Key Points
Quinine binds to quinine-dependent antibodies first and causes CDR loop reconfiguration.
A hybrid paratope consisting of quinine and reconfigured CDR recognizes its target epitope.
Abstract
Drug-induced immune thrombocytopenia (DITP) is caused by antibodies that react with specific platelet-membrane glycoproteins when the provoking drug is present. More than 100 drugs have been implicated as triggers for this condition, quinine being one of the most common. The cause of DITP in most cases appears to be a drug-induced antibody that binds to a platelet membrane glycoprotein only when the drug is present. How a soluble drug promotes binding of an otherwise nonreactive immunoglobulin to its target, leading to platelet destruction, is uncertain, in part because of the difficulties of working with polyclonal human antibodies usually available only in small quantities. Recently, quinine-dependent murine monoclonal antibodies were developed that recognize a defined epitope on the β-propeller domain of the platelet integrin αIIb subunit (GPIIb) only when the drug is present and closely mimic the behavior of antibodies found in human patients with quinine-induced thrombocytopenia in vitro and in vivo. Here, we demonstrate specific, high-affinity binding of quinine to the complementarity-determining regions (CDRs) of these antibodies and define in crystal structures the changes induced in the CDR by this interaction. Because no detectable binding of quinine to the target integrin could be demonstrated in previous studies, the findings indicate that a hybrid paratope consisting of quinine and reconfigured antibody CDR plays a critical role in recognition of its target epitope by an antibody and suggest that, in this type of drug-induced immunologic injury, the primary reaction involves binding of the drug to antibody CDRs, causing it to acquire specificity for a site on a platelet integrin.
Introduction
More than 100 drugs including quinine have been implicated as causes of immune thrombocytopenia (DITP), a relatively common, sometimes life-threatening disorder.1,2 Quinine, originally used as a prophylactic against malaria, is used at lower concentrations to impart the bitter flavor to tonic water and is still used occasionally for the prevention of nocturnal leg cramps.
For unknown reasons, quinine is much more likely to cause DITP than other drugs, with the exception of heparin, which acts by a distinctly different mechanism.3 The hallmark of DITP caused by drugs other than heparin is an antibody that is nonreactive in the absence of the sensitizing drug but binds tightly to a platelet glycoprotein, usually integrin αIIbβ3 (GPIIb/IIIa), when a drug is present.1,4,5 In contrast to drugs that act as a hapten to induce hypersensitivity, drugs that cause DITP appear not to bind covalently to the target antigen and do not inhibit antibody binding at high concentration.1,6 Nor has it been possible to show that a drug binds noncovalently to an autologous target and somehow primes it for antibody binding. A mechanism recently proposed to explain drug-dependent antibody (DDAb) binding in DITP proposes that DDAbs are derived from a pool of naturally occurring immunoglobulins that react weakly with autologous targets7 and that the drug reacts at the antibody-antigen interface to increase the Ka for binding (“sandwich model”).1,8
Studies to define mechanism(s) responsible for DDAb binding to platelets have been handicapped by the requirement to use human antibodies, which are polyclonal and often in short supply. Recently developed murine monoclonal antibodies (mAbs) 314.1 and 314.3, which are specific for the β-propeller domain of αIIb integrin and closely mimic the behavior of antibodies found in patients with quinine-induced thrombocytopenia in vitro9 and in vivo,10 have provided new tools for characterizing this interaction at a molecular level. In this report, we show that quinine binds with high affinity to a deep pocket in the complementarity-determining regions (CDRs) of these monoclonals. Previous crystallographic studies failed to demonstrate a binding site for quinine in the αIIbβ3 head structure.11 We propose that quinine-induced structural modifications of mAbs 314.1 and 314.3 result in high-affinity binding to the epitope they recognize on the αIIb integrin. Quinine selects specific CDR loop conformations and becomes part of the antibody paratope to form a hybrid paratope. This previously undescribed mechanism for DDAb-target interaction could have implications for other types of drug sensitivity.
Methods
Monoclonal antibody and Fab generation
Quinine-dependent murine mAbs 314.1 and 314.3 were described previously.9 Briefly, ascites fluid containing mAb was diluted with protein G binding buffer (Thermo Scientific) and loaded onto a protein G column, which was washed with protein G binding buffer. Immunoglobulin G (IgG) was eluted with protein G elution buffer (Thermo Scientific). After exchange into Tris-buffered saline, IgG was digested by papain at a mass ratio of 1:30 (papain:IgG) in the presence of 10 mM EDTA and 10 mM cysteine at 37°C for 16 hours. The mixture was passed through a protein A column and subjected to a Superdex 200 size exclusion chromatography in 20 mM Tris HCl (pH 7.5) and 150 mM NaCl.
