Key Points
TFL is most commonly of the germinal center B-cell-like phenotype, but a significant minority of cases is of the ABC phenotype (16%).
The absence of BCL2 translocation in FL at diagnosis is associated with transformation into ABC-like large cell lymphoma.
Abstract
Follicular lymphoma (FL) is an indolent disease but transforms in 2% to 3% of patients per year into aggressive, large cell lymphoma, a critical event in the course of the disease associated with increased lymphoma-related mortality. Early transformation cannot be accurately predicted at the time of FL diagnosis and the biology of transformed FL (TFL) is poorly understood. Here, we assembled a cohort of 126 diagnostic FL specimens including 40 patients experiencing transformation (<5 years) and 86 patients not experiencing transformation for at least 5 years. In addition, we assembled an overlapping cohort of 155 TFL patients, including 114 cases for which paired samples were available, and assessed temporal changes of routinely available biomarkers, outcome after transformation, as well as molecular subtypes of TFL. We report that the expression of IRF4 is an independent predictor of early transformation (Hazard ratio, 13.3; P < .001). We also show that composite histology at the time of transformation predicts favorable prognosis. Moreover, applying the Lymph2Cx digital gene expression assay for diffuse large B-cell lymphoma (DLBCL) cell-of-origin determination to 110 patients with DLBCL-like TFL, we demonstrate that TFL is of the germinal-center B-cell–like subtype in the majority of cases (80%) but that a significant proportion of cases is of the activated B-cell–like (ABC) subtype (16%). These latter cases are commonly negative for BCL2 translocation and arise preferentially from BCL2 translocation-negative and/or IRF4-expressing FLs. Our study demonstrates the existence of molecular heterogeneity in TFL as well as its relationship to the antecedent FL.
Introduction
Follicular lymphoma (FL) is the most common indolent lymphoma and the median survival for newly diagnosed patients is currently in excess of 10 years.1-3 However, outcomes are heterogeneous and a fraction of patients are at risk of early relapse/progression. Histologic transformation to aggressive lymphoma occurs in 2% to 3% of patients per year and is associated with treatment resistance, progression, and lymphoma-related mortality.4-7 Transformation results most commonly in a histology that cannot be distinguished from de novo diffuse large B-cell lymphoma (DLBCL). More rarely, the transformed biopsy contains a mixture in varying proportions of FL and DLBCL in the same biopsy (composite histology), resembles gray-zone lymphoma (a situation referred to as unclassifiable B-cell lymphoma with features intermediate between DLBCL and Burkitt lymphoma [or BCLU for the remainder of this study]).8 Although transformation was historically considered to be an event associated with dismal prognosis, treatment outcomes vary and prediction of survival after transformation needs to be individualized as outcome is dependent on factors such as the extent of the disease at time of transformation, prior therapies, and time to transformation.4,5,9
It remains challenging to predict subsequent transformation at the time of FL diagnosis although certain clinical parameters, such as elevated lactate dehydrogenase levels, advanced stage, and high Follicular Lymphoma International Prognostic Index (FLIPI) scores are correlated with a higher risk of transformation in some, but not all studies.4-6,10 Similarly, certain genomic features have been associated with an increased likelihood of transformation. These include BCL6 translocations,11 FAS mutations,12,13 or deletions of 1p36.3.14 Taken together, these parameters do not predict transformation with sufficient sensitivity or specificity to predicate altering patient management.
Transformation occurs by clonal evolution from a common mutated precursor, as shown by sequencing of rearranged VDJ sequences,15 or whole exomes or genomes.13,16 Recurrent genetic alterations that evolve during the process of transformation include: CDKN2A deletions and/or loss of heterozygosity13,17 ; translocations, gains, amplifications, or mutations involving MYC13,18,19 ; mutations, deletions, and loss of heterozygosity affecting TP5313,20-24 ; and mutations or deletions of B2M and CD58.13 Increased proliferation resulting from cell cycle deregulation, defective DNA damage response, and escape from immune surveillance therefore emerge as critical elements of histologic progression of FL to an aggressive lymphoma.
Cell-of-origin (COO) assignment is increasingly important to predict prognosis and response to targeted therapy in de novo DLBCL, yet information on COO of transformed FL (TFL) is scarce in the literature. In a prior study, Davies et al performed subtyping of 25 TFL specimens using the Lymphochip platform and the previously reported Bayesian class predictor.23,25 All classifiable cases (18 out of 25) were of germinal center B-cell–like (GCB) phenotype and none were of the activated B-cell–like (ABC) phenotype, although 3 cases out of 35 (9%) were assigned to the non-GCB phenotype using immunohistochemistry (IHC) in an independent cohort.23 In the study reported by Bouska et al, 42 out of 59 (71%) TFL cases were reported to be of the GCB phenotype and 17 out of 59 cases (29%) of a non-GCB phenotype.26 In addition, whereas assessment of translocations involving both the MYC and either the BCL2 or BCL6 genes (“double-hit lymphoma”) allows for identification of a poor-risk category of de novo GCB-type DLBCL,27-29 similar prognostic implications are less well studied in TFL.
In this study, we sought to assess whether transformation can be predicted at the time of FL diagnosis using clinical annotation and information on histologic grade, fluorescence in situ hybridization (FISH) break-apart assays for recurrently rearranged genes (BCL2 and BCL6), and IHC for CD10, BCL6, and IRF4. Furthermore, as subtype-specific efficacy of novel agents is actively pursued in de novo DLBCL,30-32 we also asked whether TFL could be similarly divided into distinct transcriptional phenotypes, using a large cohort of formalin-fixed and paraffin-embedded tissue (FFPET) samples. Finally, we sought to determine whether features present at the time of initial diagnosis correlate with molecular classes of TFL, potentially delineating distinct paths to transformation.
Methods
Patient samples and definition of end points
For prediction of transformation, we assembled a cohort of 126 patients who were diagnosed with FL (grades 1, 2, or 3A) between 1997 and 2013 and were either observed or treated frontline with rituximab alone or in combination with chemotherapy. This cohort was enriched for clinical extremes, consisting of 40 patients who experienced transformation within 5 years of diagnosis and 86 patients who experienced neither transformation nor progression or death, for at least 5 years from the time of initial diagnosis. For all other analyses, we assembled a cohort of 155 patients who were diagnosed with FL (grades 1, 2, or 3A) between 1970 and 2013 and subsequently experienced transformation to large cell lymphoma, as defined in the “Introduction.” Cases with discordant histology at diagnosis were excluded but cases with composite histology at the time of transformation were included. In this cohort, samples were available for both the FL and the TFL time points for 114 patients. In a further 14 and 27 patients, samples were available only at the time of FL or TFL, respectively. The Lymph2Cx assay was restricted to those 110 TFL samples in which the histology was consistent with DLBCL. The overlap between the study cohorts is shown in supplemental Figure 1 on the Blood Web site.
