Key Points
Using genetically modified mice, we identified the crucial role of Id1 in t(8;21) leukemogenesis through regulating AKT signaling.
Id1 inhibitor has a significant therapeutic effect in the mouse model of t(8;21) leukemia.
Abstract
Transcriptional regulators are recurrently altered through translocations, deletions, or aberrant expression in acute myeloid leukemia (AML). Although critically important in leukemogenesis, the underlying pathogenetic mechanisms they trigger remain largely unknown. Here, we identified that Id1 (inhibitor of DNA binding 1) plays a pivotal role in acute myeloid leukemogenesis. Using genetically modified mice, we found that loss of Id1 inhibited t(8;21) leukemia initiation and progression in vivo by abrogating protein kinase B (AKT)1 activation, and that Id1 interacted with AKT1 through its C terminus. An Id1 inhibitor impaired the in vitro growth of AML cells and, when combined with an AKT inhibitor, triggered even greater apoptosis and growth inhibition, whereas normal hematopoietic stem/progenitor cells were largely spared. We then performed in vivo experiments and found that the Id1 inhibitor significantly prolonged the survival of t(8;21)+ leukemic mice, whereas overexpression of activated AKT1 promoted leukemogenesis. Thus, our results establish Id1/Akt1 signaling as a potential therapeutic target in t(8;21) leukemia.
Introduction
An important impetus for identifying new pathways that drive cancer is the opportunity to inhibit these pathways instead of, or in addition to, conventional cancer therapies. This is especially true for acute myeloid leukemia (AML), a disease primarily affecting older individuals, for whom few therapies are capable of eradicating the disease. AML is the most common acute leukemia affecting adults,1 and standard therapy for AML is highly toxic and poorly tolerated in older patients.2 The development of novel therapeutics in AML is currently based on exploiting the newly understood pathophysiological events that are critical for maintaining leukemia cell proliferation and/or survival.3 In acute leukemia, transcriptional regulators are often altered through aberrant expression4 ; these abnormal transcriptional regulators play a critical role in leukemogenesis and represent potentially novel leukemia treatments. The inhibitor of DNA binding/differentiation (Id) family protein (Id1-4) is known to inhibit the activity of the E protein basic helix-loop-helix transcription factors (such as the E2A and transcription factor 12 [HEB]) and regulate their target genes by forming heterodimers with these proteins, blocking their ability to bind DNA.5,6 Id1 has been identified as a common downstream target of several constitutively activated oncogenic tyrosine kinases, such as FLT3/internal tandem duplication (ITD) and breakpoint cluster region (BCR)–Abelson murine leukemia viral oncogene homolog (ABL), and thus may represent a therapeutic target for leukemias associated with oncogenic tyrosine kinases.7 It has been shown that overexpression of Id1 immortalizes myeloid progenitors and leads to a myeloproliferative disease in vivo.8 High Id1 expression is associated with poor prognosis in AML, independently predicting for shorter disease-free survival and overall survival.9 In fact, high Id1 expression is seen in ∼60% of patients with M2-subtype AML, which includes patients with t(8;21), which generates the AML1-ETO fusion gene and protein. Thus, high Id1 expression may not only contribute to the initiation of AML but also represent a potential therapeutic target.
We, and others, have shown that Id1 is a key regulator of hematopoietic stem cell (HSC) behavior, as the absence of Id1 compromises the self-renewal capacity of HSCs in adult bone marrow and increases their tendency to differentiate toward the myeloid (vs the lymphoid) lineage.10,11 This impairment is associated with changes in gene expression, including the increased expression of p21, a well-established target of Id1-mediated repression.12 Id1 and Id3 are negative regulators of the transition of human pluripotent stem cells to a committed hematopoietic cell fate13 ; their expression may be required to maintain the immaturity needed for leukemia cell growth, as Id1 is frequently overexpressed in leukemia cells.14-16 However, the functional significance of this is not known.
We recently found that acetylation of AML1-ETO (and AML1-ETO9a) by p300 is required for the induction of acute leukemia in human and mouse AML models. ID1 promoter activity is upregulated by (lysine 43) acetylatable AML1-ETO (but not nonacetylatable AML1-ETO) in human hematopoietic stem/progenitor cells (HSPCs), and the Id1 promoter appears to be cooccupied by AML1-ETO and p300 in vivo.17 We developed an antitumor agent that downregulates Id1 in vivo; however, this inhibitor has not yet been adapted to penetrate myeloid cells,18 so it cannot target Id1 in AML1-ETO–driven AML. A second Id1 inhibitor, cannabidiol (CBD), has been reported to downregulate Id1 expression at both the messenger RNA (mRNA) and protein level.19 This agent has allowed us to better define the importance of Id1 in leukemia initiation, maintenance, and progression. Although ID1 inhibits the activity of E proteins, such as E2A and HEB,20 we have uncovered an unexpected but important regulatory role of ID1 in protein kinase B (AKT)1 signaling.
Materials and methods
See supplemental Materials and Methods (available on the Blood Web site) for additional methods.
Fetal liver transplantation
Fetal liver cells were harvested from embryonic day (E) 14.5 embryos. Subsequently, the E14.5 fetal liver cells were infected with retroviruses by spinoculation, with transduction efficacies of ∼10%. The fetal liver cells were cultured in X-VIVO medium with 10 ng/mL interleukin 3, 10 ng/mL interleukin 6, and 100 ng/mL stem cell factor. The efficiency of transduction by the various the enhanced green fluorescent protein-expressing retroviral vector (MIGR1)–based viruses was determined on the basis of green fluorescent protein (GFP) positivity, using flow cytometry. The C57Bl/6.SJL recipient mice were lethally irradiated with 950 cGy, given in a split dose separated by 4 hours. The transduced fetal liver cells were transplanted into recipient mice by tail-vein injection. Cell morphology was evaluated by Wright’s staining of cells prepared by cytospin centrifugation. Tissue samples were fixed in formaldehyde and further processed for paraffin embedding.17,21,22 To induce Cre activity, mice were treated with tamoxifen (160 mg/kg) orally per day for 5 days beginning 3 days posttransplantation; corn oil was used as the control.
Results
Deletion of Id1 abrogates leukemia initiation in vivo
The expression of the exon9a isoform of AML1-ETO (AE9a) in the mouse E14.5 fetal liver cells leads to the rapid development of leukemia, whose clinical characteristics are suggestive of AML without maturation.23,24 We transplanted lethally irradiated mice with AE9a transduced E14.5 fetal liver cells isolated from wild-type (WT) or Id1−/− mice and examined the peripheral blood cells of the recipient mice 15 weeks after transplantation. The white blood cell (WBC) counts of the mice in the AE9a/Id1−/− group are significantly lower than the AE9a/WT group, reflecting impaired leukemogenesis (Figure 1A). The peripheral blood and bone marrow cells of the mice in the AE9a/Id1−/− group contained far fewer GFP+, C-Kit+ leukemia blast cells (Figure 1C and supplemental Figure 3A-B), and more GFP−, Gr1+ (ie, normal) cells compared with the AE9a/WT group in peripheral blood (Figure 1C and supplemental Figure 3A). As we and others previously observed, AE9a leukemia blast cells lose CD45.2/CD45 cell surface expression, which is expressed on normal mouse peripheral blood cells (Figure 1C). Peripheral blood smears showed far fewer leukemic blast cells in the AE9a/Id1−/− group compared with the AE9a/WT group (Figure 1D), and there was less infiltration of blast cells in the spleen, with preservation of the normal architecture in the absence of Id1 (Figure 1E). Thus, loss of Id1 slowed the initiation of leukemia and markedly prolonged the median survival of the AE9a mice (267 days vs 137 days, Figure 1B). Moreover, deletion of Id1 did not affect the frequencies of Lin−Sca1+C-Kit+ (LSK), HSC, granulocyte-macrophage progenitors (GMP), common myeloid progenitors (CMP), or MEP cells (supplemental Figure 4A) nor the homing ability of the HSPCs present in E14.5 mouse fetal liver (supplemental Figure 4B), indicating that lack of Id1 function has little effect on the number of preleukemic stem or progenitor cells.