Integrin αIIbβ3 headpiece generation
The αIIbβ3 headpiece construct contains residues 1 to 600 of αIIb and 1 to 472 of β3. The αIIb headpiece was fused with a tobacco etch virus (TEV) protease site, basic coiled-coil, and His6 tag at the C terminus and then inserted into the pEF1 vector with the puromycin resistance gene. The β3 headpiece was fused C terminally with a TEV protease site, acidic coiled-coil, and StrepII tag and inserted into the pcDNA3.1 vector with the hygromycin resistance gene.12 The αIIbβ3 headpiece construct was stably expressed in HEK293 GNT1−/− cells13 cultured in Dulbecco’s modified Eagle medium (CELLGRO) with 10% serum. Culture supernatant (2 L) was concentrated to 200 mL by tangential flow filtration and buffer exchanged into 20 mM Tris HCl (pH 7.5) and 500 mM NaCl. The αIIbβ3 headpiece was purified by a Ni-NTA matrix column (QIAGEN) followed by a Strep-Tactin column (IBA, St. Louis, MO). The yield was ∼3.0 mg/L. To remove the coiled-coil and tags, material was digested at a mass ratio of 1:3 (TEV protease: αIIbβ3 headpiece) in 20 mM Tris HCl (pH 8.0) and 150 mM NaCl, plus 1 mM MgCl2 and 1 mM CaCl2, at room temperature for 16 hours. The αIIbβ3 headpiece after TEV protease digestion was passed through a Ni-NTA column and then finally subjected to Superdex 200 chromatography in 20 mM Tris HCl (pH 7.5), 150 mM NaCl, 1 mM MgCl2, and 1 mM CaCl2.
Crystallization and crystallography
Crystallization was at 20°C by hanging-drop vapor diffusion with 1:1 vol/vol of protein and crystallization solutions. Fab 314.1 was crystallized in 15% PEG 6000, 10 mM MgCl2, and 50 mM KCl. Fab 314.1 with quinine was crystallized in 20% PEG 4000, 150 mM ammonium sulfate, and 100 mM HEPES (pH 7.0). Fab 314.3 was crystallized in 40% PEG 400, 200 mM MgCl2, and 100 mM sodium citrate (pH 5.5). Fab 314.3 with quinine was crystallized in 25% PEG 1500. Crystals were transferred to stabilization solutions, ie, corresponding crystallization solutions with a 5% higher PEG concentration.
Glycerol was added as the cryoprotectant in 5% increments up to a 20% final concentration, and crystals were vitrified in liquid nitrogen. X-ray diffraction data were collected at APS GM/CA CAT ID-23. The structure of Fab 314.1 was solved by molecular replacement by PHASER14 using the structure of Fab 10E515 as the search model. The structures of Fab 314.1 with quinine, Fab 314.3, and Fab 314.3 with quinine were solved by molecular replacement using the crystal structure of Fab 314.1 as the search model. All structural refinement was with PHENIX.REFINE.16
Gel filtration
The integrin αIIbβ3 headpiece was incubated with Fab 314.1 in the presence of 0 or 1 mM quinine on ice for 15 minutes and subjected to Superdex 200 chromatography in Tris-buffered saline with 0 or 0.1 mM quinine, respectively.
Isothermal titration calorimetry
Isothermal titration calorimetry experiments were carried out using a MicroCal iTC200 (GE Healthcare). Briefly, 200 µM quinine solution was titrated into 20 µM IgG 314.1 solution (40 µM H+L) with 2.4 µL per injection after an initial injection of 0.4 µL. The interval between injections was 150 s. A total of 100 µM quinine solution was titrated into 8 µM IgG 314.3 solution (16 µM H+L) with 4.98 µL per injection after an initial injection of 0.4 µL. The interval between injections was 180 s. Quinine and IgGs were in 20 mM Tris HCl (pH 7.5), 150 mM NaCl, 1 mM CaCl2, and 1 mM MgCl2. Heats were analyzed by the manufacturer’s software.