All patients described within this study as having experienced transformation had histologic proof of transformation. Time to transformation was defined as the time between diagnosis of FL and the time when histologic proof of large cell transformation was obtained. Survival after transformation was defined as the time between transformation and death from any cause, with censoring at the date of last follow-up in living patients. All patient specimens were collected as part of a research project approved by the University of British Columbia–British Columbia Cancer Agency Research Ethics Board.
Tissue microarray, FISH, and IHC
A tissue microarray was constructed utilizing duplicate 1.0 mm cores of FFPET biopsies from all aforementioned patients. FISH assays were performed using commercially available break-apart probes for the BCL2, BCL6, and MYC loci (supplemental Table 1). Images were acquired using a Metafer CoolCube 1 camera and Metafer software (version 3.11.3). At least 100 nuclei for each sample were assessed for break-apart status by two independent scorers (R.K. and S.B.-N.). Discrepant cases were further reviewed by a third person (A.M.) to reach consensus. At least 10% of nuclei were required to have a break-apart signal for a case to be called positive. Double-hit lymphoma was defined as a case having translocations involving the MYC locus and either the BCL2 or the BCL6 loci. Four micrometer sections of the tissue microarray were stained with antibodies against human CD10, BCL6, IRF4, and TP53 on a Ventana BenchMark XT automated slide-staining system (supplemental Table 2). The number of positive tumor cells was independently assessed by two expert hematopathologists (A.M. and P.F.) and discrepant results were discussed at a multi-head microscope. For CD10, BCL6, and IRF4, at least 30% of tumor cells needed to be stained for a case to be called positive. For TP53, only cases with uniform, strong staining were called positive. The Hans algorithm was used for COO assignment by IHC.33
Lymph2Cx assay
RNA was extracted from 10 μm FFPET sections and cut to a surface of ≥1 cm2 using the AllPrep DNA/RNA FFPE Kit (Qiagen). Two hundred nanograms of FFPET-derived RNA were used for the Lymph2Cx assay, a digital gene expression (NanoString)-based test that assesses the expression of 20 genes (8 genes that are overexpressed in ABC-DLBCL, 7 genes that are overexpressed in GCB-DLBCL, and 5 housekeeping genes), as described by Scott et al.34 For each case, the linear predictor score (LPS) was calculated and the likelihood of that sample belonging to either COO subgroup was determined using Bayes’ rule, as described in Wright et al.25 Calibration between the lot of NanoString reagents used in this study and the lot used in Scott et al34 was achieved by determining the LPS for 30 RNA samples from that study on the current reagent lot.
Targeted sequencing of CARD11, CD79B, and MYD88
Adapter ligated libraries were constructed following the 100 ng SureSelect XT2 Custom Target Enrichment System for Illumina Multiplexed Sequencing Protocol version D.2. Indexed libraries were pooled and sequenced on an Illumina MiSeq instrument generating 150 base-pair paired-end reads. After demultiplexing, reads were aligned to the human reference genome (hg19) using BWA-MEM (version 0.7.5a)35 and single nucleotide variants were predicted using VarScan (version 2.3.6).36,37 All predictions with ≥5 variant-supporting reads and ≥10% variant allele frequency were taken forward for validation by Sanger sequencing, performed as previously described.38
Statistical analysis
All associations between translocation status, IHC staining result, and pathological or clinical characteristics were assessed using Fisher’s exact test. For multivariable prediction of transformation, we fitted a weighted Cox regression model, after verification of the proportional hazards assumption, as the patient cohort was enriched for transformation events.39 Assuming a transformation rate of 2.5% per year in an unselected patient cohort, the weight for events was adjusted from 1 to 0.4 and the weight of controls from 1 to 1.3. To estimate survival after transformation, we plotted survival curves according to the Kaplan–Meier method and compared survival differences between patient groups using the log-rank test. P values <.05 were considered significant.
Results
Association of clinical and pathological characteristics with transformation
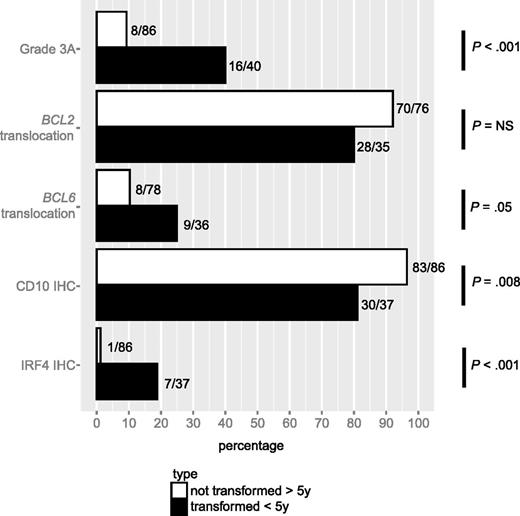
The cohort used for this analysis consisted of 40 patients who experienced transformation within 5 years after being diagnosed, and a control group of 86 patients who experienced neither transformation nor progression for at least 5 years after diagnosis (Table 1). The two groups were balanced with respect to clinical characteristics, except that the proportion of patients with elevated lactate dehydrogenase (LDH) at diagnosis was higher in the transformation group than in the control group (23% vs 5%; P = .01). However, the analysis of pathological features revealed multiple significant differences between the two groups (Figure 1). In comparison with the control group, cases in the transformation group were more likely to have grade 3A FL (40% vs 9%; P < .001), to harbor a BCL6 translocation (25% vs 10%; P = .05), to be negative for CD10 staining (81% vs 97%; P = .008), and to express IRF4 (19% vs 1%; P < .001). The proportion of cases harboring a BCL2 translocation was lower in the transformed cases than in the control cases but this difference did not reach statistical significance (80% vs 92%; P = .11). There was no difference in BCL6 staining between the two groups (97% vs 98%; P = 1.00). As some of the pathological features under study tend to co-occur, we fitted a weighted, multivariate Cox proportional hazard model that included grade 3A, BCL2, and BCL6 translocation, as well as staining for CD10 and IRF4 (Table 2). Whereas in univariate Cox regression analyses, grade 3A, BCL6 translocation, and IRF4 staining were positively associated, and CD10 staining was negatively associated, with time to transformation, in the multivariate model, only IRF4 staining predicted a higher risk of transformation when positive at diagnosis (HR 13.3; 95% CI, 3.7-48.4; P < .001).