Deletion of Id1 abrogates the initiation of leukemia in the AE9a fetal liver transplantation model. (A) Lethally irradiated recipient mice were injected with WT or Id1−/− mouse E14.5 fetal liver cells transduced with AE9a. The WBC counts of the transplanted mice in Id1−/− AE9a group are significant lower than the WT AE9a mice group 15 weeks after transplant (P = .0288). (B) Loss of Id1 prolongs the survival time of recipient mice (267 days vs 137 days, n = 15 per group, P < .001). (C) Loss of Id1 decreases the frequency of GFP+CD45/CD45.2−C-Kit+ leukemia blast cells and increases the frequency of normal GFP−Gr1+ myeloid cells in the peripheral blood of recipient mice (compared with the WT group) 15 weeks after transplantation. (D) The peripheral blood shows less leukemia blast cells in the Id1−/− AE9a group compared with the WT AE9a mice group. (E) Pathological sections of the spleen stained with hematoxylin-eosin are shown 15 weeks after transplantation (the scale bar indicates 25 μM). The leukemic blast cells are indicated by black arrows. (F) The columns represent the numbers of colonies in each plating of WT or Id1−/− E14.5 fetal liver cells transduced with AE9a (± standard deviation [SD], n = 3, P < .01). (G) The frequency of donor-derived Id1−/− CD45.2+ cells is significantly lower in the peripheral blood of recipient mice, compared with the WT CD45.2+ cells (P < .01). (H) Loss of Id1 abrogates the repopulating ability of hematopoietic cells isolated from E14.5 fetal liver cells.
Deletion of Id1 abrogates the initiation of leukemia in the AE9a fetal liver transplantation model. (A) Lethally irradiated recipient mice were injected with WT or Id1−/− mouse E14.5 fetal liver cells transduced with AE9a. The WBC counts of the transplanted mice in Id1−/− AE9a group are significant lower than the WT AE9a mice group 15 weeks after transplant (P = .0288). (B) Loss of Id1 prolongs the survival time of recipient mice (267 days vs 137 days, n = 15 per group, P < .001). (C) Loss of Id1 decreases the frequency of GFP+CD45/CD45.2−C-Kit+ leukemia blast cells and increases the frequency of normal GFP−Gr1+ myeloid cells in the peripheral blood of recipient mice (compared with the WT group) 15 weeks after transplantation. (D) The peripheral blood shows less leukemia blast cells in the Id1−/− AE9a group compared with the WT AE9a mice group. (E) Pathological sections of the spleen stained with hematoxylin-eosin are shown 15 weeks after transplantation (the scale bar indicates 25 μM). The leukemic blast cells are indicated by black arrows. (F) The columns represent the numbers of colonies in each plating of WT or Id1−/− E14.5 fetal liver cells transduced with AE9a (± standard deviation [SD], n = 3, P < .01). (G) The frequency of donor-derived Id1−/− CD45.2+ cells is significantly lower in the peripheral blood of recipient mice, compared with the WT CD45.2+ cells (P < .01). (H) Loss of Id1 abrogates the repopulating ability of hematopoietic cells isolated from E14.5 fetal liver cells.
To examine the effect of Id1 on the self-renewal of transformed E14.5 fetal liver cells, we performed serial replating assays using sorted GFP+ AE9a–transduced E14.5 fetal liver cells, isolated from WT or Id1−/− mice. Colony numbers were counted weekly, and the Id1−/− AE9a cells had less repopulating capacity than WT AE9a cells at weeks 3 and 4 (Figure 1F). To examine the role of Id1 in HSPC biology, we performed competitive transplantation assays, using E14.5 fetal liver cells isolated from either WT or Id1−/− mice. The Id1−/− HSPCs have less repopulating capacity than WT HSPCs (Figure 1G-H), with far fewer CD45.2+/Gr1+, CD45.2+/B220+, and CD45.2+/CD3e+ cells in the peripheral blood cells of the Id1−/− recipient mice (Figure 1G). These results suggest that Id1 controls the initiation of myeloid transformation by regulating the self-renewal properties of normal and transformed HSPCs.
Conditional deletion of Id1 inhibits leukemia progression in vivo
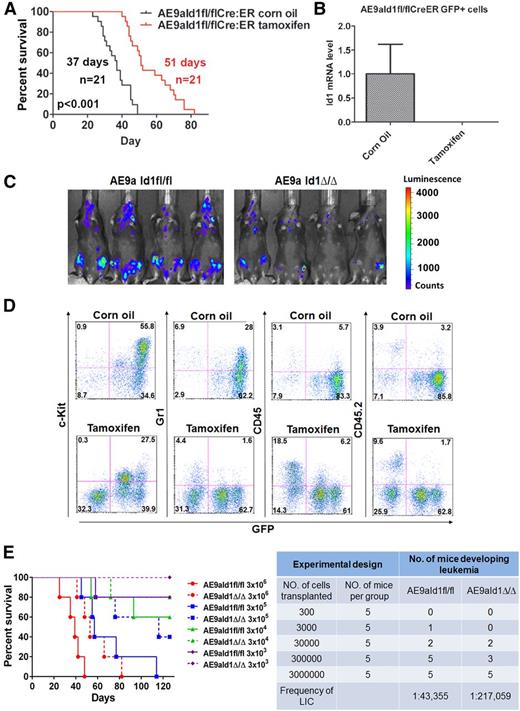
We transduced Cre-ER–expressing, Id1-floxed fetal liver cells with AE9a-expressing lentiviral vectors and transplanted them into lethally irradiated recipient mice. AE9aId1fl/flCreER leukemia cells were isolated from the spleens of these mice, and to determine whether Id1 is required for leukemia maintenance, we secondarily transplanted these leukemia cells into sublethally irradiated recipient mice and then induced the conditional deletion of Id1 by adding tamoxifen. The survival of AE9a AML–bearing mice, although shorter than the primary leukemia mice, was significantly prolonged by treatment with tamoxifen, compared with the vehicle-treated mice (Figure 2A). The in vivo treatment of mice transplanted with AE9aId1fl/flCreER cells with tamoxifen nearly eliminated Id1 expression in the AE9a-expressing GFP+ cells (Figure 2B). Similarly in vitro treatment of the GFP+bright, GFP+middle, and GFP+low populations of AE9aId1fl/flCreER cells with 0.1 µM 4-hydroxytamoxifen (4-OHT) for 3 days completely eliminated Id1 expression but did not affect Id1 expression or the survival time of the mice transplanted with AE9aId1+/+CreER cells (supplemental Figure 5A-B and data not shown).
