Abstract
Acute megakaryoblastic leukemia (AMKL) comprises between 4% and 15% of newly diagnosed pediatric acute myeloid leukemia patients. AMKL in children with Down syndrome (DS) is characterized by a founding GATA1 mutation that cooperates with trisomy 21, followed by the acquisition of additional somatic mutations. In contrast, non–DS-AMKL is characterized by chimeric oncogenes consisting of genes known to play a role in normal hematopoiesis. CBFA2T3-GLIS2 is the most frequent chimeric oncogene identified to date in this subset of patients and confers a poor prognosis.
Introduction
Acute megakaryoblastic leukemia (AMKL) is a subtype of acute myeloid leukemia (AML) characterized by abnormal megakaryoblasts that express platelet-specific surface glycoprotein. Bone marrow biopsy frequently demonstrates extensive myelofibrosis, often making aspiration in these patients difficult. AMKL is extremely rare in adults, occurring in only 1% of AML patients.1 This is in contrast to children, where it comprises between 4% and 15% of AML patients.2,3 In pediatrics, the disease is divided into 2 major subgroups: AMKL in patients with Down syndrome (DS-AMKL) and AMKL in patients without DS (non–DS-AMKL). AMKL is the most frequent type of AML in children with DS, and the incidence in these patients is 500-fold higher than in the general population.4 In contrast to non–DS-AMKL, leukemic cells carry not only megakaryocytic cell-surface markers but also erythroid markers, resulting in the distinct World Health Organization classification “myeloid leukemia in Down syndrome”5 . Somatic mutations in GATA1 are found in almost all cases of DS-AMKL and precede the development of leukemia, as indicated by their presence in patients with transient myeloproliferative disease (TMD) in the neonatal period.6-11 DS-AMKL is both biologically and clinically distinct, with superior outcomes compared with non–DS-AMKL.12-15 Pediatric non–DS-AMKL is a heterogenous group of patients, a significant proportion of whom carry chimeric oncogenes including RBM15-MKL1, CBFA2T3-GLIS2, NUP98-KDM5A, and MLL gene rearrangements.16,17 Unfortunately, the outcome of non–DS-AMKL is generally poor, with lower event-free survival than DS-AMKL and pediatric AML, even in the face of intensified treatment.2,18
DS-AMKL
TMD
DS-AMKL is associated with TMD, a hematologic disorder in infancy. In this disorder, a clonal population of megakaryoblasts accumulates in the peripheral blood. These blasts are phenotypically indistinguishable from AMKL leukemic blasts, and in the majority of cases, remission is spontaneous within 3 months in the absence of treatment. In ∼20% of TMD cases, patients will go on to develop myelodysplastic syndrome and/or AMKL.19 TMD is thought to originate in utero, as an identical mutation in GATA1, the genetic lesion associated with TMD, was found to be present at birth in twins with TMD.20 Further evidence came with the analysis of archived autopsy specimens from DS patients that identified GATA1 mutations in 2 fetal liver specimens.21 A subsequent study screening Guthrie cards from 585 DS infants identified GATA1 mutations in 3.8% of their cohort, confirming the presence of this lesion in a subset of patients at birth.22 The frequency of this lesion in newborn DS patients was significantly higher in a study that used next-generation sequencing, which has a greater sensitivity, to screen 200 neonates with DS.23 In this analysis, GATA1 mutations were detected in 29% of patients. The spontaneous resolution of TMD suggests that despite the presence of blasts in the peripheral blood that appear phenotypically indistinguishable from full-blown leukemia, they are in fact functionally different as they fail to persist. When TMD and AMKL blasts from patients with DS are injected into immunodeficient mice, this difference becomes apparent. Approximately 50% of DS-AMKL engraft into NOD/SCID mice, leading to widespread dissemination and the ability to propagate in secondary and tertiary recipients.24 In contrast, blasts from TMD patients very rarely engraft, fail to disseminate outside the bone marrow, and are unable to propagate disease in secondary and tertiary recipients.24 Exome sequencing of TMD has revealed that non–silent mutations in these blasts are primarily limited to the GATA1 gene.25 In contrast, AMKL blasts carry a higher burden of mutations, with additional lesions in epigenetic and kinase-signaling genes leading to progression of the disease. Collectively, these findings support a model whereby TMD blasts arise secondary to GATA1 mutations in the setting of trisomy 21, acquiring this so-called first hit, and persist in the bone marrow. Additional lesions can then occur providing the cooperating events that are necessary for full-blown leukemia to develop (Figure 1). Although sequencing studies have demonstrated the genetic lesions that are required for progression of TMD to AMKL, they do not provide any information on how to predict the 20% of patients that will go on to develop AMKL. An extensive analysis of germline DNA, including pathologic mutations in cancer-predisposition genes as well as genome-wide association studies to identify polymorphisms that may predispose an individual to developing AMKL, may provide clues. If predisposing factor(s) are identified, they have the potential to significantly impact clinical care, as the identification of those patients at high risk of developing AMKL would allow for early treatment of the premalignant cells with decreased intensity chemotherapy while maintaining the high cure rates.