Clinical characteristics of 40 patients with early transformation vs 86 patients without transformation or progression for at least 5 years
| . | Transformed <5 y . | Not transformed >5 y . | . | ||
|---|---|---|---|---|---|
| . | (No.) . | (%) . | (No.) . | (%) . | P* . |
| Age | |||||
| ≤60 | 20 | 50 | 49 | 57 | .56 |
| >60 | 20 | 50 | 37 | 43 | |
| Gender | |||||
| Female | 17 | 43 | 42 | 49 | .57 |
| Male | 23 | 58 | 44 | 51 | |
| B symptoms | |||||
| Absent | 35 | 90 | 73 | 87 | .77 |
| Present | 4 | 10 | 11 | 13 | |
| N/A | 1 | — | 2 | — | |
| ECOG | |||||
| 0-1 | 36 | 95 | 78 | 96 | .65 |
| 2-4 | 2 | 5 | 3 | 4 | |
| N/A | 2 | — | 5 | — | |
| Stage | |||||
| I-II | 11 | 28 | 20 | 23 | .66 |
| III-IV | 28 | 72 | 66 | 77 | |
| N/A | 1 | — | 0 | — | |
| Extranodal sites | |||||
| 0-1 | 38 | 95 | 77 | 90 | .50 |
| ≥1 | 2 | 5 | 9 | 10 | |
| Nodal areas | |||||
| ≤4 | 15 | 56 | 35 | 47 | .50 |
| >4 | 12 | 44 | 40 | 53 | |
| N/A | 13 | — | 11 | — | |
| Tumor mass | |||||
| <7 cm | 26 | 81 | 60 | 73 | .47 |
| ≥7 cm | 6 | 19 | 22 | 27 | |
| N/A | 8 | — | 4 | — | |
| LDH | |||||
| ≤ ULN | 24 | 77 | 76 | 95 | .01 |
| > ULN | 7 | 23 | 4 | 5 | |
| N/A | 9 | — | 6 | — | |
| Hemoglobin | |||||
| <12 g/dL | 5 | 16 | 4 | 5 | .11 |
| ≥12 g/dL | 27 | 84 | 80 | 95 | |
| N/A | 8 | — | 2 | — | |
| FLIPI | |||||
| Low/intermediate | 21 | 70 | 59 | 78 | .46 |
| High | 9 | 30 | 17 | 22 | |
| N/A | 10 | — | 10 | — | |
| Primary treatment of stage III/IV | |||||
| Observation or local treatment | 15 | 54 | 26 | 39 | .26 |
| Systemic treatment | 13 | 46 | 40 | 61 | |
| . | Transformed <5 y . | Not transformed >5 y . | . | ||
|---|---|---|---|---|---|
| . | (No.) . | (%) . | (No.) . | (%) . | P* . |
| Age | |||||
| ≤60 | 20 | 50 | 49 | 57 | .56 |
| >60 | 20 | 50 | 37 | 43 | |
| Gender | |||||
| Female | 17 | 43 | 42 | 49 | .57 |
| Male | 23 | 58 | 44 | 51 | |
| B symptoms | |||||
| Absent | 35 | 90 | 73 | 87 | .77 |
| Present | 4 | 10 | 11 | 13 | |
| N/A | 1 | — | 2 | — | |
| ECOG | |||||
| 0-1 | 36 | 95 | 78 | 96 | .65 |
| 2-4 | 2 | 5 | 3 | 4 | |
| N/A | 2 | — | 5 | — | |
| Stage | |||||
| I-II | 11 | 28 | 20 | 23 | .66 |
| III-IV | 28 | 72 | 66 | 77 | |
| N/A | 1 | — | 0 | — | |
| Extranodal sites | |||||
| 0-1 | 38 | 95 | 77 | 90 | .50 |
| ≥1 | 2 | 5 | 9 | 10 | |
| Nodal areas | |||||
| ≤4 | 15 | 56 | 35 | 47 | .50 |
| >4 | 12 | 44 | 40 | 53 | |
| N/A | 13 | — | 11 | — | |
| Tumor mass | |||||
| <7 cm | 26 | 81 | 60 | 73 | .47 |
| ≥7 cm | 6 | 19 | 22 | 27 | |
| N/A | 8 | — | 4 | — | |
| LDH | |||||
| ≤ ULN | 24 | 77 | 76 | 95 | .01 |
| > ULN | 7 | 23 | 4 | 5 | |
| N/A | 9 | — | 6 | — | |
| Hemoglobin | |||||
| <12 g/dL | 5 | 16 | 4 | 5 | .11 |
| ≥12 g/dL | 27 | 84 | 80 | 95 | |
| N/A | 8 | — | 2 | — | |
| FLIPI | |||||
| Low/intermediate | 21 | 70 | 59 | 78 | .46 |
| High | 9 | 30 | 17 | 22 | |
| N/A | 10 | — | 10 | — | |
| Primary treatment of stage III/IV | |||||
| Observation or local treatment | 15 | 54 | 26 | 39 | .26 |
| Systemic treatment | 13 | 46 | 40 | 61 | |
ECOG, Eastern Cooperative Oncology Group performance status; LDH, lactate dehydrogenase; N/A, not available; ULN, upper limit of normal.
N/A cases were excluded for contingency analyses.
Association of pathological characteristics with transformation. Prevalence of grade 3A, translocations involving the BCL2 or BCL6 loci, and staining by IHC for CD10, BCL6, and IRF4 in tissue specimens of patients who presented with transformation within 5 years after diagnosis vs those who presented with neither transformation nor progression for at least 5 years after diagnosis.
Association of pathological characteristics with transformation. Prevalence of grade 3A, translocations involving the BCL2 or BCL6 loci, and staining by IHC for CD10, BCL6, and IRF4 in tissue specimens of patients who presented with transformation within 5 years after diagnosis vs those who presented with neither transformation nor progression for at least 5 years after diagnosis.
Weighted Cox regression model for time to transformation
| . | Univariate . | Multivariate . | ||||
|---|---|---|---|---|---|---|
| . | HR . | 95% CI . | P . | HR . | 95% CI . | P . |
| Grade 3A | 5.3 | 2.4-11.8 | <.001 | 2.5 | 0.7-8.3 | .14 |
| BCL2 translocation | 0.4 | 0.1-1.1 | .07 | 1.0 | 0.3-4.1 | .96 |
| BCL6 translocation | 2.6 | 1.0-6.4 | .05 | 1.5 | 0.3-6.3 | .60 |
| CD10 IHC | 0.2 | 0.1-0.6 | .004 | 1.0 | 0.2-4.5 | 1.00 |
| IRF4 IHC | 11.5 | 3.1-42.4 | <.001 | 13.3 | 3.7-48.4 | <.001 |
| . | Univariate . | Multivariate . | ||||
|---|---|---|---|---|---|---|
| . | HR . | 95% CI . | P . | HR . | 95% CI . | P . |
| Grade 3A | 5.3 | 2.4-11.8 | <.001 | 2.5 | 0.7-8.3 | .14 |
| BCL2 translocation | 0.4 | 0.1-1.1 | .07 | 1.0 | 0.3-4.1 | .96 |
| BCL6 translocation | 2.6 | 1.0-6.4 | .05 | 1.5 | 0.3-6.3 | .60 |
| CD10 IHC | 0.2 | 0.1-0.6 | .004 | 1.0 | 0.2-4.5 | 1.00 |
| IRF4 IHC | 11.5 | 3.1-42.4 | <.001 | 13.3 | 3.7-48.4 | <.001 |
CI, confidence interval; HR, hazard ratio.