Results
Structures of the quinine-dependent Fabs and their complexes with quinine
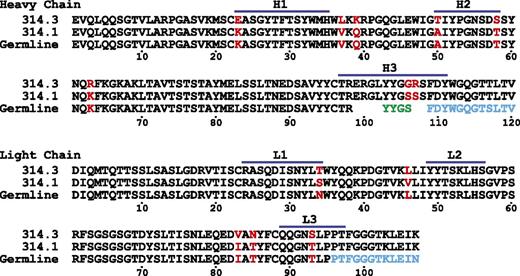
Crystals of the quinine-dependent Fabs and their complexes with quinine diffracted to resolutions ranging from 2.0 to 2.85 Å (Table 1). All have 1 molecule per asymmetric unit except the crystal of Fab 314.3 alone, which has 4 molecules per asymmetric unit. Each Fab contains four immunoglobulin domains, ie the heavy-chain variable (VH) domain and constant domain and light-chain variable (VL) and constant domains. Fab 314.1 and Fab 314.3 are closely related to one another with 5 CDR loop differences in VH, 2 CDR loop differences in VL, and additional framework differences (Figure 1).9 In Fab-quinine complex structures, electron densities for quinine were evident after 1 round of rigid body refinement. Quinine binds in a tightly fitting pocket formed by CDR loops H1, H2, H3, and L3 (Figure 2).
Crystallographic statistics
| . | Fab314.1 . | Fab314.1 + quinine . | Fab314.3 . | Fab314.3 + quinine . |
|---|---|---|---|---|
| Wavelength (Å) | 0.97936 | 0.97946 | 1.00695 | 1.03321 |
| Space group | P312 | P41212 | P21 | P21 |
| Cell | ||||
| a, b, c (Å) | 73.8,73.8,190.3 | 62.3,62.3,232.9 | 67.3,145.2,101.5 | 54.2,55.5,72.2 |
| α, β, γ (°) | 90.0,90.0,120.0 | 90.0, 90.0, 90.0 | 90.0, 90.9, 90.0 | 90.0, 99.0, 90.0 |
| Resolution (Å) | 50 - 2.0 | 50 - 2.85 | 50 - 2.7 | 50 - 2.5 |
| Reflections (total/unique) | 221 231/40 094 (10 840/2,412)* | 57 965/10 563 (4,865/816) | 218 092/53 487 (15 958/3,905) | 53 941/14 747 (3,050/1,025) |
| I/σ | 18.2 (8.7) | 17.2 (2.6) | 7.4 (0.9) | 9.6 (1.5) |
| CC1/2 (%) | 99.8 (97.0) | 99.9 (91.6) | 97.5 (36.4) | 98.9 (58.2) |
| Completeness (%) | 96.6 (80.4) | 92.6 (100) | 99.9 (100) | 99.3 (95.6) |
| Rmerge (%)† | 6.4 (16.9) | 6.4 (84.8) | 25.5 (174) | 13.0 (76.3) |
| Refinement | ||||
| Rwork‡ | 0.188 | 0.247 | 0.222 | 0.196 |
| Rfree§ | 0.219 | 0.304 | 0.260 | 0.248 |
| RMSD | ||||
| Bond (Å) | 0.007 | 0.004 | 0.005 | 0.003 |
| Angle (°) | 1.149 | 0.856 | 1.018 | 0.882 |
| MolProbity percentile | 99th | 99th | 97th | 98th |
| PDB code | 4UIK | 4UIL | 4UIM | 4UIN |
| . | Fab314.1 . | Fab314.1 + quinine . | Fab314.3 . | Fab314.3 + quinine . |
|---|---|---|---|---|
| Wavelength (Å) | 0.97936 | 0.97946 | 1.00695 | 1.03321 |
| Space group | P312 | P41212 | P21 | P21 |
| Cell | ||||
| a, b, c (Å) | 73.8,73.8,190.3 | 62.3,62.3,232.9 | 67.3,145.2,101.5 | 54.2,55.5,72.2 |
| α, β, γ (°) | 90.0,90.0,120.0 | 90.0, 90.0, 90.0 | 90.0, 90.9, 90.0 | 90.0, 99.0, 90.0 |
| Resolution (Å) | 50 - 2.0 | 50 - 2.85 | 50 - 2.7 | 50 - 2.5 |
| Reflections (total/unique) | 221 231/40 094 (10 840/2,412)* | 57 965/10 563 (4,865/816) | 218 092/53 487 (15 958/3,905) | 53 941/14 747 (3,050/1,025) |
| I/σ | 18.2 (8.7) | 17.2 (2.6) | 7.4 (0.9) | 9.6 (1.5) |
| CC1/2 (%) | 99.8 (97.0) | 99.9 (91.6) | 97.5 (36.4) | 98.9 (58.2) |
| Completeness (%) | 96.6 (80.4) | 92.6 (100) | 99.9 (100) | 99.3 (95.6) |
| Rmerge (%)† | 6.4 (16.9) | 6.4 (84.8) | 25.5 (174) | 13.0 (76.3) |
| Refinement | ||||
| Rwork‡ | 0.188 | 0.247 | 0.222 | 0.196 |
| Rfree§ | 0.219 | 0.304 | 0.260 | 0.248 |
| RMSD | ||||
| Bond (Å) | 0.007 | 0.004 | 0.005 | 0.003 |
| Angle (°) | 1.149 | 0.856 | 1.018 | 0.882 |
| MolProbity percentile | 99th | 99th | 97th | 98th |
| PDB code | 4UIK | 4UIL | 4UIM | 4UIN |
RMSD, root mean square deviation; PDB, Protein Data Bank.