DS-AMKL pathogenesis. In utero truncating mutations in GATA1 lead to a TMD in the neonatal period that resolves in the absence of treatment. Residual cells either undergo apoptosis or acquire additional cooperating mutations leading to overt AMKL with an average latency of 3 years. Recurrently targeted genes include but are not limited to cohesin complex components, CTCF, the PRC2 complex, and kinase-signaling genes. Of the 26 sequenced DS-AMKL cases that carry mutations in cohesin, 6 contained mutations in a PRC2 complex gene as well as a kinase as shown in this example.25 Cohesin mutation, ●; GATA1 mutation, ★; kinase mutation, ▲; PRC2 mutation, ▪; Trisomy 21, ×××.
DS-AMKL pathogenesis. In utero truncating mutations in GATA1 lead to a TMD in the neonatal period that resolves in the absence of treatment. Residual cells either undergo apoptosis or acquire additional cooperating mutations leading to overt AMKL with an average latency of 3 years. Recurrently targeted genes include but are not limited to cohesin complex components, CTCF, the PRC2 complex, and kinase-signaling genes. Of the 26 sequenced DS-AMKL cases that carry mutations in cohesin, 6 contained mutations in a PRC2 complex gene as well as a kinase as shown in this example.25 Cohesin mutation, ●; GATA1 mutation, ★; kinase mutation, ▲; PRC2 mutation, ▪; Trisomy 21, ×××.
GATA1
The GATA family of proteins consists of transcription factors, 3 of which are expressed principally in hematopoietic cells (GATA1, GATA2, and GATA3). The GATA1 protein is typically present in cells of erythroid, megakaryocytic, mast, and eosinophilic lineages, whereas GATA2 is expressed in early hematopoietic progenitors.26 GATA1 is required for the development of erythrocytes, megakaryocytes, eosinophils, and mast cells. Mutations in GATA1 have been associated with thrombocytopenia, familial dyserythropoietic anemia, thalassemia, porphyria, Diamond-Blackfan anemia, TMD, and DS-AMKL.26-31 The mutations found in nonmalignant diseases either weaken or eliminate the interaction of GATA1 with its cofactor FOG1 or interfere with DNA binding.28-32 In contrast, the mutations detected in DS patients consist of short deletions, insertions, and point mutations within exon 2 that introduce a premature stop codon.7 This shorter mutant protein retains the ability to bind DNA and interact with its cofactor, but it lacks the transcriptional activation domain and hence has reduced transactivation potential.7 To model TMD, a knockin line of mice expressing a truncated form of GATA1 was generated and found to result in hyperproliferative megakaryocytic progenitors in the yolk sac and fetal liver that disappeared by the end of gestation.33 A separate group crossed mice transgenic for a truncated form of GATA1 to the GATA1 knockout strain.34 During the neonatal period, mice accumulate immature megakaryocytic progenitors in the liver that disappear during weaning of the pups. Regardless of the difference in timing, these models serve to validate that a truncated GATA1 protein is able to confer a proliferative advantage, generating a pool of precursors that have the potential to develop into a leukemic population. The mechanism whereby truncated GATA1 is able to induce a preleukemic state is not fully elucidated, although genome-wide chromatin immunoprecipitation sequencing of genes bound by GATA1 merged with expression profiling revealed a large number of activated and repressed genes, respectively, that were occupied by the GATA1 protein.35 Further studies have shown that GATA1 is able to activate lineage specific genes and repress progenitor maintenance genes depending on the cofactors present.36 It is therefore plausible that deregulation of these targets contributes to the differentiation arrest seen with the truncated GATA1 that is no longer able to transactivate transcription of lineage specific genes. A second mechanism proposed is the upregulation of genes by mutant GATA1 that promote self-renewal, as has been demonstrated for the microRNA miR-486-5p.37 Additionally, it is possible that the extra gene dosage of chromosome 21 contributes to this process; in fact, trisomy 21 has an impact on fetal hematopoiesis in and of itself.38-40 Fetal livers from DS patients have a two- to threefold increase in megakaryocyte erythroid progenitors, and trisomic stem cells exhibit alterations of hematopoiesis in vitro with an increase in multilineage colony-forming potential, an indicator of increased self-renewal.39-41 Supporting this cooperativity between GATA1 mutations and trisomy 21 is the specificity of GATA1 mutations: almost without exception, GATA1 mutations are not found outside the context of trisomy 21.26 Even in rare cases of non-DS-AMKL that carry GATA1 mutations, somatic copy number amplifications in the DS critical region of chromosome 21 are found to be present.16