Conditional deletion of Id1 inhibits leukemia progression in vivo. (A) Conditional deletion of Id1 by tamoxifen treatment significantly prolongs the survival time of recipient mice transplanted with AE9aId1fl/flCreER cells compared with the vehicle-treated (corn oil) group (51 days vs 37 days, n = 21, P < .01). (B) The expression of Id1 is minimal in the sorted GFP+ cells isolated from the bone marrow of the mice in the tamoxifen treatment group described in panel A. (C) In vivo luciferase imaging shows that deletion of Id1 by tamoxifen impairs leukemia progression in recipient mice transplanted with AE9aId1fl/flCreER/MSCV-luciferase cells (compared with the corn oil control group). (D) Conditional deletion of Id1 significantly decreases the frequencies of C-Kit+GFP+ leukemia blast cells and GFP+Gr1+ leukemia cells and increases the frequencies of normal GFP−CD45/CD45.2+ cells in the peripheral blood of mice transplanted with AE9aId1fl/flCreER cells. (E) Survival of mice receiving different dilutions of AE9a-transduced Id1fl/fl or Id1Δ/Δ fetal liver cells (n = 5 per group, left panel). Conditional deletion of Id1 significantly decreases the frequency of leukemia-initiating cells, in the limiting dilution assay (right panel).
Conditional deletion of Id1 inhibits leukemia progression in vivo. (A) Conditional deletion of Id1 by tamoxifen treatment significantly prolongs the survival time of recipient mice transplanted with AE9aId1fl/flCreER cells compared with the vehicle-treated (corn oil) group (51 days vs 37 days, n = 21, P < .01). (B) The expression of Id1 is minimal in the sorted GFP+ cells isolated from the bone marrow of the mice in the tamoxifen treatment group described in panel A. (C) In vivo luciferase imaging shows that deletion of Id1 by tamoxifen impairs leukemia progression in recipient mice transplanted with AE9aId1fl/flCreER/MSCV-luciferase cells (compared with the corn oil control group). (D) Conditional deletion of Id1 significantly decreases the frequencies of C-Kit+GFP+ leukemia blast cells and GFP+Gr1+ leukemia cells and increases the frequencies of normal GFP−CD45/CD45.2+ cells in the peripheral blood of mice transplanted with AE9aId1fl/flCreER cells. (E) Survival of mice receiving different dilutions of AE9a-transduced Id1fl/fl or Id1Δ/Δ fetal liver cells (n = 5 per group, left panel). Conditional deletion of Id1 significantly decreases the frequency of leukemia-initiating cells, in the limiting dilution assay (right panel).
To better monitor the in vivo growth of the AML cells, in real time, we used a bicistronic retroviral vector (MSCV-luciferase) to express the luciferase protein in the leukemia cells. The recipient mice, transplanted with AE9aId1fl/flCreER-luciferase–expressing leukemia cells, were treated with tamoxifen or vehicle control, as described previously, and imaged 17 days posttransplantation using a bioluminescence monitoring system. The AE9a-expressing AML cells are brightly luciferase positive, which allowed us to determine that the loss of Id1 impaired leukemia progression (Figure 2C). We also compared the peripheral blood of the mice transplanted with Id1fl/flCreER AE9a leukemia cells and either treated with tamoxifen or vehicle control, by flow cytometry, and found far fewer GFP+C-Kit+ cells in the tamoxifen-treated mice (Figure 2D). Once again, the GFP+ leukemia cells did not express CD45/CD45.2 (Figure 2D). To determine the effect of Id1 on leukemia stem cell (LSC) self-renewal, we performed limiting dilution assays by transplanting sublethally irradiated mice with different numbers of AE9aId1fl/fl or AE9aId1Δ/Δ cells and observing their survival (Figure 2E, left panel). By using the limiting dilution analysis software L-Calc, we found that loss of Id1 significantly decreased the LSC frequency in AE9a-driven leukemia (1 in 43 355 vs 1 in 217 059; P < .05) (Figure 2E, right panel). Thus, loss of Id1 impairs the maintenance of AML1-ETO–driven AML, establishing Id1 as a potential target for new treatment strategies in human AML.
Knockdown of Id3 does not affect leukemia development in mouse model
There are 3 other Id family members, besides Id1, so we examined their expression in a variety of human AML cell lines, using quantitative polymerase chain reaction, and found considerable variability (supplemental Figure 1A). To determine whether AML1-ETO can regulate the expression of other Id family member genes, we performed chromatin immunoprecipitation sequencing analysis on Kasumi-1 cells, using antibodies to ETO and p300. The peak enrichment of A-E and p300 binding was found at the Id1, Id2, and Id3 promoters, but not the Id4 promoter or the gene body.17 A-E and p300 colocalize at specific regions of the Id1 and Id3 promoters (Figure 3A), and we identified Id1 as upregulated A-E target genes based on microarray analysis that we conducted on A-E–transduced primary human HSPCs. As with other A-E–upregulated genes, deletion of the p300 binding domain from A-E abrogated the upregulation of Id1 significantly (Figure 3B). The Id3 upregulation by A-E is not statistically significant (P > .05), but nonetheless, it appears to be reversed upon NHR1 deletion (Figure 3B).
Role of Id1 vs Id3 in leukemia development. (A) AML1-ETO and p300 colocalize at the promoter regions of Id3, but not Id2 and Id4, in chromatin immunoprecipitation sequencing analysis of Kasumi-1 cells. (B) The expression of Id1 and Id3 were examined in MIGR1, AML1-ETO, or AML1-ETO∆NHR1 (that lacks the nervy homology region 1 [NHR1] domain) transduced human CD34+ cord blood cells by microarray analysis (***P < .01). (C and D) In vitro doxycycline treatment decreased the expression of Id3 at both mRNA and protein levels in AE9a cells transduced with one of several inducible shRNAs against Id3. (E) The expression level of Id3 was downregulated in sorted RFP+GFP+ cells isolated from the bone marrow of the recipient mice transplanted with AE9aId1fl/flCreER cells following doxycycline or dox/tamox treatment (left panel). The expression of Id1 was downregulated in the sorted RFP+GFP+ cells isolated from the bone marrow of the recipient mice transplanted with AE9aId1fl/flCreER cells in the tamoxifen-treated groups (right panel). (F) Inducible KD of Id3 by doxycycline does not significantly affect the in vitro proliferation of AE9aId1fl/flCreER cells, with or without 4-OHT treatment (*P < .05, **P < .01). (G) Inducible KD of Id3 by doxycycline does not significantly affect the survival time of recipient mice transplanted with AE9aId1fl/flCreER cells, compared with the control shRNA (n = 10, ***P < .01).