Numbers in parentheses correspond to the last-resolution shell.
Rmerge = ∑h∑i|Ii(h) − < I(h) > |/∑h∑i|Ii(h)I, where Ii(h) and < I(h) > are the ith and mean measurement of the intensity of reflection h.
Rwork = ∑h||Fobs (h)| − |Fcalc (h)||/∑h|Fobs (h)|, where Fobs (h) and Fcalc (h) are the observed and calculated structure factors, respectively. No I/σ(I) cutoff was applied.
Rfree is the R value obtained for a test set of 1000 randomly selected reflections excluded from refinement.
Alignment of 314.1 and 314.3 V domains and corresponding germline sequences. Residues that differ between 314.1 and 314.3 are in red. Segments derived from the D and J regions are colored in green and cyan, respectively.
Alignment of 314.1 and 314.3 V domains and corresponding germline sequences. Residues that differ between 314.1 and 314.3 are in red. Segments derived from the D and J regions are colored in green and cyan, respectively.
Crystal structures of Fabs 314.1 and 314.3 and their complexes with quinine. View of the antigen-binding site of Fab 314.1 and its quinine complex (A,C,E,G) and Fab 314.3 and its quinine complex (B,D,F,H) in identical orientations. Quinine is shown in stick with carbon atoms wheat, oxygens red, and nitrogens blue (C,D) or as a wheat surface (G,H). CDR loops are colored red (H3), blue (L3), yellow (H2 and L2), and green (H1 and L1). The Fab solvent-accessible surface is transparent in panels A-D, revealing CDR loops and framework (white) shown in cartoon. In panels E-H, the solvent-accessible surfaces include quinine when present and are opaque to better show the marked differences in topography of the surfaces that occur because of both quinine binding and CDR H3 and L3 loop backbone reorientation.
Crystal structures of Fabs 314.1 and 314.3 and their complexes with quinine. View of the antigen-binding site of Fab 314.1 and its quinine complex (A,C,E,G) and Fab 314.3 and its quinine complex (B,D,F,H) in identical orientations. Quinine is shown in stick with carbon atoms wheat, oxygens red, and nitrogens blue (C,D) or as a wheat surface (G,H). CDR loops are colored red (H3), blue (L3), yellow (H2 and L2), and green (H1 and L1). The Fab solvent-accessible surface is transparent in panels A-D, revealing CDR loops and framework (white) shown in cartoon. In panels E-H, the solvent-accessible surfaces include quinine when present and are opaque to better show the marked differences in topography of the surfaces that occur because of both quinine binding and CDR H3 and L3 loop backbone reorientation.
Quinine selects a markedly different conformation of the CDR H3 loop than that seen in absence of quinine for both Fab 314.1 and 314.3 (Figure 2A-D).
Fab-quinine interactions
Quinine, with a molecular mass of 324.4 Da, consists of fused aromatic and alkyl rings (Figure 3). The quinoline heterocyclic aromatic ring is fused to the quinuclidine, bicyclic, alkylamine ring (labeled in Figure 3C-D). Quinuclidine is a strong base with a pKa of 11.0.17 Quinine has extensive interactions with both Fab 314.1 (Figure 3A,C,E) and 314.3 (Figure 3B,D,F). The binding pocket is composed of a floor and surrounding wall. The floors, almost identical in the 2 Fabs, consist of Gln89 and Pro96 in CDR L3, His35 in CDR H1, Trp47 (heavy-chain framework), Ala/Thr50 in H2, and Glu99 and Phe109 in H3. The quinine-binding pocket is mostly hydrophobic, except for the negatively charged Glu99.