Patients with trisomy 21 have, in essence, an extra copy of many genes on chromosome 21 (chr21), and overexpression of one or more has been hypothesized to provide the cellular setting that is permissible for persistence and eventual transformation of GATA1 mutant cells. Candidate genes on chr21 that contribute to a preleukemic phenotype include but are not limited to ERG, RUNX1, DYRK1A, and MIR125B2.42-45 ERG is a member of the ETS transcription gene family. Increased expression of ERG is seen in some cases of AML and it is also a translocation partner in t(16;21) myeloid leukemia.46,47 ERG has been recently shown to play a role in hematopoietic stem cells as well as the development of the megakaryocytic lineage, and furthermore, transgenic expression of ERG and a mutant GATA1 protein in murine fetal liver cells results in a TMD like disease.48-50 Additionally, overexpression of ERG in hematopoietic progenitor cells by retroviral transduction and subsequent transplantation into mice results in megakaryoblastic leukemia.44 Another candidate is the RUNX1 gene, also found on chr21. Perhaps counterintuitively, RUNX1 expression was found to be lower in DS-AMKL cases in comparison with non–DS-AMKL in 2 separate cohorts despite the increase number of genomic copies.51,52 Although the mechanism of this downregulation is not clear, in core binding factor leukemias, a decrease in RUNX1 activity either by mutation or the transdominant effect of a translocation involving RUNX1 is associated with increased leukemic potential. Thus, a downregulation of RUNX1 in DS-AMKL would be consistent with previous data that a loss of RUNX1 wild-type function enhances self-renewal and blocks differentiation. In line with this hypothesis, RUNX1 upregulation was found to precede megakaryocyte differentiation in human hematopoietic cells and downregulation was seen when cells underwent erythroid differentiation, suggesting that it functions in megakaryocytic lineage commitment.45 A decrease in RUNX1 could therefore impair differentiation allowing persistence of GATA1 mutant cells in a more immature state.
Cooperating mutations
Given that only 20% of TMD progresses to leukemia, what then are the subsequent events or alterations that promote the preleukemic state to that of a fully transformed malignancy? Exome and targeted sequencing of 46 genes has provided insight to this question, identifying recurrently mutated genes in three major categories: cohesin, epigenetic regulators, and signaling molecules.25 Core cohesin complex components including STAG2, RAD21, SMC3, SMC1A, and the cohesin complex loading protein NIPBL were mutated in 53% of the 49 DS-AMKL cases and none of the 41 TMD cases interrogated. This is significantly higher than the reported frequency of 6% to 12% in AML, suggesting these mutations may play a specific role in promoting megakaryocytic disease.53-55 Additionally, 6 cases carried mutations in CTCF, a transcriptional repressor and insulator protein. Cohesin maintains sister chromatid cohesion, allowing for faithful chromosome segregation and DNA repair.56 In addition, the complex also functions in transcriptional regulation through DNA looping. CTCF and cohesin have been found to co-localize extensively throughout mammalian genomes.57 It has been suggested that together, they play a role in the establishment and maintenance of topological domains.58 Their disruption thus has the potential to significantly disrupt chromatin architecture and, in doing so, gene expression. Interestingly, GATA1 has been found to co-occupy genes with the RAD21 cohesin component as well as CTCF in adult proerythrocytes (796 and 656 target genes, respectively), providing direct evidence for cooperative effects between these genes.59
EZH2, the catalytic subunit of the Polycomb repressive complex 2 (PRC2) was the most frequently targeted epigenetic regulator in DS-AMKL. Combined with SUZ12, PRC2 mutations were mutually exclusive and collectively occurred in 17 of 49 cases (35%), the majority of which also contained alterations in CTCF or cohesin. In erythroid cells, PRC2 is involved in epigenetic silencing of a subset of GATA1-repressed genes, some of which are associated with progenitor cells such as KIT and GATA2.60 Disruption of the repression may therefore enhance the self-renewal of cells, contributing to the differentiation block provided by the truncated GATA1 protein.