Role of Id1 vs Id3 in leukemia development. (A) AML1-ETO and p300 colocalize at the promoter regions of Id3, but not Id2 and Id4, in chromatin immunoprecipitation sequencing analysis of Kasumi-1 cells. (B) The expression of Id1 and Id3 were examined in MIGR1, AML1-ETO, or AML1-ETO∆NHR1 (that lacks the nervy homology region 1 [NHR1] domain) transduced human CD34+ cord blood cells by microarray analysis (***P < .01). (C and D) In vitro doxycycline treatment decreased the expression of Id3 at both mRNA and protein levels in AE9a cells transduced with one of several inducible shRNAs against Id3. (E) The expression level of Id3 was downregulated in sorted RFP+GFP+ cells isolated from the bone marrow of the recipient mice transplanted with AE9aId1fl/flCreER cells following doxycycline or dox/tamox treatment (left panel). The expression of Id1 was downregulated in the sorted RFP+GFP+ cells isolated from the bone marrow of the recipient mice transplanted with AE9aId1fl/flCreER cells in the tamoxifen-treated groups (right panel). (F) Inducible KD of Id3 by doxycycline does not significantly affect the in vitro proliferation of AE9aId1fl/flCreER cells, with or without 4-OHT treatment (*P < .05, **P < .01). (G) Inducible KD of Id3 by doxycycline does not significantly affect the survival time of recipient mice transplanted with AE9aId1fl/flCreER cells, compared with the control shRNA (n = 10, ***P < .01).
In many situations, Id1 and Id3 have overlapping effects on E protein activity, so targeting Id1 and Id3 together may be more potent than targeting either one alone.14,16 To examine this question, we used 4 different inducible Id3 short hairpin RNAs (shRNAs; Id3shRNA1-4) to knock down (KD) Id3 and achieved KD efficiencies of >80% at both the mRNA and protein levels (Figure 3C-D). We transduced AE9a-expressing Id1fl/flCreER cells with Id3 inducible shRNA-1, which was the least leaky among the 4 Id3 shRNAs, and transplanted the cells into sublethally irradiated recipient mice. Id3 shRNA expression was induced by doxycycline, whereas the Id1 gene was deleted by tamoxifen administration. The KD of Id3 in the AE9a-expressing leukemia cells was significant, as determined by quantitative polymerase chain reaction (Figure 3E), yet we found no significant effect on the proliferation of the leukemia cells or the survival of the leukemic mice (in the presence of Id1) (Figure 3F-G). The functions of Id1 and Id3 are biological distinguishable; for example, Id3 but not Id1 promotes the erythroid differentiation in K562 leukemia cells,25 which could explain the shorter survival of the Id1/Id3-null leukemia mice. Overall, these results indicate that Id3 does not play a distinctive role in AE9a-driven leukemia, even in the absence of Id1.
The Id1 inhibitor CBD induces leukemia cell apoptosis and blocks leukemogenesis
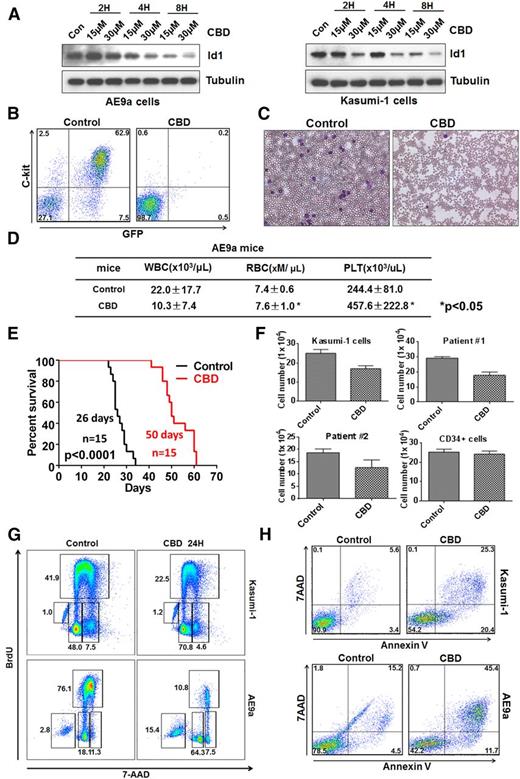
CBD has been reported to decrease Id1 expression, so we treated murine AE9a and human Kasumi-1 cells and found a dose-dependent downregulation of Id1 protein levels (Figure 4A). We collected AE9a-expressing leukemia cells from the spleen and bone marrow of lethally irradiated recipient mice and injected them into secondary recipients after treating the cells with either 15 μM CBD or the vehicle control for 12 hours. CBD treatment significantly decreased the number of C-Kit+GFP+ leukemia cells in the peripheral blood of the recipient mice (Figure 4B-C). The WBC counts of the control-treated AE9a mice were higher than the CBD-treated mice, and the red blood cell and platelet counts were significantly lower, compared with the CBD-treated group (Figure 4D). CBD treatment also significantly prolonged the median survival of the AE9a mice, compared with the control group (Figure 4E).
CBD, an Id1 inhibitor, blocks leukemogenesis and induces the apoptosis of leukemia cells. (A) CBD treatment downregulates Id1 expression in AE9a cells (left panel) and Kasumi-1 cells (right panel). (B) AE9a leukemia cells were treated with CBD at 15 μM or vehicle control for 12 hours, and 1 × 105 living cells were injected into individual sublethally irradiated recipient mice. The frequency of GFP+C-Kit+ AE9a cells in the peripheral blood was examined 3 weeks after transplantation. (C) Leukemia blast cells are found in the peripheral blood of AE9a mice in the control group but not the CBD treatment group on day 21. (D) The WBC counts of the mice that received CBD-treated AE9a cells were lower than the mice that received control vehicle-treated cells, whereas the red blood cell (RBC) and platelet (PLT) counts of the CBD-treated group were significantly higher (± SD; n = 5). (E) Twelve hours of CBD ex vivo treatment, at 15 μM, prolongs the survival time of the recipient mice transplanted with AE9a cells. (F) Forty-eight hours of CBD, at 10 μM, inhibited the growth of primary t(8;21)+ leukemia cells and Kasumi-1 cells (± SD; n = 3) but not normal human CD34+ HSPCs. (G) Twenty-four-hour treatment with CBD, at 15 μM, caused G0/G1-phase cell cycle arrest in Kasumi-1 cells (upper panel) and AE9a cells (lower panel). (H) CBD treatment induces the apoptosis of Kasumi-1 (upper panel) and AE9a (lower panel) cells, based on 5-bromo-2′-deoxyuridine (BrdU) incorporation assays.
CBD, an Id1 inhibitor, blocks leukemogenesis and induces the apoptosis of leukemia cells. (A) CBD treatment downregulates Id1 expression in AE9a cells (left panel) and Kasumi-1 cells (right panel). (B) AE9a leukemia cells were treated with CBD at 15 μM or vehicle control for 12 hours, and 1 × 105 living cells were injected into individual sublethally irradiated recipient mice. The frequency of GFP+C-Kit+ AE9a cells in the peripheral blood was examined 3 weeks after transplantation. (C) Leukemia blast cells are found in the peripheral blood of AE9a mice in the control group but not the CBD treatment group on day 21. (D) The WBC counts of the mice that received CBD-treated AE9a cells were lower than the mice that received control vehicle-treated cells, whereas the red blood cell (RBC) and platelet (PLT) counts of the CBD-treated group were significantly higher (± SD; n = 5). (E) Twelve hours of CBD ex vivo treatment, at 15 μM, prolongs the survival time of the recipient mice transplanted with AE9a cells. (F) Forty-eight hours of CBD, at 10 μM, inhibited the growth of primary t(8;21)+ leukemia cells and Kasumi-1 cells (± SD; n = 3) but not normal human CD34+ HSPCs. (G) Twenty-four-hour treatment with CBD, at 15 μM, caused G0/G1-phase cell cycle arrest in Kasumi-1 cells (upper panel) and AE9a cells (lower panel). (H) CBD treatment induces the apoptosis of Kasumi-1 (upper panel) and AE9a (lower panel) cells, based on 5-bromo-2′-deoxyuridine (BrdU) incorporation assays.