Details of quinine binding. Quinine complexes with Fab 314.1 (A) and Fab 314.3 (B). Quinine is shown in thicker stick with wheat carbons; Fab CDR loops and framework are shown in thinner stick with silver carbons. Oxygens are red and nitrogens are blue. Fabs are shown with white transparent surfaces. 2Fo-Fc quinine omit map density is shown as magenta mesh contoured at 3σ. Interactions between quinine and Fab 314.1 (C) and Fab 314.3 (D) shown schematically with Maestro (Schrodinger, New York, NY). Hydrophobic residues are shown in green, hydrophilic residues are cyan, positively charged residues are violet, negatively charged residues are pink, and glycine residues are white. Stripes show backbone contact regions. Arrows show hydrogen bonds. Green line shows π-π stacking. Red-blue line shows a salt bridge. Red curved lines show π-cation interaction. Shadows on atoms in quinine schematize their amount of exposure to solvent. Comparisons for Fab 314.1 (E) and Fab 314.3 (F) with and without quinine. Carbons for the Fabs are shown in silver (no quinine) and in cyan (with quinine).
Details of quinine binding. Quinine complexes with Fab 314.1 (A) and Fab 314.3 (B). Quinine is shown in thicker stick with wheat carbons; Fab CDR loops and framework are shown in thinner stick with silver carbons. Oxygens are red and nitrogens are blue. Fabs are shown with white transparent surfaces. 2Fo-Fc quinine omit map density is shown as magenta mesh contoured at 3σ. Interactions between quinine and Fab 314.1 (C) and Fab 314.3 (D) shown schematically with Maestro (Schrodinger, New York, NY). Hydrophobic residues are shown in green, hydrophilic residues are cyan, positively charged residues are violet, negatively charged residues are pink, and glycine residues are white. Stripes show backbone contact regions. Arrows show hydrogen bonds. Green line shows π-π stacking. Red-blue line shows a salt bridge. Red curved lines show π-cation interaction. Shadows on atoms in quinine schematize their amount of exposure to solvent. Comparisons for Fab 314.1 (E) and Fab 314.3 (F) with and without quinine. Carbons for the Fabs are shown in silver (no quinine) and in cyan (with quinine).
In Fab 314.1, the wall is made of CDR loops L3, H3, Trp33 in H1, and S59 in H2. Strong hydrophobic interaction exists between the bicyclic quinuclidine and the aromatic ring of Trp33 in CDR H1. Hydrophobic interactions also occur between the aromatic quinoline and the side chains of Leu94 and Pro96 in L3 and between the methoxy group attached to quinoline and the aromatic ring of Phe109 in H3. Furthermore, 2 charged interactions are prominent in the pocket of both 314.1 and 314.3. Strong salt bridge and hydrogen bonds link the negatively charged Glu99 in H3 to the positively charged nitrogen atom in the quinuclidine. The second charged interaction is a π-cation bond between the same positively charged nitrogen atom in the quinuclidine ring and the aromatic ring of Trp33 in H1 (Figure 3A,C).
The interactions between quinine and Fab 314.3 are similar, with a few key differences (Figure 3B,D). The S107R substitution in H3 creates a strong π-π stacking interaction between the guanidine group of Arg107 and the quinoline aromatic ring (Figure 3B). As a consequence, the quinoline moves toward residue 107 in 314.3 compared with 314.1 and loses its interaction with the L3 wall, which moves away from quinine in 314.3, compared with 314.1. L3 wall repositioning also occurs in quinine-bound 314.3 compared with free 314.3 (Figure 3F). L3 repositioning may in part be driven to accommodate torsion of the L3 backbone at the peptide bond between N92 and T93, so its carbonyl oxygen can hydrogen bond to a strongly bound water that bridges to the quinoline nitrogen (Figure 3B). Quinoline ring repositioning weakens the hydrophobic interaction between the methoxy group of quinine and the aromatic ring of Phe109 in H3. To compensate for the loss of the intimate contact of the CDR L3 main chain, the side chain of Leu94 in L3 moves closer to form a stronger hydrophobic interaction with the quinoline ring (compare Figure 3B with Figure 3A). At the same time, quinuclidine also moves closer to Glu99, resulting in stronger salt bridge and hydrogen bonds between negatively charged Glu99 and the positively charged quinuclidine nitrogen atom (compare Figure 3B with Figure 3A). The A50T substitution in H2 forces the vinyl group attached to the quinuclidine ring to change rotamer and now face L3 residue Leu94 (Figure 3A-D).
The presence of the Arg107 substitution and repositioning of Leu94 in 314.3 narrows the quinine-binding pocket and confers better shape complementarity for quinine (Figure 3A-B and Table 2). The Fab 314.3 heavy chain also has a larger interface with quinine as measured by buried surface accessible area (Table 2).18,19 The overall result of the CDR loop substitutions, reshaping of the binding pocket, and repositioning of quinine is a twofold higher binding affinity in mAb314.3 as described below. In both cases, quinine forms strong interactions with the Fab, with more than 87% of its own surface buried in the binding pocket. All of the exposed atoms of quinine, except for one oxygen atom, are hydrophobic (Figure 2C-D). The solvent-exposed portion of quinine putatively contributes to the αIIbβ3-binding surface. The 13% of quinine exposed corresponds to short edges of the quinoline and quinuclidine rings (Figure 3A-B). This surface is hydrophobic, except for the hydroxyl group in the link between the 2 rings.