Temporal patterns in translocation status and IHC staining in FL and transformed lymphoma
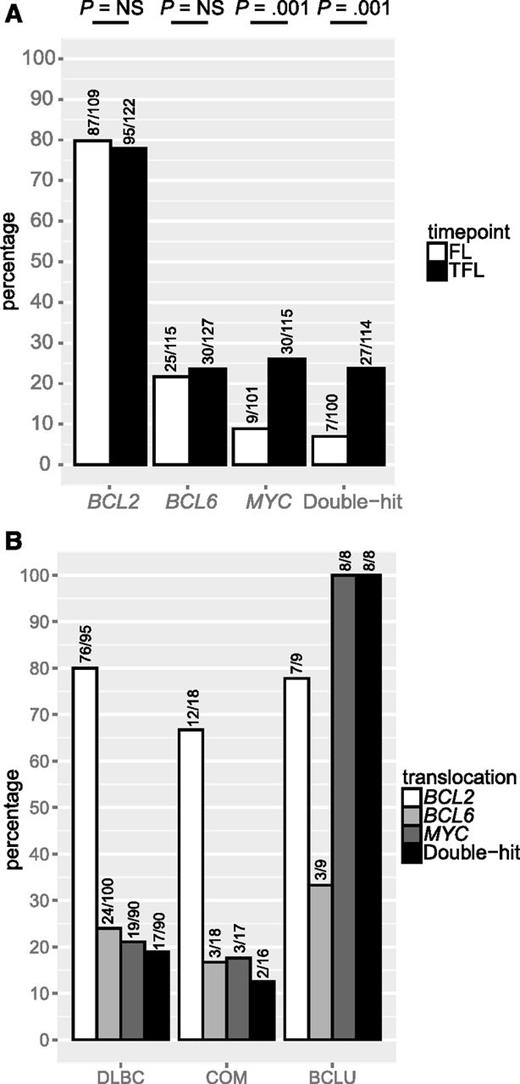
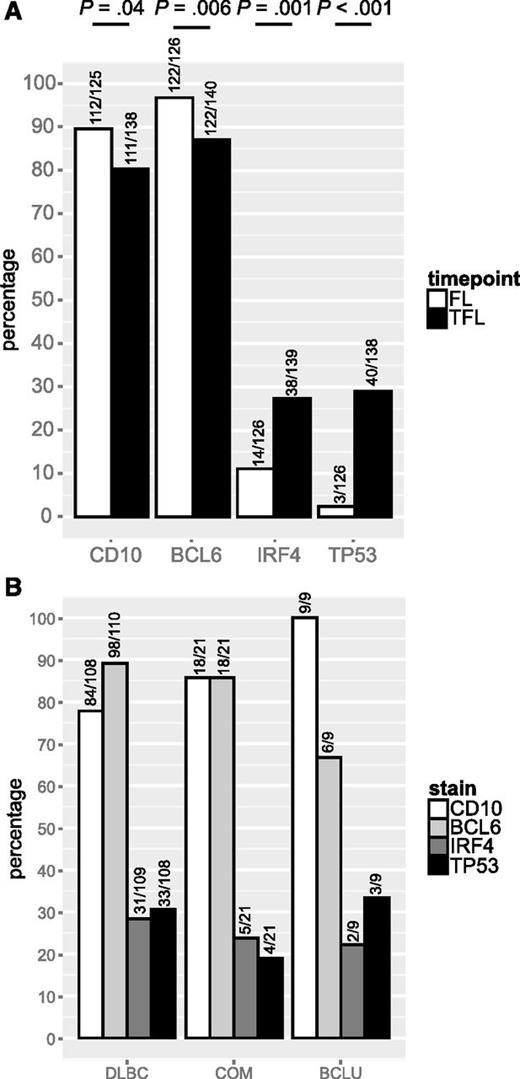
We leveraged a cohort of 128 FL and 141 TFL samples that included 114 patients for which samples from both time points were available, to assess the presence of BCL2, BCL6, and MYC translocation, as well as staining by IHC for CD10, BCL6, IRF4, and TP53. Patient characteristics are shown in Table 3. In this cohort, where all patients experienced transformation during the course of their disease, BCL2 and BCL6 translocations were found in 80% and 22% of FL samples, respectively, and these percentages were not significantly different in TFL samples. In contrast, MYC translocation was detected in 9% of FL samples but the percentage was significantly higher in TFL samples (26%; P = .001). Consequently, double-hit status was also significantly more common in TFL than in FL (24% vs 7%; P = .001; Figure 2A). MYC translocation or double-hit were present in all TFL presenting morphologically as BCLU (Figure 2B). TFL samples expressed CD10 less commonly than FL (80% vs 90%; P = .04) as well as BCL6 (87% vs 97%; P = .006), but expressed IRF4 (27% vs 11%; P = .001) and TP53 (29% vs 2%; P < .001; Figure 3A) significantly more often. IRF4 and TP53 expression were not correlated (data not shown) and were found at similar frequencies in all histologic subtypes of TFL (Figure 3B).