Close to 50% of DS-AMKL cases carry activating kinase mutations in JAK1, JAK2, JAK3, MPL, KRAS, or NRAS or loss-of-function mutations in SH2B3. These kinase genes fall broadly into 2 categories: JAK/signal transducer and activator of transcription (STAT) and RAS signaling, both of which play a role in megakaryopoiesis (Figure 2).61,62 Mutations between these 2 signaling cascades are, for the most part, mutually exclusive, although occasional cases carry a lesion in both. They result in constituitively activated signaling, leading to a gain of function as demonstrated by cytokine-independent growth in laboratory assays.63-65 Overexpression of one of the DS-AMKL–associated JAK3-activating mutations has been shown to result in a lethal megakaryocyte progenitor expansion in a subset of mice, further supporting this signaling pathway in AMKL.64
JAK signaling in megakaryopoiesis. Cytokine binding to its cellular receptor leads to dimerization and phosphorylation that in turn binds and activates JAK, leading to downstream activation of RAS signaling and phosphorylation of STAT transcription factors. Receptors and kinases with activating mutations identified in AMKL include MPL, PDGFRB, JAK1, JAK2, JAK3, NRAS, and KRAS. Mutations in SH2B3 have been identified in DS-AMKL. SHC1, adapter molecule; SH2B3, inhibitor of JAK2.
JAK signaling in megakaryopoiesis. Cytokine binding to its cellular receptor leads to dimerization and phosphorylation that in turn binds and activates JAK, leading to downstream activation of RAS signaling and phosphorylation of STAT transcription factors. Receptors and kinases with activating mutations identified in AMKL include MPL, PDGFRB, JAK1, JAK2, JAK3, NRAS, and KRAS. Mutations in SH2B3 have been identified in DS-AMKL. SHC1, adapter molecule; SH2B3, inhibitor of JAK2.
Non–DS-AMKL
RBM15-MKL1
The t(1;22) translocation and its association with AMKL in infants was initially identified in a cohort of 252 children with AML accrued over a 24-month period.66 In this report, no cases of t(1;22) were identified in a concurrent pediatric ALL cohort of 2382 cases, and the translocation was exclusively found in patients with AMKL, all of whom were <1 year of age. This fusion was very specific for infant AMKL, as the 22 other infants with AML who lacked the translocation had a different phenotypic subtype. Further, the remaining 12 non–DS-AMKL cases carried no recurring chromosomal abnormalities and were all older. Others have since confirmed this association, but it was not until 10 years after the initial report that the genes involved in the translocation were characterized.67-70 Two groups simultaneously identified the genes on chromosomes 1 and 22 involved in the translocation: RBM15 (also known as OTT) and MKL1 (also known as MAL), respectively.67,70 Since their initial cloning, much has been learned about the function of the genes, and a role of the translocation in inducing leukemia has been demonstrated in a knockin mouse model.71
MKL1 is a transcriptional coactivator for serum response factor (SRF), a transcription factor that regulates the expression of genes involved in cell growth, proliferation, and differentiation, as well as genes that control the actin cytoskeleton.72 In serum-starved cells, MKL1 associates with G actin monomers and is retained in the cytoplasm. Following serum stimulation and Rho-mediated actin polymerization, G actin pools are depleted and MKL1 translocates to the nucleus, associating with SRF to activate gene transcription.73,74 During murine megakaryocyte differentiation, Mkl1 is upregulated. Consistent with this, Mkl1-knockout mice have an increased percentage of megakaryocytic progenitors and a decrease in mature megakaryocytes as well as dysplastic megakaryocytes.75,76 RBM15 belongs to the Spen family of proteins and encodes a protein containing 3 amino-terminal RNA recognition motifs that bind to nucleic acids and a C-terminal SPOC domain that is thought to interact with the SMRT and NCoR corepressor complexes, as well as RBPJ, a transcription factor downstream of Notch signaling.77,78 Rbm15-knockout mice are embryonic lethal; thus, to evaluate the effect of this protein on hematopoiesis, conditional-knockout mice have been generated.79,80 These mice have a block in B lymphopoiesis and expansion of the myeloid, megakaryocytic, and progenitor compartments.75,79 The fusion of MKL1 to RBM15 deregulates the normal intracellular localization of MKL1 such that it is becomes constitutively localized to the nucleus, resulting in SRF activation even in the absence of stimuli.81 In addition to the SRF transcriptional program, the fusion also aberrantly activates RBPJ transcriptional targets. Although both transcription programs have been shown to be deregulated by the fusion gene, the degree to which they contribute to transformation is still unclear.