Quinine-Fab interface characteristics
| . | Fab 314.1 . | Fab 314.3 . |
|---|---|---|
| . | Solvent-accessible surface area (Å2)* . | |
| Quinine total | 502 | 509 |
| Buried on quinine | 440 (87.6%) | 452 (88.8%) |
| Buried on H chain | 154 | 202 |
| Buried on L chain | 127 | 128 |
| Buried total on Fab | 281 | 330 |
| Shape complementarity index† | 0.764 | 0.809 |
Conformational change of Fab CDR loops
Quinine binding requires substantial reorientation of Fab314.1 and Fab 314.3 CDR H3 loops and minor reorientiation of L3 loops (Figure 3E-F). The 314.3 H3 loop is in a crystal lattice contact, whereas the 314.1 H3 loop is not in a contact and therefore may be closer to the low-energy conformation of the H3 loop in solution. In absence of quinine, the 2 H3 loops move in opposite directions relative to the quinine-bound conformation: the 314.1 H3 loop moves into the quinine-binding cavity (Figure 3E), whereas the 314.3 H3 loop moves away from the cavity (Figure 3F).
The markedly different orientations for the 314.1 and 314.3 H3 loops in absence of quinine suggest that in absence of quinine, residues 100 to 107 of the H3 loop are highly flexible. In contrast, these residues adopt nearly identical orientations in presence of quinine. The restraint imposed by quinine relates to its binding to residues Tyr104, Gly105, Gly106, Arg or Ser107, and Ser108 (Figure 3C-D) in the flexible portion of the H3 loops, which form one wall of the quinine-binding site. Thus, quinine binding selects a single conformation of the H3 loop. Quinine binding thus constructs unique conformations for the Fab 314.1 and 314.3 paratopes and becomes part of the paratope itself.
Binding between quinine, quinine-dependent mAbs, and integrin αIIbβ3
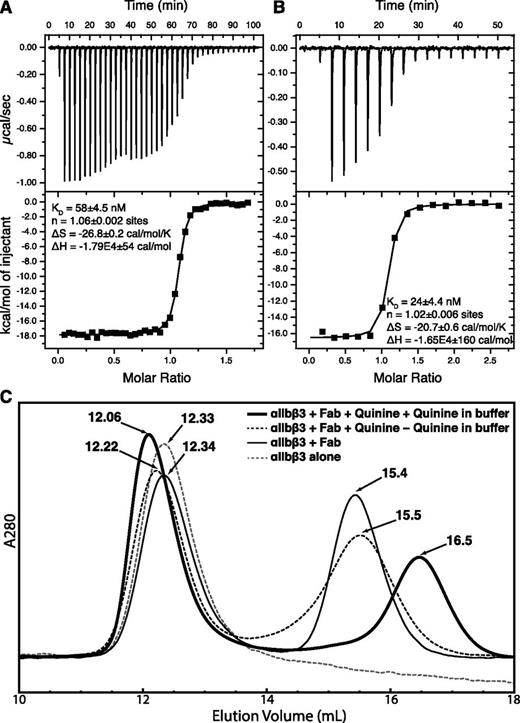
Isothermal titration calorimetry was used to determine the thermodynamics of quinine binding to IgG. Data are expressed in terms of binding site (H- and L-chain concentration) in Figure 4A-B. Close to 1 molecule of quinine (n of 1.06 or 1.02) bound per binding site. Binding is exothermic, driven by large ΔH values of −16.5 to −17.9 kcal/mol. Correlating with its CDR substitutions described above, 314.3 bound quinine more tightly (KD = 24 nM) than 314.1 (KD = 58 nM).
Quinine-Fab binding. (A-B) Isothermal titration calorimetry. Titrations of quinine into 314.1 (A) and 314.3 (B) IgG in the calorimetry cell are as described in “Methods.” Because the 2 binding sites in IgG bind quinine independently, the results are expressed in terms of the concentration of half of an IgG, ie, the concentration of heavy and light chains. (C) Gel-filtration chromatography of the αIIbβ3 headpiece, with or without Fab 314.1, with and without quinine, and with and without quinine in the running buffer.