Treatment with CBD also significantly inhibited the growth of primary leukemia cells isolated from patients with t(8;21)+ AML. In contrast, the proliferation of normal human cord blood CD34+ cells was less sensitive to CBD compared with the leukemia cells, suggesting that CBD has no major cytotoxic effect on normal hematopoietic precursors (Figure 4F). CBD treatment triggered a G1 cell cycle “arrest” in AE9a and Kasumi-1 cells (Figure 4G), which could lead to the delayed growth. It also triggered the apoptosis of AE9a and Kasumi-1 cells within 24 hours (Figure 4H). Thus, inhibition of Id1 by CBD impairs AE9a-driven leukemia cell growth ex vivo, by inducing cell cycle arrest and apoptosis.
Constitutively activated AKT1 rescues the phenotype of Id1-deficient leukemia cells
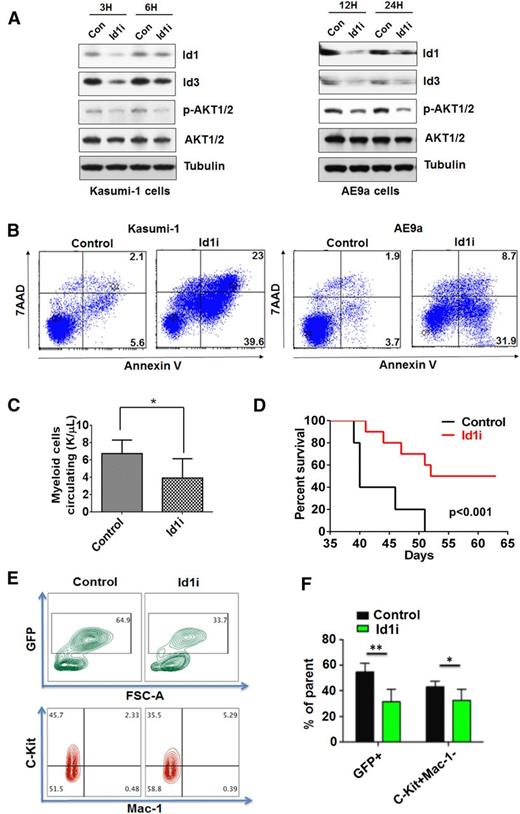
To define the role of ID1 in AML, we examined the proteins that interact with ID1 in human AML cells, using mass spectrometry analysis, and found AKT1 bound to ID1 (Figure 5A). We first verified this finding, using Kasumi-1 cells and an anti-Id1 antibody for immunoprecipitation (Figure 5B). We next performed glutathione S-transferase (GST) pull-down assays, which showed a direct interaction between ID1 and AKT1 (Figure 5C). Other ID family members, ID2 and ID3, did not bind AKT1 in Kasumi-1 cells (Figure 5D) demonstrating the specificity of this interaction. To map the AKT1 interacting domain, we made N-terminal basic helix-loop-helix (bHLH) domain and C-terminal deletion constructs in ID1. By overexpressing full-length ID1 or these ID1 deletions, in 293T cells, we found that the C terminus of ID1 is required for its interaction with AKT1 (Figure 5E). We also found that the level of phosphorylated AKT1 is lower in the 293T cells overexpressing the ID1 C-terminal–deleted protein than in cells overexpressing WT ID1 (Figure 5E), suggesting that this interaction may promote AKT1 phosphorylation. To further examine the effects of ID1 on the AKT pathway, we performed western blot analyses examining AE9aId1fl/flCreER cells treated with 15 µM CBD for 3, 6, 12, 24, and 48 hours or 0.1 μM 4-OHT for 24, 48, and 72 hours. As the level of ID1 decreases, the phosphorylation of AKT1 also significantly decreases (Figure 5F and supplemental Figure 6A). Inhibition of Id1 by CBD, or 4-OHT treatment, did not affect the level of phosphatidylinositol 3-kinase (PI3K)/p85 in the AE9a cells or the Kasumi-1 cells (Figure 5F and supplemental Figure 6A). We also found that the inhibition of Id1 by CBD decreases the phosphorylation of mTOR, the substrate of AKT1, but not the phosphorylation of P70S6K and 4EBP1 in Kasumi-1 cells (supplemental Figure 6B).
Physical and functional interaction of Id1 with AKT1 in leukemia cells. (A) Sodium dodecyl sulfate–polyacrylamide gel electrophoresis and Coomassie staining Kasumi-1-MIGR1-HA-Id1 cells using an anti-HA antibodies and normal mouse immunoglobulin (Ig) G as control. (Asterisk identifies a differentially expressed sodium dodecyl sulfate–polyacrylamide gel electrophoresis band.) (B) Immunoprecipitation was performed using anti-Id1 antibody in the Kasumi-1 cells treated with CBD or vehicle control, and IgG was used as control. The anti-AKT1 and anti-Id1 antibodies were used for western blot analysis. (C) Id1 interacts with AKT1 in vitro. Purified GST-Id1 and His-AKT1 were used for GST pull-down assays, and anti-AKT1 or anti-GST antibodies were used for western blotting. (D) Id2 or Id3 does not bind to AKT1 in Kasumi-1 cells. Immunoprecipitation was performed using anti-Id2 or Id3 antibody, and IgG was used as control. (E) The C terminus of Id1 is required for its interaction with AKT1. Also, Id1 lacking its C terminus does not increase the level of phosphorylated AKT1 in 293T cells. (F) AKT pathway regulators expression in Kasumi-1 cells treated with Id1 inhibitor (15 µM) was examined by doing western blot analysis. (G) In vivo luciferase imaging showed that constitutively activated AKT1 (myristoylated AKT1) can promote leukemia progression in recipient mice transplanted with AE9aId1Δ/Δ cells. (H) Constitutively activated AKT1 (myristoylated [myris] AKT1) promotes leukemia progression in mice transplanted with AE9aId1Δ/Δ cells (n = 10, ***P < .01).
Physical and functional interaction of Id1 with AKT1 in leukemia cells. (A) Sodium dodecyl sulfate–polyacrylamide gel electrophoresis and Coomassie staining Kasumi-1-MIGR1-HA-Id1 cells using an anti-HA antibodies and normal mouse immunoglobulin (Ig) G as control. (Asterisk identifies a differentially expressed sodium dodecyl sulfate–polyacrylamide gel electrophoresis band.) (B) Immunoprecipitation was performed using anti-Id1 antibody in the Kasumi-1 cells treated with CBD or vehicle control, and IgG was used as control. The anti-AKT1 and anti-Id1 antibodies were used for western blot analysis. (C) Id1 interacts with AKT1 in vitro. Purified GST-Id1 and His-AKT1 were used for GST pull-down assays, and anti-AKT1 or anti-GST antibodies were used for western blotting. (D) Id2 or Id3 does not bind to AKT1 in Kasumi-1 cells. Immunoprecipitation was performed using anti-Id2 or Id3 antibody, and IgG was used as control. (E) The C terminus of Id1 is required for its interaction with AKT1. Also, Id1 lacking its C terminus does not increase the level of phosphorylated AKT1 in 293T cells. (F) AKT pathway regulators expression in Kasumi-1 cells treated with Id1 inhibitor (15 µM) was examined by doing western blot analysis. (G) In vivo luciferase imaging showed that constitutively activated AKT1 (myristoylated AKT1) can promote leukemia progression in recipient mice transplanted with AE9aId1Δ/Δ cells. (H) Constitutively activated AKT1 (myristoylated [myris] AKT1) promotes leukemia progression in mice transplanted with AE9aId1Δ/Δ cells (n = 10, ***P < .01).