Clinical characteristics of TFL cohort
| . | All TFL patients (n = 155) . | |
|---|---|---|
| . | (No.) . | (%) . |
| Year of FL diagnosis | ||
| <2003 | 104 | 67 |
| ≥2003 | 51 | 33 |
| Year of TFL diagnosis | ||
| <2003 | 59 | 38 |
| ≥2003 | 96 | 62 |
| Antecedent FL grade | ||
| Grade 1 or 2 | 135 | 87 |
| Grade 3A | 20 | 13 |
| Time to transformation (y) | ||
| ≤5 | 98 | 63 |
| 5-10 | 39 | 25 |
| >10 | 18 | 12 |
| Age at TFL diagnosis (y) | ||
| ≤60 | 65 | 42 |
| >60 | 90 | 58 |
| First line treatment of FL | ||
| Single-agent chemo | 34 | 24 |
| Multi-agent chemo* | 22 | 15 |
| Multi-agent chemo† + rituximab | 14 | 10 |
| Radiation or surgery | 21 | 15 |
| Observation | 52 | 36 |
| N/A | 12 | — |
| Initial treatment after transformation | ||
| Multi-agent chemo‡ | 46 | 32 |
| Multi-agent chemo§ + rituximab | 82 | 56 |
| Other | 18 | 12 |
| N/A | 9 | — |
| Hematopoietic stem cell transplant | ||
| Autologous | 12 | 8 |
| Allogeneic | 3 | 2 |
| None | 133 | 90 |
| N/A | 7 | — |
| Rituximab before transformation | ||
| No | 120 | 79 |
| Yes | 32 | 21 |
| N/A | 3 | — |
| Rituximab after transformation | ||
| No | 61 | 40 |
| Yes | 91 | 60 |
| N/A | 3 | — |
| . | All TFL patients (n = 155) . | |
|---|---|---|
| . | (No.) . | (%) . |
| Year of FL diagnosis | ||
| <2003 | 104 | 67 |
| ≥2003 | 51 | 33 |
| Year of TFL diagnosis | ||
| <2003 | 59 | 38 |
| ≥2003 | 96 | 62 |
| Antecedent FL grade | ||
| Grade 1 or 2 | 135 | 87 |
| Grade 3A | 20 | 13 |
| Time to transformation (y) | ||
| ≤5 | 98 | 63 |
| 5-10 | 39 | 25 |
| >10 | 18 | 12 |
| Age at TFL diagnosis (y) | ||
| ≤60 | 65 | 42 |
| >60 | 90 | 58 |
| First line treatment of FL | ||
| Single-agent chemo | 34 | 24 |
| Multi-agent chemo* | 22 | 15 |
| Multi-agent chemo† + rituximab | 14 | 10 |
| Radiation or surgery | 21 | 15 |
| Observation | 52 | 36 |
| N/A | 12 | — |
| Initial treatment after transformation | ||
| Multi-agent chemo‡ | 46 | 32 |
| Multi-agent chemo§ + rituximab | 82 | 56 |
| Other | 18 | 12 |
| N/A | 9 | — |
| Hematopoietic stem cell transplant | ||
| Autologous | 12 | 8 |
| Allogeneic | 3 | 2 |
| None | 133 | 90 |
| N/A | 7 | — |
| Rituximab before transformation | ||
| No | 120 | 79 |
| Yes | 32 | 21 |
| N/A | 3 | — |
| Rituximab after transformation | ||
| No | 61 | 40 |
| Yes | 91 | 60 |
| N/A | 3 | — |
Chemo, chemotherapy; N/A, not available.
Ten patients out of 22 were treated with anthracycline-containing regimens.
Three patients out of 14 were treated with cyclophosphamide-doxorubicin-vincristine-prednisone and rituximab (or CHOP-R), and 11 out of 14 with cyclophosphamide-vincristine-prednisone and rituximab.
Forty-one patients out of 46 were treated with anthracycline-containing regimens.
Seventy-seven patients out of 82 were treated with CHOP-R.
BCL2, BCL6, and MYC translocation in FL and TFL. (A) Prevalence of BCL2, BCL6, and MYC translocation in FL and TFL. (B) Prevalence of BCL2, BCL6, and MYC translocation in TFL by histology. NS, not significant.
BCL2, BCL6, and MYC translocation in FL and TFL. (A) Prevalence of BCL2, BCL6, and MYC translocation in FL and TFL. (B) Prevalence of BCL2, BCL6, and MYC translocation in TFL by histology. NS, not significant.
CD10, BCL6, IRF4 and TP53 expression in FL and TFL. (A) Prevalence of CD10, BCL6, IRF4, and TP53 expression in FL and TFL by IHC. (B) Prevalence of CD10, BCL6, IRF4, and TP53 expression in TFL by histology.
CD10, BCL6, IRF4 and TP53 expression in FL and TFL. (A) Prevalence of CD10, BCL6, IRF4, and TP53 expression in FL and TFL by IHC. (B) Prevalence of CD10, BCL6, IRF4, and TP53 expression in TFL by histology.
Survival after transformation by histology and double-hit status
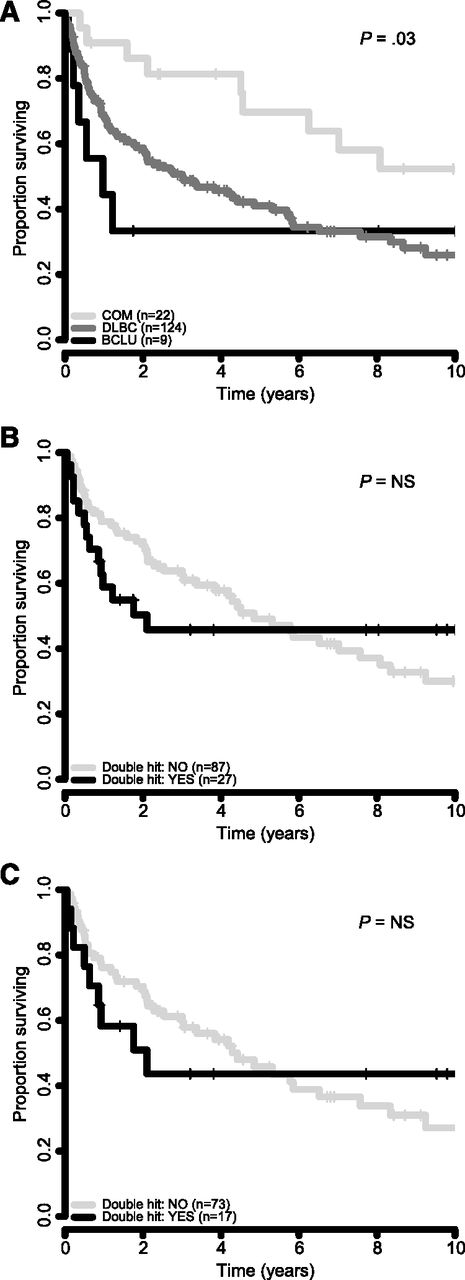
Next, we sought to determine outcome after transformation, based on TFL morphology and double-hit status. Cases with a composite histology at the time of transformation had the best survival, whereas cases with BCLU morphology had the worst outcome (2-year survival from time of transformation was 86%, 59%, and 33% for cases with composite, DLBCL, and BCLU histology, respectively (log-rank test, P = .03; Figure 4A). Double-hit status was associated with an inferior survival at 2 years after transformation as compared with cases without double-hit in the entire cohort (50% vs 73%) and in only those cases with DLBCL morphology (51% vs 70%), but these differences were not statistically significant (Figure 4B-C).