Quinine-Fab binding. (A-B) Isothermal titration calorimetry. Titrations of quinine into 314.1 (A) and 314.3 (B) IgG in the calorimetry cell are as described in “Methods.” Because the 2 binding sites in IgG bind quinine independently, the results are expressed in terms of the concentration of half of an IgG, ie, the concentration of heavy and light chains. (C) Gel-filtration chromatography of the αIIbβ3 headpiece, with or without Fab 314.1, with and without quinine, and with and without quinine in the running buffer.
The quinine-dependent mAbs studied here only bind to cell-surface integrin αIIbβ3 in the presence of quinine.9 In agreement, in absence of quinine, the integrin αIIbβ3 headpiece eluted in the same position in gel filtration in the absence and presence of Fab 314.1 (12.33 and 12.34 mL; Figure 4C). In contrast, when the headpiece, Fab, and quinine were incubated with one another, and quinine was present in running buffer, earlier elution (12.06 mL; Figure 4C) demonstrated complex formation. If headpiece, Fab, and quinine were incubated with one another, but quinine was omitted from running buffer, partial Fab dissociation occurred during gel filtration, as shown by the intermediate elution position of 12.22 mL (Figure 4C).
Discussion
Here, we show how quinine binds to 2 Fabs, remodels their paratopes, and enables binding to the platelet integrin αIIbβ3. Our study provides a model of how drug binding may reprogram the specificity of an antibody and provoke DITP (Figure 5). Conformational changes in the CDR loops of the 2 Fabs studied here are required for quinine binding. Quinine either induces conformational changes of CDR loops by fitting between them or selects among preexisting conformations in equilibrium for the conformation that can bind. Either way, the resulting binding pockets fit quinine with high shape complementarity. Although the 314.1 and 314.3 CDR H3 loops differ slightly in sequence, they have almost identical conformations when bound to quinine. The differences in conformation between the 314.1 and 314.3 H3 loops in absence of quinine, and between the H3 loops in absence of quinine and presence of quinine, demonstrate marked flexibility of residues 100 to 107 of the CDR H3 loop.
Models for DDAb binding to a cell-surface molecule. (A) The sandwich model. (B) The hybrid paratope model. The antigen could represent any cell-surface molecule, including integrin αIIbβ3.
Models for DDAb binding to a cell-surface molecule. (A) The sandwich model. (B) The hybrid paratope model. The antigen could represent any cell-surface molecule, including integrin αIIbβ3.
We examined canonical structures and length distributions for CDR loops20 to determine if 314.1 and 314.3 have any unusual features. The structures of many CDR can be predicted from their sequences; these are known as canonical CDR structures. 314.1 and 314.3 are identical in CDR loop length and canonical portions of their CDR loops; therefore, we discuss them together as “314.” L3 loops vary in length from 7 to 13 residues; most have 9 residues, as is the case with Fab 314 (Figure 1). The L3 loop of 314 belongs to canonical cluster L3-9-cis7-1, which is characterized by a cis-proline at position 7, ie, cis-Pro95 in Fab 314. The entire length of this loop is canonical in structure. CDR H3 loops range from 5 to 26 residues in length, with most from 6 to 18 residues. The length of 314 CDR H3 is 15 residues (Figure 1), and like 92% of H3 loops of this length, it belongs to cluster H3-anchor-1.20 However, only the first 3 and last 4 residues of H3 loops have canonical structures; ie, structures that are predictable from their sequence. The canonical portions of the H3 loops of unbound and quinine-bound 314.1 and 314.3 have identical, canonical structures. Most interestingly, the flexible portion of their H3 loops corresponds precisely to the 8 noncanonical residues, 100 to 107.
The previous 50% effective concentration (EC50) value for bivalent, quinine-dependent binding of 314.1 IgG to platelets is ∼5 nM.9 The monovalent KD values for quinine binding to 314.1 and 314.3 IgG measured in this study are 58 nM and 24 nM, respectively. In a previous study in which crystals of the integrin αIIbβ3 headpiece were soaked with 0.2 mM quinine, no electron density representing quinine could be identified in the crystal structure.11 Together, the findings argue that quinine binds to the antibodies first and the quinine-antibody complexes then recognize integrin αIIbβ3.