To determine whether AKT1 is important for the function of Id1 in leukemia development, we overexpressed constitutively activated myrisAkt1 (myristoylated AKT1) in AE9aId1Δ/Δ cells and transplanted them into sublethally irradiated recipient mice. In vivo luciferase imaging (Figure 5G) and monitoring the survival time of the transplanted mice (Figure 5H) showed that overexpression of a constitutively activated form of AKT1 rescued Id1 null leukemia cell growth (Figure 5G-H). These results suggested that inhibition of Id1 might impair the self-renewal of AML1-ETO+ leukemia cells by blocking AKT signaling.
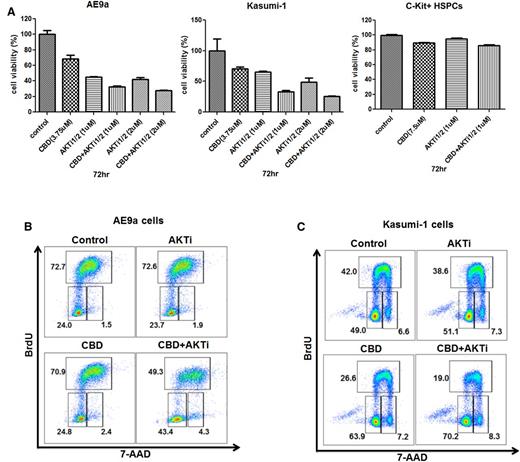
Targeting the same pathway with 2 different approaches can be additive, synergistic, or even antagonistic, so we treated AE9a and Kasumi-1 cells with different concentrations of CBD and an AKT inhibitor (alone or in combination). The combination treatment had greater growth inhibitory effects on the leukemia cells at 72 hours with no effect on the growth of normal C-Kit+ HSPCs (Figure 6A). This effect was seen using relatively low concentrations of CBD (3.75 μM) and the AKT inhibitor (1 μM). The AKT inhibitor itself had no effect on the cell cycle of AE9a or Kasumi-1 cells at 1 or 2 μM, whereas the AE9a cells and the Kasumi-1 cells treated with the combination of CBD and AKT inhibitor exhibited a marked increase in G1-phase cells (from 23% to 24% to 43% to 48%) and a decrease in S-phase cells (from 72% to 45% to 49%) (Figure 6B-C), respectively. Taken together, these results suggest that targeting Id1 and AKT could be more effective than targeting either alone in t(8;21) AML patients.
The combined effect of CBD and an AKT inhibitor on t(8;21) leukemia cell growth and cell cycle. (A) Growth inhibition of AE9a cells, Kasumi-1 cells, and C-Kit+ HSPCs treated at different concentrations of CBD or AKT inhibitor, alone or together, at 72 hours. Values represent the mean ± SD for 3 separate experiments. (B and C) Cell cycle analysis on AE9a and Kasumi-1 cells treated with CBD alone, AKT inhibitor alone, or both together at 24 hours. BrdU was added during the last hour of incubation. Cells were then processed for double staining with anti-BrdU–allophycocyanin (APC) and 7 amino actinomycin D (7-AAD) and analyzed by flow cytometry.
The combined effect of CBD and an AKT inhibitor on t(8;21) leukemia cell growth and cell cycle. (A) Growth inhibition of AE9a cells, Kasumi-1 cells, and C-Kit+ HSPCs treated at different concentrations of CBD or AKT inhibitor, alone or together, at 72 hours. Values represent the mean ± SD for 3 separate experiments. (B and C) Cell cycle analysis on AE9a and Kasumi-1 cells treated with CBD alone, AKT inhibitor alone, or both together at 24 hours. BrdU was added during the last hour of incubation. Cells were then processed for double staining with anti-BrdU–allophycocyanin (APC) and 7 amino actinomycin D (7-AAD) and analyzed by flow cytometry.
The Id1 inhibitor in vivo treatment prolongs the survival time of the AE9a leukemia mice
Pimozide can promote Id1 degradation by targeting USP1, the deubiquitinase of Id1. We treated human Kasumi-1 cells and murine AE9a cells with 8 μM Id1i and found downregulation of Id1 and Id3 protein levels (Figure 7A). The phosphorylation of AKT1 also decreased after Id1i treatment of Kasumi-1 and AE9a cells (Figure 7A). Because Id1i treatment triggered the apoptosis of Kasumi-1 and AE9a cells within 48 hours (Figure 7B), and because CBD is a controlled substance that is difficult to obtain in sufficient quantities to treat mice, we used Id1i to treat mice with AE9a leukemia. Mice were treated with either Id1i (at 7.5 mg/kg) or vehicle for 2 weeks. Five weeks after transplantation, the CBC analysis showed fewer myeloid cells circulating in the Id1i treatment group (Figure 7C), with far fewer GFP+C-Kit+Mac-1− leukemia blast cells (Figure 7E-F). The Id1i also significantly prolonged the median survival of the AE9a mice, compared with the control group (Figure 7D), demonstrating its therapeutic potential in t(8;21)+ AML.
Id1i, pimozide, induces the apoptosis of leukemia cells and prolongs the survival time of AE9a leukemia mice. (A) Id1i (8 μM) treatment downregulates Id1 expression and the level of phosphorylated AKT1 in Kasumi-1 cells (left panel) and AE9a cells (right panel). (B) Id1i (8 μM) treatment induces the apoptosis of Kasumi-1 (left panel) and AE9a (right panel) cells at 48 hours, based on Annexin-V/7-AAD staining. (C) AE9a leukemia mice were treated with Id1i (7.5 mg/kg) or vehicle control for 2 weeks, beginning on day 3. More myeloid cells circulating are found in the peripheral blood of AE9a mice in the control group compared with the Id1i treatment group 5 weeks after transplantation. (D) Id1i (7.5 mg/kg) in vivo treatment prolongs the survival of the recipient mice transplanted with AE9a leukemia cells (53 days vs 40 days, n = 10 for each group, P < .001). (E and F) The frequencies of GFP+ and C-Kit+ Mac-1 cells in the peripheral blood of AE9a mice in the control or Id1i treatment group were examined 6 weeks after transplantation (n = 10, **P < .01, *P < .05).