Survival correlates from time of transformation. (A) Survival by morphology of TFL in 155 patients. (B) Survival by double-hit translocation status in all 114 TFL cases in which BCL2 and MYC translocation status could be ascertained. (C) Survival by double-hit translocation status in 90 TFL cases with DLBCL morphology.
Survival correlates from time of transformation. (A) Survival by morphology of TFL in 155 patients. (B) Survival by double-hit translocation status in all 114 TFL cases in which BCL2 and MYC translocation status could be ascertained. (C) Survival by double-hit translocation status in 90 TFL cases with DLBCL morphology.
COO assignment of transformed lymphoma
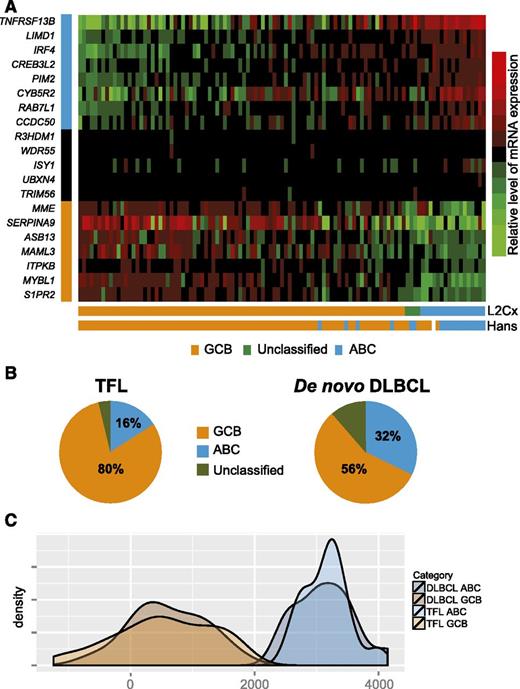
We applied the Lymph2Cx assay to 110 TFL samples that had morphology features of DLBCL. Sufficient gene counts to assign COO were obtained in 107 out of 110 samples (97%). Out of these 107 TFL samples, 86 were of the GCB subtype (80%), whereas 17 were of the ABC subtype (16%) and 4 were unclassified (4%) (Figure 5A). We also determined COO using the Hans classifier and, excluding 1 case for which IHC failed and the 4 cases that were unclassifiable by gene expression profiling, the concordance rate between the Lymph2Cx assay and the Hans algorithm was 92%. Using the Lymph2Cx assay, the proportion of GCB cases was significantly higher (80% vs 56%), and the proportion of ABC cases conversely lower (16% vs 32%; P < .001) in TFL than in a cohort of 335 de novo DLBCL that we have previously reported (Figure 5B).40 As the transcriptomic and genomic landscape of TFL is distinct from de novo DLBCL, we compared the distribution of the LPS between molecular subtypes of TFL (n = 103) and de novo DLBCL (n = 30). These distributions were superimposable using low-density gene expression data from the Lymph2Cx assay, suggesting that the assignment of TFL into GCB and ABC classes identifies transcriptional subtypes as distinct from each other as in de novo DLBCL (Figure 5C). Furthermore, as most prior literature described TFL as resembling GCB-DLBCL,13,23 the finding that 16% of TFL cases were of the ABC subtype raised the question of whether these ABC cases had truly transformed from underlying FL or had arisen de novo. We therefore determined clonotypes of the rearranged variable region of the immunoglobulin heavy chain locus in 3 cases, in which high quality DNA was available, through polymerase chain reaction amplification and sequencing, as per the BIOMED-2 protocol.41 A clonal relationship could indeed be documented in all 3 cases (data not shown).
Lymph2Cx assay in 107 TFL cases with DLBCL morphology. (A) Heatmap showing the relative expression of 20 genes (8 genes that are overexpressed in ABC-DLBCL, 5 housekeeping genes, and 7 genes that are overexpressed in GCB-DLBCL). (B) Pie charts showing the relative proportions of ABC and GCB large cell lymphoma in TFL and de novo DLBCL, respectively. (C) Kernel density plot of the distribution of the LPS in molecular subtypes of TFL and de novo DLBCL, respectively. mRNA, messenger RNA.
Lymph2Cx assay in 107 TFL cases with DLBCL morphology. (A) Heatmap showing the relative expression of 20 genes (8 genes that are overexpressed in ABC-DLBCL, 5 housekeeping genes, and 7 genes that are overexpressed in GCB-DLBCL). (B) Pie charts showing the relative proportions of ABC and GCB large cell lymphoma in TFL and de novo DLBCL, respectively. (C) Kernel density plot of the distribution of the LPS in molecular subtypes of TFL and de novo DLBCL, respectively. mRNA, messenger RNA.
Correlation of COO with outcome and cytogenetics
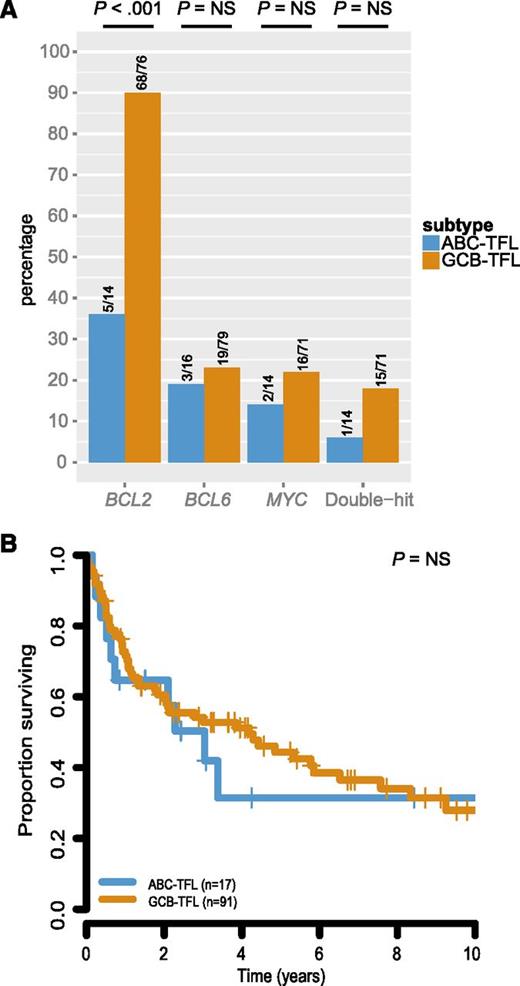
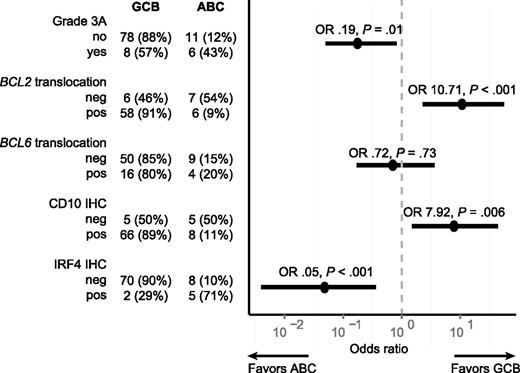
To further evaluate whether GCB-TFL and ABC-TFL represented distinct entities, we assessed translocation status of BCL2, BCL6, and MYC within these subtypes. BCL2 translocation was found in 68 out of 76 GCB-TFL cases (89%) but only in 5 out of 14 ABC-TFL cases (36%; P < .001; Figure 6A). BCL6 and MYC translocations were also more commonly present in GCB-TFL than in ABC-TFL, but this enrichment did not reach statistical significance. As molecular subtyping in de novo DLBCL has important prognostic implications,42 we compared survival after transformation between GCB-TFL and ABC-TFL, but could not detect any significant survival difference in TFL (Figure 6B). The finding that ABC-TFL cases were commonly negative for the BCL2 translocation raised the question of whether pathological findings within FL samples correlated with molecular subtypes in subsequent TFL (Figure 7). Indeed, FL cases that were graded 3A or expressed IRF4 preferentially transformed into the ABC subtype of TFL. On the other hand, the presence of a BCL2 translocation or expression of CD10 was associated with subsequent transformation into GCB-type TFL. Taken together, these findings highlight that the underlying genetic and phenotypic heterogeneity in FL translates into distinct TFL subtypes.
