPCR, quantitative PCR.
Bcl-xL promotes survival of eosinophils with constitutively active NF-κB. (A) RNA was isolated from sort-purified eosinophils of EpxCre/R-RFP/IκBF/+ (gray bars) and EpxCre/R-RFP/IκBF/F (black bars) mice. Expression of bcl-2 and bcl-xL was determined by RT-qPCR. Porphobilinogen deaminase was used as housekeeping control. Bar graphs show the mean + SEM of 4 mice per group. *P < .05. (B) Eosinophils (Siglec-F+SSChi) from spleen and peritoneum of IL-5 transgenic mice were stained intracellularly for expression of Bcl-2 and Bcl-xL (black outline) and compared to isotype control staining (gray shade). (C) RT-qPCR for bcl-xL in BMDEs stimulated for 3 hours with IL-5 (left graph) or GM-CSF (right graph) in the presence or absence of the NF-κB inhibitor (inh.) Bay11-7085. Bar graphs show the mean + SEM of 6 separate samples per group from 2 independent experiments. *P < .05; **P < .01; ***P < .001. (D) BMDEs from wild-type mice were retrovirally transduced with either mock or bcl-xL–expressing retroviruses. Dot plots show frequencies of transduced eosinophils (reporter+Siglec-F+) before (day 0) and after (day 4) IL-5 withdrawal. Similar results were obtained in 2 other independent experiments. 2−ΔCt, the difference of Ct values obtained by RT-PCR analysis between the gene of interest and the house-keeping gene; GFP, green fluorescent protein; qPCR, quantitative PCR.
To further investigate whether Bcl-xL is sufficient for prevention of apoptosis after growth factor deprivation, we transduced BMDEs with a bcl-xL–encoding retroviral vector and removed IL-5 from the culture medium. The transduction efficiency of mock vector and bcl-xL vector was comparable. Within 4 days, all mock vector–transduced eosinophils died, and only eosinophils that expressed Bcl-xL, as indicated by the reporter signal, survived (Figure 5D). This result demonstrates that Bcl-xL is required and sufficient to prevent apoptosis caused by growth factor deprivation.
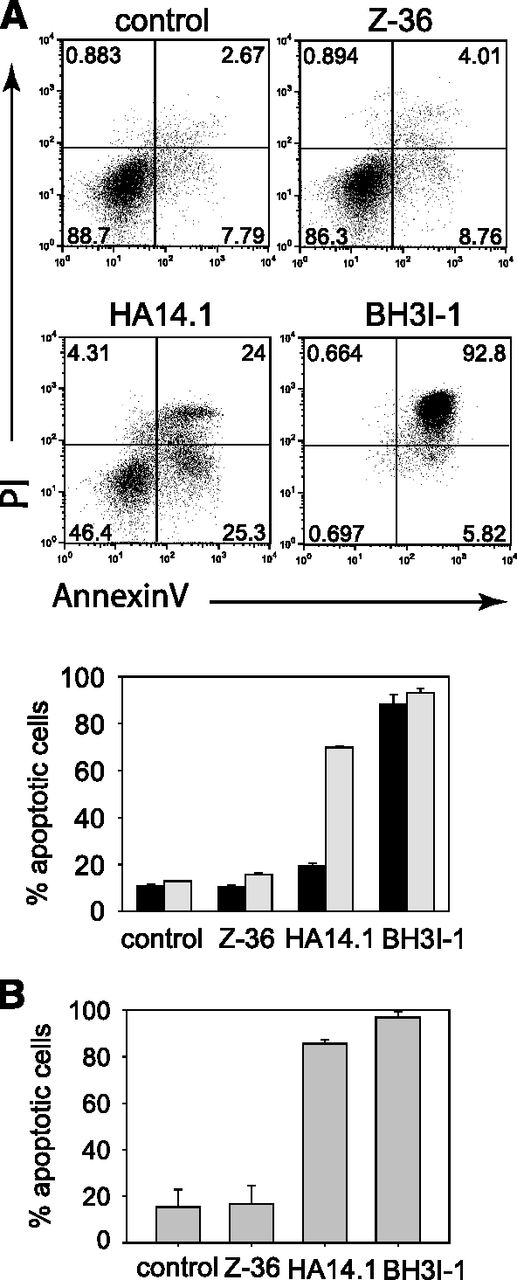
Bcl-xL can block the mitochondria-dependent apoptosis pathway by direct binding to the BH3 domain of proapoptotic Bcl-2 family members or by preventing their binding to BH3-only proteins. In addition, Bcl-xL has been demonstrated to bind to Beclin-1, another BH3 domain–containing protein that regulates the early steps of autophagy and causes apoptosis in the presence of Bax or Bak.40 Hence, we tested 3 different Bcl-xL inhibitors for their proapoptotic activity in eosinophils. Z36 blocks the binding between Bcl-xL and Beclin-1.41 HA14.1 binds the BH1 and BH3 domains in Bcl-xL and Bcl-2 but also decreases Bcl-xL and Bcl-2 expression while increasing p53 and Puma in HeLa cells.42 BH3I-1 blocks the interaction of Bcl-xL with Bak, Bax, Bad, or Bim and thereby causes apoptosis.43 More than 80% of eosinophils became apoptotic within 4 hours after adding BH3I-1 to the culture, whereas the other 2 inhibitors did not affect survival at that time point (Figure 6A). However, HA14.1 and Z-36 induced apoptosis at 24 hours after culture setup, especially at higher concentrations of these inhibitors, and apoptosis could not be prevented by higher IL-5 concentrations in the cultures (supplemental Figure 4).