In studies done to address the role of the RBM15-MKL1 chimeric gene in AMKL, knockin mice were engineered to express the chimeric oncogene under control of the endogenous Rbm15 promoter.71 These mice display abnormal fetal and adult hematopoiesis, with a small fraction developing AMKL between 18 and 24 months of age.71 Using this mouse model, the authors present data to support RBM15-MKL1–activated RBPJ mediated transcriptional activity that leads to upregulation of the Notch pathway.71 Consistent with this, Rbm15 has been shown to modulate Notch-induced transcription in a cell-type–specific manner.82 Given that only a fraction of mice developed overt AMKL at a late age, the authors reasoned that cooperating oncogenic events were required to induce AMKL. The identification of such cooperating mutations has proved elusive due to a paucity of clinical samples with high tumor content for next-generation sequencing analysis. Nonetheless, careful analysis of one patient specimen along with a matched germline specimen revealed 12 high confidence mutations, one of which occurred in MMP8, a matrix metalloproteinase gene that is expressed in megakaryocyte-erythroid progenitors.83 Further studies are necessary to determine if this mutation is able to cooperate with the RBM15-MKL1 oncogene.
CBFA2T3-GLIS2
Until recently, with the exception of the RBM15-MKL1 fusion, the genetic etiology of non–DS-AMKL had remained elusive. A high-resolution study of DNA copy-number abnormalities and loss of heterozygosity on pediatric de novo AML samples demonstrated a very low burden of genomic alterations in all pediatric AML subtypes with the exception of AMKL.84 AMKL cases were characterized by complex chromosomal rearrangements and a high number of copy-number abnormalities. We predicted that these lesions would have functional consequences and therefore performed transcriptome and exome sequencing on diagnostic leukemia samples from 14 pediatric non–DS-AMKL cases as part of the St. Jude Children’s Research Hospital–Washington University Pediatric Cancer Genome Project.16 Indeed, we detected structural variations that resulted in the expression of chimeric transcripts in 12 of 14 samples. Remarkably, in 7 of 14 cases, a cryptic inversion on chromosome 16 [inv(16)(p13.3q24.3)] was detected that resulted in the joining of CBFA2T3, a member of the ETO family of nuclear corepressors, to GLIS2, a member of the GLI family of transcription factors.16 The gene expression profile of CBFA2T3-GLIS2 AMKL was distinct from that of AMKL cells lacking this chimeric transcript and from other genetic subtypes of pediatric AML.16 Furthermore, the CBFA2T3-GLIS2 fusion gene conferred a poor prognosis, a finding that has since been confirmed.16,17,85 This fusion was subsequently reported to also occur at a low frequency in pediatric cytogenetically normal AML.85 Expression of CBFA2T3-GLIS2 in Drosophila and murine hematopoietic cells induced bone morphogenic protein (BMP) signaling, a pathway not previously implicated in AML, and resulted in a marked increase in the self-renewal capacity of hematopoietic progenitors.16 The contribution of BMP signaling to self-renewal in CBFA2T3-GLIS2 modified murine hematopoietic cells has since been confirmed in colony-formation assays utilizing Bmp2 and Bmp4 conditional-knockout marrow (unpublished data).