Survival and cytogenetic correlates of molecular subtypes in TFL. (A) Prevalence of BCL2, BCL6, and MYC translocation in TFL by COO subtype. (B) Survival from time of transformation by COO subtype in TFL. NS, not significant.
Survival and cytogenetic correlates of molecular subtypes in TFL. (A) Prevalence of BCL2, BCL6, and MYC translocation in TFL by COO subtype. (B) Survival from time of transformation by COO subtype in TFL. NS, not significant.
Association of pathological findings in FL and transformation into either the GCB or ABC subtype of TFL. The figure shows a forest plot of OR (circles) and corresponding 95% CIs (horizontal lines). neg, negative; OR, odds ratios; pos, positive.
Association of pathological findings in FL and transformation into either the GCB or ABC subtype of TFL. The figure shows a forest plot of OR (circles) and corresponding 95% CIs (horizontal lines). neg, negative; OR, odds ratios; pos, positive.
Mutations in CARD11, CD79B, and MYD88
Given that a subset of TFL is of the ABC subtype, we asked the question whether ABC-TFL is characterized, similarly to de novo ABC-DLBCL, by gene mutations that activate or amplify B-cell–receptor signaling and/or nuclear factor-κB signaling. To that effect, we performed next-generation sequencing of bait-captured coding sequences of CARD11, CD79B, and MYD88 in all 17 ABC-TFLs. In order to maximize discovery, we included all cases in the analysis, regardless of coverage (supplemental Table 3). We identified 9 mutations in 5 patients (Table 4), suggesting that genetic alterations involving the B-cell–receptor and/or nuclear factor-κB signaling pathways are common in ABC-TFL. For 2 of these patients (FL1005 and FL1161), germline DNA was available and all 4 variants present in these two cases were confirmed to be somatic. Mutations affecting the described amino acid residues of CARD11 and CD79B (Table 4) had been previously described in ABC-DLCBL.43,44 For MYD88, we identified a total of 4 mutations of which one corresponded to the classical hotspot mutation situated in the toll/interleukin-1 receptor domain (L265P), whereas the other 3 mutations (P141S, V144L, and S149I) were located in the intermediary domain of MYD88. Mutations affecting two of the latter amino acid residues (P141 and S149) have been previously reported in de novo DLBCL.45,46
Mutations in CARD11, CD79B, and MYD88
| Sample . | Gene . | Chromosomal position . | Nucleotide change . | Variant allele frequency (%) . | Predicted amino acid change . |
|---|---|---|---|---|---|
| FL1012T2 | CD79B | chr17:62006798 | T>G | 31 | Y196S* |
| FL1012T2 | MYD88 | chr3:38181433 | G>T | 17 | S149I† |
| FL1101T2 | MYD88 | chr3:38182259 | T>C | 59 | M232T† |
| FL1101T2 | CARD11 | chr7:2984162 | C>T | 76 | G123D‡ |
| FL1161T2 | CD79B | chr17:62006680 | A>T | 50 | L199Q* |
| FL1161T2 | MYD88 | chr3:38182641 | T>C | 42 | L265P† |
| FL1227T2 | MYD88 | chr3:38181408 | C>T | 23 | P141S† |
| FL1227T2 | MYD88 | chr3:38181417 | G>T | 24 | V144L† |
| FL1242T2 | CD79B | chr17:62006680 | A>G | 75 | L199P* |
| Sample . | Gene . | Chromosomal position . | Nucleotide change . | Variant allele frequency (%) . | Predicted amino acid change . |
|---|---|---|---|---|---|
| FL1012T2 | CD79B | chr17:62006798 | T>G | 31 | Y196S* |
| FL1012T2 | MYD88 | chr3:38181433 | G>T | 17 | S149I† |
| FL1101T2 | MYD88 | chr3:38182259 | T>C | 59 | M232T† |
| FL1101T2 | CARD11 | chr7:2984162 | C>T | 76 | G123D‡ |
| FL1161T2 | CD79B | chr17:62006680 | A>T | 50 | L199Q* |
| FL1161T2 | MYD88 | chr3:38182641 | T>C | 42 | L265P† |
| FL1227T2 | MYD88 | chr3:38181408 | C>T | 23 | P141S† |
| FL1227T2 | MYD88 | chr3:38181417 | G>T | 24 | V144L† |
| FL1242T2 | CD79B | chr17:62006680 | A>G | 75 | L199P* |
NCBI protein ID NP_000617.1.
NCBI protein ID NP_002459.2.
NCBI protein ID NP_115791.3.