Quinine and its congeners cinchonidine, quinidine, and cinchonine showed similar variation in potency in triggering antibody binding to platelets for 314.1 and for 2 patients with quinine-dependent DITP.9 Although human DDAbs require ∼1000-fold higher concentrations of quinine than 314.1 for binding to platelets, the similar variation in potency among congeners indicates qualitative similarity among 314.1, 314.3, and DITP patient antibodies. Our structures explain the order of potency (quinine > cinchonidine > quinidine = cinchonine) for DDAb binding. Cinchonidine has a structure identical to quinine, except it lacks the methoxy group attached to the quinoline ring. This methoxy group forms strong hydrophobic interactions with the aromatic ring of CDR H3 residue Phe109 in both Fab 314.1 and 314.3 (Figure 3). Missing this important interaction explains the lower EC50 for cinchonine stimulation of mAb 314.1 binding to platelets.9 Quinidine and cinchonine differ from quinine in the chirality of both carbons 8 and 9 (see Figure 3C-D) and therefore have different 3-dimensional structures with a different orientation between the quinoline and quinuclidine rings. These differences explain the much lower EC50 values for stimulation of antibody binding to platelets for both quinidine and cinchonine, which is identical to quinidine but lacks the methoxy group. A previous sandwich model proposed that structural elements of quinine contact both antibody and target protein to increase the Ka for antibody binding (Figure 5A).1,8 Our findings show that 88% to 89% of the quinine surface is buried by the 314.1 and 314.3 Fabs, leaving only 11% to 12% available for binding to integrin αIIbβ3. Furthermore, quinine binding alters the conformation of the CDR H3-loop markedly and the CDR L3 loop modestly.
“Hybrid paratope” may be an apt term for the type of DDAb recognition revealed here in which the drug interacts so intimately with antibody (Figure 5B). In the unbound 314.1 and 314.3 Fab conformations, the antigen-binding site is highly concave (Figure 2E-F). Concave antibody binding sites are common for binding small ligands such as haptens. Quinine binding reconfigures the CDR H3 and L3 loops. Moreover, quinine essentially becomes part of the antibody surface, ie, the paratope that binds the epitope. The hybrid paratope surface is irregular but is overall planar, as generally seen with antibodies that recognize proteins.21 The paratope is hybrid in the sense both that it is a complex with a foreign drug molecule and that its CDR loops are reshaped when a drug binds. Although the studies described here were necessarily conducted with murine monoclonal antibodies, the close similarity between in vitro and in vivo behavior of mAbs 314.1 and 314.3 and their human counterparts9,10 and previous studies with human quinine- and quinidine-dependent DDAbs hinting at the possibility that the drug may react first with antibody to promote binding to its target8,22-24 favor the possibility that specific binding of human DDAbs results from drug-induced remodeling of antibody CDR similar to that described here for monoclonals 314.1 and 314.3.
Many forms of drug sensitivity are initiated by binding of a drug, usually via covalent linkage, to an autologous protein, rendering it capable of triggering an immune response.25,26 Our findings suggest that, at least in the case of antibodies that cause drug-dependent thrombocytopenia, the first step in sensitization could be fortuitous binding of drug to a B-cell receptor (BCR) that is very weakly autoreactive7,27 and remodeling of its CDR so that it binds with high affinity to an autologous protein expressed on platelets. In some circumstances, eg, in an inflammatory state, proliferation and affinity maturation of such a B cell could lead to expression of antibodies capable of binding to and destroying platelets when the drug is present. Thus, drug-induced immunologic injury could begin with chance recognition by BCR of a drug that modifies CDR structure and specificity rather than through linkage of a chemically reactive form of the drug to an autologous protein to create a haptenic determinant, widely thought to be the starting point for most “idiosyncratic” drug-sensitivity reactions.28 Many types of drug sensitivity are T-cell mediated.26 Because the T-cell receptor is structurally homologous to BCR, it can be speculated that a similar mechanism could trigger proliferation of drug-dependent T cells capable of causing immune injury when a drug is present. Further studies are indicated to define the extent to which direct binding of a drug to BCR or T-cell receptor initiates sensitivity reactions that have pathologic consequences.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
This work was supported by grants from the National Institutes of Health, National Heart, Lung, and Blood Institute (HL-103526 and HL-13629).
Authorship
Contribution: Jianghai Zhu and Jieqing Zhu performed research; T.A.S. designed research; Jianghai Zhu and T.A.S. analyzed data and wrote the paper; and D.W.B. and R.H.A. oversaw development of monoclonals 314.1 and 314.3, consulted on studies while they were in progress, and assisted in manuscript preparation and editing.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
The current affiliation for Jianghai Zhu is Frederick National Laboratory for Cancer Research, National Institutes of Health, National Cancer Institute, Frederick, MD. The current affiliations for Jieqing Zhu are Blood Research Institute, BloodCenter of Wisconsin, Milwaukee, WI, and Department of Biochemistry, Medical College of Wisconsin, Milwaukee, WI.
Correspondence: Timothy A. Springer, Boston Children’s Hospital, 3 Blackfan Circle, Room 3103, Boston, MA 02115; e-mail: timothy.springer@childrens.harvard.edu.