CBFA2T3-GLIS2–expressing cells remained growth factor dependent in vitro, suggesting that cooperating mutations in growth factor signaling pathways are likely required for full leukemic transformation. Moreover, transplantation of CBFA2T3-GLIS2–transduced bone marrow cells into syngeneic recipients failed to induce overt leukemia, consistent with a requirement for cooperative mutations. Failure to induce leukemia in mice as a single lesion has been previously reported for other chimeric genes that confer the ability to serially replate in colony-forming assays, including AML1-ETO.86 Overall, the total burden of somatic mutations in our cohort was significantly lower in the CBFA2T3-GLIS2–expressing cases for which germline DNA was available than in non–DS-AMKL that lacked this fusion gene (7.2 ± 3.6 vs 16.6 ± 5.1, P = .009).16 Of the 15 CBFA2T3-GLIS2–positive cases analyzed to date, 5 carried lesions in either a Janus kinase (JAK) gene and/or a somatic amplification of the DS critical region on chromosome 21. However, the majority of cases do not contain an identifiable cooperating lesion (unpublished data).16 As these cases have been interrogated by single-nucleotide polymorphism arrays, exome, and/or transcriptome sequencing, a more thorough whole-genome approach may help to further delineate the additional events required by this fusion oncogene. Whole-genome sequencing would allow the identification of somatic mutations in noncoding intergenic regions that are oncogenic. Examples of these types of lesions include TERT promoter mutations and superenhancer formation upstream of the TAL1 oncogene, as identified in melanoma and T-cell acute lymphoblastic leukemia, respectively.87,88
Lower-frequency fusion events
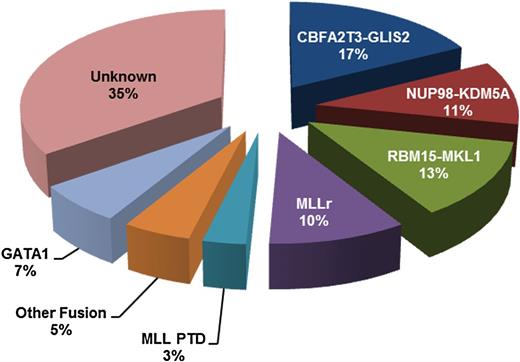
In addition to CBFA2T3-GLIS2, ∼8% of our pediatric cohort carried the previously described NUP98-KDM5A fusion gene (Figure 3).16 In parallel with our efforts, de Rooij and colleagues evaluated a separate non–DS-AMKL cohort for NUP98 fusion events by split-signal fluorescence in situ hybridization and found a similar frequency of 11%.17 NUP98, a nucleoporin family member with transactivation activity, fused to KDM5A, an H3K4me3-binding PHD finger, was initially described in adult AML.89,90 When introduced into murine bone marrow, this fusion oncogene induces a myeloid differentiation arrest and mice develop AML with an average latency of 69 days.91 Wang and colleagues demonstrated this fusion to be bound to H3K4me3 mononucleosomes, showing the PHD finger plays a role in targeting the fusion to the genome.91 Interestingly, microarray analysis identified several polycomb proteins carrying H3K4me3 marks to be transcriptionally upregulated in response to the fusion, whereas housekeeping genes with constitutive H3K4me3 marks remained unchanged. Affected polycomb targets confirmed by chromatin immunoprecipitation include genes upregulated in MLL rearranged leukemia such as HOXA5, HOXA7, HOXA9, HOXA10, MEIS1, and PBX1.91 Furthermore, the authors demonstrate a block in PRC2 binding, the complex that antagonizes polycomb proteins through transcriptional repression of target genes. Therefore, the NUP98-KDM5A fusion is able to prevent silencing of critical transcription factors that play a role in maintaining hematopoietic progenitor status, similar to MLL gene rearrangements. It is perhaps not surprising then that MLL-AF9 and MLL-AF10 fusion events have also been detected in non–DS-AMKL.17 As these lesions are also found in other subtypes of AML, there are likely additional factors contributing to the development of megakaryoblastic disease. Cooperating mutations, the target cell, and the microenvironment all have the potential to direct lineage during the process of transformation.