Id1i, pimozide, induces the apoptosis of leukemia cells and prolongs the survival time of AE9a leukemia mice. (A) Id1i (8 μM) treatment downregulates Id1 expression and the level of phosphorylated AKT1 in Kasumi-1 cells (left panel) and AE9a cells (right panel). (B) Id1i (8 μM) treatment induces the apoptosis of Kasumi-1 (left panel) and AE9a (right panel) cells at 48 hours, based on Annexin-V/7-AAD staining. (C) AE9a leukemia mice were treated with Id1i (7.5 mg/kg) or vehicle control for 2 weeks, beginning on day 3. More myeloid cells circulating are found in the peripheral blood of AE9a mice in the control group compared with the Id1i treatment group 5 weeks after transplantation. (D) Id1i (7.5 mg/kg) in vivo treatment prolongs the survival of the recipient mice transplanted with AE9a leukemia cells (53 days vs 40 days, n = 10 for each group, P < .001). (E and F) The frequencies of GFP+ and C-Kit+ Mac-1 cells in the peripheral blood of AE9a mice in the control or Id1i treatment group were examined 6 weeks after transplantation (n = 10, **P < .01, *P < .05).
Discussion
AML is characterized by the increased self-renewal, proliferation, and impaired differentiation of HSPCs.26 To define the role of Id1 in LSC biology, we have studied its relevance in the initiation, self-renewal, and maintenance of the AML model. We have found that loss of Id1 delays leukemia initiation in the AE9a fetal liver transplantation model. Limiting dilution assays demonstrate impaired LSC self-renewal, consistent with ex vivo data and with impaired in vivo cancer progression. Our observations provide critical genetic evidence for an important role of Id1 in AML1-ETO–driven leukemia initiation and progression and suggest that targeting LSC self-renewal by pharmacologic inhibition of Id1 may be useful in the therapy of human AML.
We have identified unanticipated effects of Id1 on AKT signaling, which suggest that activation of this pathway may be a key mechanism for Id1, given that a constitutively activated form of AKT1 can reverse the effects of Id1 loss on leukemogenesis. Although constitutively active AKT signaling triggers the depletion of normal HSCs over time, AKT activation contributes to the induction of acute leukemia in mice. Indeed, enhanced AKT activation is an important mechanism of transformation in AML,27 and in t(8;21)-driven AML, given that PI3K-dependent activation of AKT has been observed in Kasumi-1 cells.28 Id1 has also been shown to enhance endothelial progenitor cell–driven angiogenesis in ovarian cancer, an effect mediated largely by the PI3K/AKT signaling pathway.29,30 We have determined that Id1 physically interacts with AKT1, through its C-terminal region, and promotes AKT1 phosphorylation; this is consistent with the decrease in PI3K/p110γ phosphorylation induced by loss of Id1, and it contrasts with the transcriptional effects of Id1, which depend on its bHLH domain, a domain not required for its interaction with AKT1. Moreover, Id2 and Id3, which both have bHLH domains, do not bind AKT1, together indicating that Id1 regulates AKT signaling through mechanisms distinct from its transcriptional effects. Given that myristoylated AKT1 rescues the Id1 null AML mouse model, targeting AKT1 might be a useful therapeutic strategy for t(8;21) leukemia.
Multiple studies have shown that the Janus kinase (JAK)/signal transducer and activator of transcription (STAT)/mitogen-activated protein kinase pathway is highly activated in AML1-ETO–expressing cells; thus, its inactivation could profoundly inhibit the proliferation of these cells. For example, AML1-ETO and its alternatively spliced variant AML1-ETO9a have recently been shown to enhance JAK/STAT signaling via downregulation of CD45, a protein tyrosine phosphatase and a negative regulator of JAK/STAT signaling.31-33 The constitutive activation of STAT transcription factors, particularly STAT5, is a common event in myeloid leukemia and is seen in Kasumi-1 cells.28 The JAK2 inhibitor TG101209 inhibited the proliferation and promoted the apoptosis of t(8;21) leukemia cells,29 significantly impaired the leukemia-initiating potential of AE9a leukemia cells, and prolonged the survival of AE9a leukemia–bearing mice.31 Triptolide, a compound isolated from the traditional Chinese medicinal herb, inhibits cell proliferation and induces apoptosis of t(8;21)-bearing cells via downregulation of C-KIT and inhibition of JAK/STAT signaling.34 Lastly, the mitogen-activated protein kinase pathway is activated by AML1-ETO, and EriB, the diterpenoid isolated from Labiatae family herb, triggers apoptosis by downregulating extracellular signal-regulated kinase 1/2 phosphorylation and AP-1 activation in AML1-ETO–expressing cells. 21,35 The phosphorylation levels of JAK2, STAT5, MEK1/2, extracellular signal-regulated kinase 1/2, and AKT1 were significantly downregulated by inhibiting Id1, suggesting a mechanism for the impaired proliferation of AML1-ETO+ LSCs that we observed. Thus, we have identified a novel mechanistic role of Id1 in leukemogenesis.
Although Id proteins may be considered difficult to target, given their lack of enzymatic activity or ligand-binding sites, CBD appears to be a relatively nontoxic cannabinoid that can significantly decrease Id1 expression. Alternatively, small-molecule inhibitors of USP1 have been shown to promote Id1 degradation and kill leukemic cells,36 and curcumin, the active ingredient of turmeric, can reduce Id1 expression and induce apoptosis.37 Inhibiting BMP signaling also decreases Id1 (and Id3) protein expression; thus, small-molecule BMP type I receptor antagonists may represent a novel way of targeting Id-expressing cancer cells.38 Regardless of which strategy is pursued, the successful clinical development of an Id1 inhibitor will require much additional work. Given that CBD made AML cells more sensitive to an AKT inhibitor, and vice versa, combining Id1 inhibitors with AKT inhibitors or other inhibitors of cell signaling could be a promising therapeutic strategy for AML patients.
The data reported in this article have been deposited in the Gene Expression Omnibus database (accession number GSE69900).
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank Dr Yan Liu for providing AKT inhibitor and the University of Miami Division of Veterinary Resources, Flow Cytometry Core Facility and the Epigenomics Core of Weill Cornell Medical College for their help.
This work was supported by funding from an Institutional Research grant (IRG-98-277-13) from the American Cancer Society (L.W.), Gabrielle’s Angel Foundation for Cancer Research (L.W.), the Women’s Cancer Association of the University of Miami (L.W.), Leukemia Research Foundation (L.W.), Stanley J. Glaser Foundation Research Award (L.W.), an Empire State Stem Cell Scholar award (L.W.), a Leukemia Lymphoma Society fellowship (L.W.), and a grant from the National Institutes of Health, National Cancer Institute R01CA166835 (S.D.N.).
Authorship
Contribution: L.W. and S.D.N. designed the research, analyzed data, and wrote the manuscript; M.G.C. and R.B. contributed new reagents; L.W., N.M., X.-J.S., Y.T., M.G.C., F.L., M.H., H.X., G.H., and M.M. performed research and analyzed data; and A.M. and E.R. analyzed data.
Conflict-of- interest disclosure: The authors declare no competing financial interests.
Correspondence: Lan Wang, University of Miami Miller School of Medicine, 1501 NW 10th Ave, BRB 701, Miami, FL 33136; e-mail: l.wang30@med.miami.edu.