The Bcl-xL inhibitors HA14.1 and BH3I-1 promote apoptosis of murine and human eosinophils. (A) Murine BMDEs were cultured in presence of IL-5 and the indicated Bcl-xL inhibitors for 4 hours (black bars) or 24 hours (gray bars) and analyzed for apoptosis by flow cytometry. Dot plots show representative samples at 24 hours after addition of the inhibitors. (B) Human eosinophils isolated from the peripheral blood of healthy volunteers were cultured for 24 hours in the presence of IL-5 and the indicated Bcl-xL inhibitors. Bars show the mean + SEM (n = 3) of 3 independent experiments.
The Bcl-xL inhibitors HA14.1 and BH3I-1 promote apoptosis of murine and human eosinophils. (A) Murine BMDEs were cultured in presence of IL-5 and the indicated Bcl-xL inhibitors for 4 hours (black bars) or 24 hours (gray bars) and analyzed for apoptosis by flow cytometry. Dot plots show representative samples at 24 hours after addition of the inhibitors. (B) Human eosinophils isolated from the peripheral blood of healthy volunteers were cultured for 24 hours in the presence of IL-5 and the indicated Bcl-xL inhibitors. Bars show the mean + SEM (n = 3) of 3 independent experiments.
To address whether this effect can be confirmed with human eosinophils, we purified untouched eosinophils from peripheral blood of healthy volunteers and cultured them for 24 hours in 10 ng/mL of IL-5 and 50 μM BH3I-1, HA14.1, or Z36. Similar to the results with murine eosinophils, we observed pronounced apoptosis in the presence of BH3I-1 and HA14.1, whereas Z36 did not induce apoptosis (Figure 6B). Therefore, we conclude that Bcl-xL–mediated inhibition of growth factor deprivation–induced apoptosis in eosinophils acts mainly by direct interaction with proapoptotic proteins at the mitochondrial membrane.
Discussion
Although high eosinophil numbers are not detrimental to tissues per se, eosinophils can cause severe damage to organs when they get activated. Therefore, it is critical to investigate the regulation of eosinophil development and homeostasis in order to identify potential target structures for therapeutic intervention that reduce the number of eosinophils. Here, we found that the cytokines IL-3, IL-5, and GM-CSF could all prevent growth factor withdrawal–induced apoptosis, but only IL-5 was able to cause eosinophil development from bone marrow precursors. Consistent with this observation, previous studies have shown that IL-3 and GM-CSF are not sufficient for eosinophil development from human CD34+ bone marrow cells.44,45 This finding suggests that only the IL-5 receptor is expressed on eosinophil-committed precursors or that certain signals from the IL-5 receptor α chain are required for eosinophil development. Of interest, IL-3 and GM-CSF appeared to be sufficient for eosinophil development from human peripheral blood precursors.46 Most likely, the precursors in the blood are already more committed to the eosinophil lineage as compared to their more immature counterparts in the bone marrow.