Key genomic events in non–DS-AMKL. A total of 142 pediatric non–DS-AMKL cases were analyzed for the presence of fusion gene events by transcriptome sequencing, reverse-transcription polymerase chain reaction (RT-PCR), or split-signal fluorescence in situ hybridization. A total of 96 samples were evaluated for the presence of the MLL-PTD by RT-PCR and 46 samples were evaluated for the presence of somatic GATA1 single-nucleotide variations and insertion/deletion by exome and/or Sanger sequencing. The proportion of MLL-PTD and GATA1 mutant cases was calculated based on the total number of samples evaluated for each of the lesions. Patients carrying GATA1 mutations did not have stigmata of DS or evidence of mutant reads in germline DNA, suggesting they are not mosaics. Cases that did not undergo transcriptome sequencing and were negative by RT-PCR for CBFA2T3-GLIS2, NUP98-KDM5A, RBM15-MKL1, and MLL rearrangements (MLLr) are designated as unknown. “Other fusion” includes single cases of each of the following: GATA2-HOXA9, NIPBL-HOXB9, MN1-FLI1, HLXB9-ETV6, FUS-ERG, and RUNX1-CBFA2T3. Data compiled from Gruber et al16 and de Rooij et al.17
Key genomic events in non–DS-AMKL. A total of 142 pediatric non–DS-AMKL cases were analyzed for the presence of fusion gene events by transcriptome sequencing, reverse-transcription polymerase chain reaction (RT-PCR), or split-signal fluorescence in situ hybridization. A total of 96 samples were evaluated for the presence of the MLL-PTD by RT-PCR and 46 samples were evaluated for the presence of somatic GATA1 single-nucleotide variations and insertion/deletion by exome and/or Sanger sequencing. The proportion of MLL-PTD and GATA1 mutant cases was calculated based on the total number of samples evaluated for each of the lesions. Patients carrying GATA1 mutations did not have stigmata of DS or evidence of mutant reads in germline DNA, suggesting they are not mosaics. Cases that did not undergo transcriptome sequencing and were negative by RT-PCR for CBFA2T3-GLIS2, NUP98-KDM5A, RBM15-MKL1, and MLL rearrangements (MLLr) are designated as unknown. “Other fusion” includes single cases of each of the following: GATA2-HOXA9, NIPBL-HOXB9, MN1-FLI1, HLXB9-ETV6, FUS-ERG, and RUNX1-CBFA2T3. Data compiled from Gruber et al16 and de Rooij et al.17
In addition to the previously described NUP98-KDM5A fusion, we identified 3 novel fusion genes expressed in a single case each: GATA2-HOXA9, MN1-FLI1, and NIPBL-HOXB9 (Figure 3). Each of these chimeric transcripts are predicted to encode a fusion protein that would alter signaling pathways known to play a role in normal hematopoiesis, suggesting that these lesions are “driver” mutations that directly contribute to the development of leukemia. Several of the genes involved in these translocations play a direct role in normal megakaryocytic differentiation (GATA2 and FLI1), have been previously shown to be involved in leukemogenesis (HOXA9, MN1, and HOXB9), or are highly expressed in hematopoietic stem cells or myeloid/megakaryocytic progenitors.91-96 Genome-wide approaches in a larger AMKL cohort are necessary to determine if these fusion genes are recurrent. Current efforts in our laboratories include experiments to determine the ability of these fusion genes to enhance self-renewal, block differentiation, and induce leukemia in murine model(s) with a focus on the mechanism whereby these processes take place.
Conclusion
Pediatric AMKL is a heterogeneous disease comprising chimeric oncogenes or truncating GATA1 mutations that enhance self-renewal and block myeloid differentiation. Cooperating mutations that contribute to transformation include amplifications of chromosome 21 (either somatic or constitutional) as well as single-nucleotide variations and insertion/deletion in cohesin complex genes, CTCF, epigenetic regulators, and kinase genes. In ∼35% of pediatric non–DS-AMKL cases, the genetic alterations leading to the malignancy are unknown, warranting further comprehensive genomic studies (Figure 3). CBFA2T3-GLIS2 is the most frequent fusion event with a distinct biology in addition to a poor prognosis, occurring in 18% of patients. Development of targeted agents that inhibit the fusion directly, or critical self-renewal pathways upregulated as a result of the fusion, such as BMP, may provide therapeutic benefit. The diversity of CBFA2T3-GLIS2–negative non–DS-AMKL cases suggest that alternative less targeted approaches, such as the promotion of megakaryoblast differentiation, should be evaluated in an attempt to improve outcomes across patients with a wide spectrum of mutations.97,98 The presence of JAK/STAT- and RAS-pathway–activating mutations provides a rationale for the use of kinase inhibitors, although their role as cooperating hits warrants caution, as these agents may be additive to existing treatment but not sufficient to eliminate disease on their own.
Acknowledgments
This work was supported by grants from the Eric Trump Foundation, Gabrielle Angel Foundation, and the American Lebanese Syrian Associated Charities of St. Jude Children’s Research Hospital.
Authorship
Contribution: T.A.G. and J.R.D. wrote the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Tanja A. Gruber, Department of Oncology, St. Jude Children’s Research Hospital, Memphis, TN 38105; e-mail: tanja.gruber@stjude.org.



This feature is available to Subscribers Only
Sign In or Create an Account Close Modal