![Figure 1. Deletion of Id1 abrogates the initiation of leukemia in the AE9a fetal liver transplantation model. (A) Lethally irradiated recipient mice were injected with WT or Id1−/− mouse E14.5 fetal liver cells transduced with AE9a. The WBC counts of the transplanted mice in Id1−/− AE9a group are significant lower than the WT AE9a mice group 15 weeks after transplant (P = .0288). (B) Loss of Id1 prolongs the survival time of recipient mice (267 days vs 137 days, n = 15 per group, P < .001). (C) Loss of Id1 decreases the frequency of GFP+CD45/CD45.2−C-Kit+ leukemia blast cells and increases the frequency of normal GFP−Gr1+ myeloid cells in the peripheral blood of recipient mice (compared with the WT group) 15 weeks after transplantation. (D) The peripheral blood shows less leukemia blast cells in the Id1−/− AE9a group compared with the WT AE9a mice group. (E) Pathological sections of the spleen stained with hematoxylin-eosin are shown 15 weeks after transplantation (the scale bar indicates 25 μM). The leukemic blast cells are indicated by black arrows. (F) The columns represent the numbers of colonies in each plating of WT or Id1−/− E14.5 fetal liver cells transduced with AE9a (± standard deviation [SD], n = 3, P < .01). (G) The frequency of donor-derived Id1−/− CD45.2+ cells is significantly lower in the peripheral blood of recipient mice, compared with the WT CD45.2+ cells (P < .01). (H) Loss of Id1 abrogates the repopulating ability of hematopoietic cells isolated from E14.5 fetal liver cells.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/126/5/10.1182_blood-2015-03-635532/4/m_640f1.jpeg?Expires=1767717772&Signature=DA74aSHFemUSn7Xl0xw4z-FquPKFy9ygHn4RBW-uAkm0Z-JJPqI59IikeW9wQ~rv5Ct0urdKE1MiEdFvn7vCdzRbzbtruwsPMpHDSa-p1HXeS5lRkAqGZmQ5ibmpJAm5GBPbXkLsLM6aDTUCw3W1I~ZpaA~~Zp7cQwXHAOYPoGe9PKWYxj7GIyviV7cU1lTi4oMJkSFKVz6dKItQImmuOuqX6TsW8sktvZyNXwCcMnOCH-JmCjMuQ82f7GIYanGndWdsAqPjfErCUbUGHUyOH7DHGHGXgOydoK8KMDjmhOtqtsdpsNasDGCzLwqb1~bxyA9QdbfaLxInrFlFXDCGGQ__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
![Figure 3. Role of Id1 vs Id3 in leukemia development. (A) AML1-ETO and p300 colocalize at the promoter regions of Id3, but not Id2 and Id4, in chromatin immunoprecipitation sequencing analysis of Kasumi-1 cells. (B) The expression of Id1 and Id3 were examined in MIGR1, AML1-ETO, or AML1-ETO∆NHR1 (that lacks the nervy homology region 1 [NHR1] domain) transduced human CD34+ cord blood cells by microarray analysis (***P < .01). (C and D) In vitro doxycycline treatment decreased the expression of Id3 at both mRNA and protein levels in AE9a cells transduced with one of several inducible shRNAs against Id3. (E) The expression level of Id3 was downregulated in sorted RFP+GFP+ cells isolated from the bone marrow of the recipient mice transplanted with AE9aId1fl/flCreER cells following doxycycline or dox/tamox treatment (left panel). The expression of Id1 was downregulated in the sorted RFP+GFP+ cells isolated from the bone marrow of the recipient mice transplanted with AE9aId1fl/flCreER cells in the tamoxifen-treated groups (right panel). (F) Inducible KD of Id3 by doxycycline does not significantly affect the in vitro proliferation of AE9aId1fl/flCreER cells, with or without 4-OHT treatment (*P < .05, **P < .01). (G) Inducible KD of Id3 by doxycycline does not significantly affect the survival time of recipient mice transplanted with AE9aId1fl/flCreER cells, compared with the control shRNA (n = 10, ***P < .01).](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/126/5/10.1182_blood-2015-03-635532/4/m_640f3.jpeg?Expires=1767717772&Signature=IUGm5PlgZQKm~yXJnVPIsJy42IhSM34B3vC-JfWL1Z7uLx8jAwcy8FYSUYX0KiyqT4y16gnxZUN8MXwXzFk6twLaiAh2-Xa2firnB7QQp5zAjxvh31YdnABw80m9apiggWFxEjkEsG~YRNb90d8xN7QvGOsA2e0uz9hTautBa2StwWLU56UD1zXzaV48MfA-SOGB0qwQ8cxWsAuXSI8Hh9nACiY~yCA4anruc2qC4dK8JOZHPJoTgcPfmCIxcitTS175hoZUgvFhvL2Un8kfTK3AhJkhUE-vTz10WZ3-MkcsCE2b6cKTKmYo45n64uoGKoirWUKEPH~XzZhNV5Bc2g__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
![Figure 5. Physical and functional interaction of Id1 with AKT1 in leukemia cells. (A) Sodium dodecyl sulfate–polyacrylamide gel electrophoresis and Coomassie staining Kasumi-1-MIGR1-HA-Id1 cells using an anti-HA antibodies and normal mouse immunoglobulin (Ig) G as control. (Asterisk identifies a differentially expressed sodium dodecyl sulfate–polyacrylamide gel electrophoresis band.) (B) Immunoprecipitation was performed using anti-Id1 antibody in the Kasumi-1 cells treated with CBD or vehicle control, and IgG was used as control. The anti-AKT1 and anti-Id1 antibodies were used for western blot analysis. (C) Id1 interacts with AKT1 in vitro. Purified GST-Id1 and His-AKT1 were used for GST pull-down assays, and anti-AKT1 or anti-GST antibodies were used for western blotting. (D) Id2 or Id3 does not bind to AKT1 in Kasumi-1 cells. Immunoprecipitation was performed using anti-Id2 or Id3 antibody, and IgG was used as control. (E) The C terminus of Id1 is required for its interaction with AKT1. Also, Id1 lacking its C terminus does not increase the level of phosphorylated AKT1 in 293T cells. (F) AKT pathway regulators expression in Kasumi-1 cells treated with Id1 inhibitor (15 µM) was examined by doing western blot analysis. (G) In vivo luciferase imaging showed that constitutively activated AKT1 (myristoylated AKT1) can promote leukemia progression in recipient mice transplanted with AE9aId1Δ/Δ cells. (H) Constitutively activated AKT1 (myristoylated [myris] AKT1) promotes leukemia progression in mice transplanted with AE9aId1Δ/Δ cells (n = 10, ***P < .01).](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/126/5/10.1182_blood-2015-03-635532/4/m_640f5.jpeg?Expires=1767717772&Signature=NBeK7J8SbtKNXEuioc4XYL7PmeM2dMDl-wuqv-MhfmKUblFluOuvkUxiMufWu2XClK3-h9c46sG83z8j6zz4gEMz9gDupgfCeyXENcUWzh4Ognmjv1d4QeLTDqWqP6QKzmPtOsES7guhIxQg8jVsOxVJJ528AFT7X6hn2mQlMM4o14R3A5pRMVfwWF7ylfKxOlFgAzQJc8285R6ZyvOFAFcPewx4OV-nSu0irTNLxvIGAHEZ2aDAbe4sYzQ~8ZDwT2sC5wQ2KsolCENrHqZm9kPnEZUqCFwN5wbWVLCBkJzI5pPzFNFkKh3LuaH9flBGsiyhSDprrefYrK0KpviQxQ__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
This feature is available to Subscribers Only
Sign In or Create an Account Close Modal