The efficient inhibition of apoptosis by all 3 cytokines shows their potential to promote eosinophilia by increasing the lifespan of eosinophils. The prevention of apoptosis by βc chain–mediated signals is poorly understood. The βc chain is linked to the STAT5, p38-MAPK, PI3K, and NF-κB pathways, which could all be involved in preventing apoptosis in eosinophils. p38-MAPK has been shown to prevent human eosinophil apoptosis but only in the absence of IL-5.47 We demonstrate here that inhibition of STAT5 or p38-MAPK with 5 μM concentrations of inhibitors had no effect on growth factor deprivation–induced apoptosis, whereas the apoptosis rate was increased 10-fold after inhibition of NF-κB. Generally, the NF-κB pathway can be induced by a variety of different mechanisms, including PI3K-activated kinase Akt, which phosphorylates IκB kinase followed by IκB kinase–mediated phosphorylation and degradation of IκBα to release NF-κB for translocation into the nucleus. Phosphorylated Ser585 in βc is a docking site for 14-3-3 adaptor proteins, which recruit and activate PI3K to promote IL-3– or GM-CSF–induced survival of a T-cell line transduced with βc constructs.35,48 However, the PI3K and MAPK pathways were reported to play no major role for GM-CSF–mediated inhibition of apoptosis in human peripheral blood eosinophils and murine airway eosinophils.49,50 In contrast, PI3K was involved in prevention of apoptosis in murine eosinophils isolated from the pleural cavity of allergic mice.51 Similar effects were observed after engagement of β-adrenergic receptors on murine eosinophils, and this process appeared to be mediated by inhibition of the transcription factor forkhead in rhabdomyosarcoma.50 The NF-κB pathway is also induced by receptors of the TNF family. Although signaling through the receptors for TNF-α or FasL mainly induces active apoptosis of eosinophils in vitro,52 it was also reported that FasL- or TNF-α–mediated signals can promote survival of eosinophils by activation of NF-κB–dependent target genes.52,53 Similarly, TNF-α was shown to activate the NF-κB pathway in ex vivo isolated human eosinophils, and it was proposed that a NF-κB–induced protein protects the cell from caspase-dependent apoptosis.54,55 In addition, it was shown that NF-κB–induced secretion of IL-6 promotes autocrine activation of PI3K and subsequent expression of the antiapoptotic protein Mcl-1 in neutrophils56 . Hence, NF-κB–regulated antiapoptotic genes can be induced via the βc chain and even by TNF-family receptors, which are generally considered to mediate proapoptotic signals. Consistent with our results, it was shown that a cell-permeable version of IκBα (TAT-IκBα) prevented nuclear translocation of NF-κB and thereby promoted apoptosis in vitro.57 However, studies with selective and constitutive activation of the NF-κB pathway in normal or helminth-infected mice have not been reported. Using newly generated EpxCre mice, we demonstrate that specific deletion of IκBα in eosinophils did increase the expression of Bcl-xL and enhanced their survival. Of interest, this effect was observed only in helminth-infected mice, indicating that NF-κB–induced genes play a minor role for eosinophil survival under steady-state conditions. This finding could be due to the fact that only helminth-induced eosinophilia, but not steady-state development of eosinophils, requires IL-5.19 By retroviral transduction and selective inhibition, we show that Bcl-xL is indeed required and sufficient to prevent growth factor deprivation–induced apoptosis in murine and human eosinophils.
In conclusion, our results demonstrate that Bcl-xL plays a critical role for regulation of eosinophil survival. Hence, therapeutic interventions focusing on inhibition of Bcl-xL function in vivo may indeed reduce eosinophilia associated with allergic inflammation and hypereosinophilic syndromes.
The online version of this article contains a data supplement.
There is an Inside Blood Commentary on this article in this issue.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank Isabell Schiedewitz and Kirstin Castiglione for technical assistance;
Manfred Kirsch and Lisa Gundel for animal care; Hans-Jörg Fehling for providing R-RFP mice; Erwin Strasser for collecting human PBMCs; and Jonathan Jantsch and members of the D.V. laboratory for helpful discussion.
This work was supported by Deutsche Forschungsgemeinschaft grants Vo944/2-2 and Vo944/7-1, National Institutes of Health National Institute of Allergy and Infectious Diseases grant AI26918, and the Howard Hughes Medical Institute.
Authorship
Contribution: C.S., R.W., S.H., and D.W. performed the experiments; R.L. and R.A.R. provided critical reagents and gave advice on the manuscript; and C.S., R.W., and D.V. designed the experiments and wrote the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
The current affiliation for S.H. is BSL BIOSERVICE Scientific Laboratories, Munich, Germany.
Correspondence: David Voehringer, Department of Infection Biology, Universitätsklinikum Erlangen, Wasserturmstrasse 3-5, 91054 Erlangen, Germany; e-mail: david.voehringer@uk-erlangen.de.
References
Author notes
C.S. and R.W. contributed equally to this study.



This feature is available to Subscribers Only
Sign In or Create an Account Close Modal