Discussion
In this study, we associated routinely available biomarkers with risk of transformation, assessed temporal genetic and phenotypic changes in sequential specimens, performed molecular subtyping of TFL, and correlated transformation into either COO subtype with pathological features present in the initial FL sample. Whereas multiple features, including grade 3A, BCL6 translocation, IRF4 expression, as well as the lack of CD10 staining were associated with early transformation, only IRF4 positivity maintained independent predictive value in a weighted, multivariate Cox regression model. IRF4 is a transcription factor of the interferon regulatory factor family that exerts critical functions in germinal center formation and plasma cell differentiation.47-49 It can be detected in 14% to 17% of FL cases50,51 by IHC and expression was shown to correlate with poor overall survival in two phase 2 studies, but not in a larger trial from the Southwest Oncology Group.51 IRF4 expression has also been associated with poor progression-free survival in the FL2000 and PRIMA trials of the Lymphoma Study Association.52 The expression of IRF4 has been linked to higher proliferation rates, as assessed by Ki-67 staining, and to FL grades 3A and 3B. Our study adds texture to the literature by documenting an increased risk of early transformation in IRF4-expressing FL. Although the absolute number of IRF4-positive FL is small in our study, the reproducible association of IRF4 expression with poor outcome suggests that these patients might benefit from alternative treatment approaches. In this regard, lenalidomide is an enticing candidate as it was shown to downregulate IRF4 in the ABC subtype of DLBCL.53
Once transformation is suspected on clinical grounds, a repeat biopsy provides histologic documentation required for the management of TFL, in addition to important correlative information. Our data illustrate that the outcome of patients with a composite histology at time of transformation is better than the outcome of patients with a morphology that is in keeping with DLBCL or BCLU, in line with a prior study.54 Double-hit lymphoma usually refers to de novo DLBCL with concurrent MYC and BCL2 (or more rarely BCL6) translocations and has been associated with poor outcome following chemotherapy alone or when combined with rituximab in most studies.27-29,55 The prognostic implication of double-hit status at time of transformation from underlying FL is not as clearly established. In our series, double-hit status was found in 24% of all TFLs, and in 19%, 13%, and 100% of TFLs with a DLBCL, composite, or BCLU morphology, respectively. We observed a trend toward inferior survival posttransformation in double-hit TFL that did not reach statistical significance, possibly due to the relatively small sample size or heterogeneity in pre- and posttransformation treatments. Future studies are therefore needed to refine our understanding of the prognostic implication of double-hit status by itself and in the context of genome-wide alterations in TFL.
De novo DLBCL can be dissected into COO subtypes that differ by gene-expression profiles, activation of differential pathways, patient outcomes, and response to therapy.42,56-59 As TFL with a DLBCL morphology cannot be readily distinguished from de novo DLBCL based on microscopy, it is relevant to ask whether TFL can be similarly divided into molecular subtypes. Historically, TFL was considered to be related to the GCB subtype of de novo DLBCL, based on its transcriptional profile and the landscape of genetic alterations.13,23 In keeping with the report by Bouska et al,26 our study shows that a majority (80%) of TFL cases are of the GCB phenotype, but that a significant minority (16%) can nonetheless be identified as being of the ABC phenotype using the Lymph2Cx assay. In our series, ABC TFL cases did not have a poorer outcome after transformation than GCB TFL cases, although a definitive conclusion cannot be drawn due to the relatively small number of ABC cases and treatment heterogeneity. Importantly, we show that ABC TFLs have a low prevalence of BCL2 translocations and that the absence of a BCL2 translocation at the FL stage is associated with transformation into the ABC subtype of TFL. This observation mirrors the finding that ABC-related genes are enriched in the gene expression profile of BCL2 translocation-negative FL.60 In addition, IRF4 expression is a key contributor to the ABC subtype, and it is therefore not surprising that IRF4 expression in the antecedent FL similarly increases the likelihood for transformation into this subtype. In FL, negative staining for CD10, positive staining for IRF4, and lack of BCL2 translocation tend to co-occur, as shown by Karube et al,61 delineating cases with distinct molecular characteristics from the more common BCL2 translocation-positive cases. Based on our study, we can conclude that such molecular heterogeneity in FL is tied to molecular heterogeneity at the time of transformation and that the lymphoma phenotype at transformation is largely determined by underlying characteristics in the preceding FL. However, the relationship between findings in FL and at subsequent transformation is imperfect as, for example, 6 out of 13 BCL2 translocation-negative FLs transformed into the GCB subtype in our study, testifying to the complexity of paths to transformation.
In conclusion, our study shows that IRF4 expression in diagnostic FL samples is associated with early transformation. With regard to temporal changes over the course of transformation, MYC is more commonly translocated, and IRF4 more commonly expressed, in TFL than in antecedent FL. Furthermore, we show that composite histology at the time of transformation is associated with better outcome than DLBCL or BCLU morphologies and that 80% and 16% of DLBCL-like TFLs are of the GCB or the ABC subtype, respectively. These latter findings are important as they document inter-patient heterogeneity in TFL and suggest that treatment approaches may need to be tailored to underlying biology in order to reverse the adverse outcome that is currently associated with transformation. The finding that a subset of TFL is of the ABC subtype is particularly intriguing, as it raises the question of whether these patients would be candidates for targeting of the B-cell–receptor signaling pathway. In this regard, our finding that mutations in CD79B and MYD88 are recurrent in ABC-TFL provides further rationale to consider investigating the clinical benefit of targeted agents such as ibrutinib and lenalidomide in ABC-TFL. Future studies will, however, need to complement our results by adding whole transcriptome and broader mutational context in order to comprehensively elucidate the genomic makeup of TFL and provide insight into additional druggable alterations.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
This study was supported by a Program Project Grant from the Terry Fox Research Institute (Grant No. 1023). R.K. is supported by fellowships from the Ligue Genevoise Contre le Cancer et Fondation Dr Henri Dubois-Ferrière Dinu Lipatti, the Canadian Institutes for Health Research, the Michael Smith Foundation for Health Research, and the University of British Columbia. A.M. is supported by a fellowship award from the Dr Mildred Scheel Cancer Foundation (Deutsche Krebshilfe).
Authorship
Contribution: R.K., A.M., D.W.S., and R.D.G. designed and performed the research, analyzed and interpreted data, and wrote the paper; P.F. and K.T. performed pathological review of cases; S.B.-N. and D.E. performed FISH; Y.Z. performed statistical analysis; E.A.C. performed hybrid-capture, library construction, and the sequencing run; H.P.S. analyzed next-generation sequencing data; M.B., B.M., and A.T. performed RNA and DNA extractions; F.C.C. provided bioinformatics assistance; L.H.S. and J.M.C. supervised assembly of clinical data, reviewed the manuscript, and provided editorial input; C.S. participated in the original design of the project, reviewed the manuscript, and provided editorial input; and M.A.M. and S.P.S. participated in the original design of the project.
Conflict-of-interest disclosure: D.W.S., J.M.C., and R.D.G. are inventors on a patent held by the National Cancer Institute that has been licensed by NanoString Technologies. The remaining authors declare no competing financial interests.
Correspondence: Randy D. Gascoyne, BC Cancer Agency and BC Cancer Research Centre, 675 W 10th Ave, Vancouver, BC V5Z 1L3, Canada; e-mail: rgascoyn@bccancer.bc.ca.
References
Author notes
R.K. and A.M. contributed equally to this study.
D.W.S. and R.D.G. contributed equally to this study.