In this issue of Blood, Wilson et al generate and analyze a treasure trove of epigenetic data, such as transcription factor occupancy, histone modifications, and chromatin interaction frequencies, genome-wide (ie, epigenomic data), in a cell line model of hematopoietic stem/progenitor cells (HSPCs).1 To appreciate the importance of these data, consider an analogy of gene expression being a song or symphony (transcripts) played by musicians (transcription factors and transcriptional machinery) reading the score encoded in the genome sequence. Previous studies2 revealed the positions of a few transcription factors across the genome, so we only knew about, for example, the violinists and oboists. No wonder we did not understand how the music was being generated (how expression was regulated). By mapping the sites of occupancy of many more transcription factors (now a total of 29), as well as positions of 4 histone modifications and DNase hypersensitive sites, Wilson et al1 reveal many more of the players and their partners. Furthermore, their data on 3-dimensional interaction frequencies of chromatin show how groups of musicians (protein complexes) come together in an orchestra to read the score and perform a symphony.
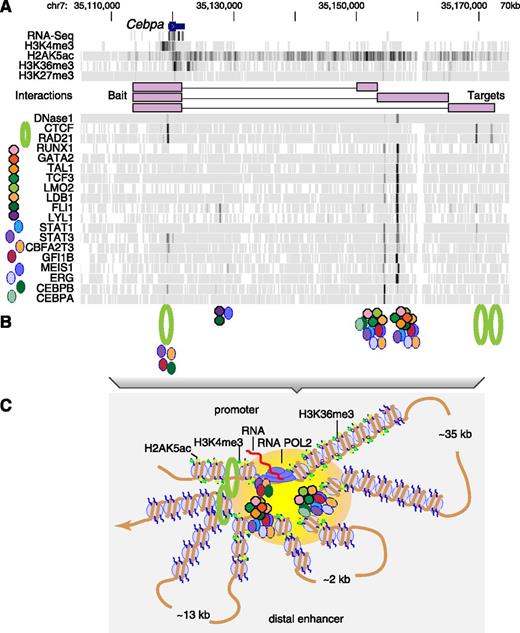
A landscape of diverse regulatory features coupled with chromatin interaction data leads to descriptive models of gene regulation. (A) As illustrated for the Cebpa locus (gene on top line), the strength of signals (proportional to the density of the gray along each track) from sequencing RNA, locations of cleavage by DNase, and chromatin immunoprecipitation of modified histones and transcription factors (named on the left and marked by distinctive icons) reveals where along the locus all these players in gene regulation are located. This panel was generated from data displayed at http://tinyurl.com/E-MTAB-3954. (B) Colocation of the transcription factors indicates the positions of at least 3 categories of protein complexes. Members of an octameric complex (a previously described heptamer2 plus TCF3) are shown as octagons of distinctive colors, members of other complexes are shown as ovals, and the RAD21 component of cohesin plus CTCF are shown as a green circle. (C) Maps of the frequency of chromatin interactions (shown as connections between purple rectangles representing HindIII fragments in the promoter Hi-C experiment in A) show that at least 3 complexes of proteins bound downstream of Cebpa interact with complexes at the promoter. Thus, one can surmise a structure with the promoter and enhancers juxtaposed in a region with transcriptional activity (indicated by the yellow-orange oval), anchored on cohesin complexes, and with intervening DNA in 3 loops of widely differing sizes.
A landscape of diverse regulatory features coupled with chromatin interaction data leads to descriptive models of gene regulation. (A) As illustrated for the Cebpa locus (gene on top line), the strength of signals (proportional to the density of the gray along each track) from sequencing RNA, locations of cleavage by DNase, and chromatin immunoprecipitation of modified histones and transcription factors (named on the left and marked by distinctive icons) reveals where along the locus all these players in gene regulation are located. This panel was generated from data displayed at http://tinyurl.com/E-MTAB-3954. (B) Colocation of the transcription factors indicates the positions of at least 3 categories of protein complexes. Members of an octameric complex (a previously described heptamer2 plus TCF3) are shown as octagons of distinctive colors, members of other complexes are shown as ovals, and the RAD21 component of cohesin plus CTCF are shown as a green circle. (C) Maps of the frequency of chromatin interactions (shown as connections between purple rectangles representing HindIII fragments in the promoter Hi-C experiment in A) show that at least 3 complexes of proteins bound downstream of Cebpa interact with complexes at the promoter. Thus, one can surmise a structure with the promoter and enhancers juxtaposed in a region with transcriptional activity (indicated by the yellow-orange oval), anchored on cohesin complexes, and with intervening DNA in 3 loops of widely differing sizes.
The breadth and diversity of epigenomic data on HPC-7 cells now are on a par with those for a small number cell lines studied intensively in multiple laboratories (such as embryonic stem cells) and major consortia (such as K562, HepG2, and GM12857 cells in the ENCODE Project Consortium3 ). Although data from transformed cells such as K562 and HepG2 are useful for deducing some general principles, data from primary cells are the most relevant to specific issues. Although a specialized approach has generated histone modification maps in HSPCs,4 the scarcity of these cells precludes application of most genome-wide assays. This large collection of epigenomic data in HPC-7 cells is a great boon to hematology, as this multipotent cell line is capable of differentiating into several myeloid lineages,5 and thus serves as a model for HSPCs.
The 3-dimensional chromatin interaction maps generated by Wilson et al1 turn the static landscape inferred from the maps of nuclease accessibility, transcription factor occupancy, and histone modifications into a snapshot of regulatory regions working together (see figure). The complex interactions among regulatory regions first revealed in studies of hemoglobin genes also are found for many, if not most, genes regulated in a stage- and/or tissue-specific manner. Multiple candidate enhancers, as predicted by patterns of histone modifications and factor occupancy, can be identified for most genes, but the epigenomic maps do not reveal the target genes for the candidate enhancers. This is especially problematic in gene-dense regions. Although proximity and correlations of epigenomic signals can be used to infer targets, direct information about interactions between regulatory regions currently is the best guide. Generating maps of interaction frequency across an entire genome at high resolution6 requires a staggering number of sequencing reads, well beyond the budget or capacity of most investigators. Thus, Wilson et al1 adopted the promoter Hi-C approach7 to reveal a highly informative subset of interactions: those between promoters and distal regions.
To illustrate the power of the new data, consider the gene Cebpa (see figure). The detailed maps (see figure panel A) identify the locations of transcription factors, which is analogous to knowing the locations of musicians resolved by instrument played (violin, oboe, flute, percussion, etc). Aligning the maps reveals groups of colocated proteins (summarized in figure panel B) defining a complex of hematopoietic transcription factors (analogous to the string section in an orchestra), other complexes of transcription factors (analogous to the woodwinds), and components of cohesin (analogous to the percussion section). The 3-dimensional interaction maps show that all these components are close together physically, with the components (separated along genomic coordinates) coming together in an orchestra of regulatory molecules (see figure panel C). Wilson et al1 show that this candidate enhancer activates reporter gene expression in a tissue-specific manner in transgenic mice embryos.
To facilitate access to and use of this information, Wilson et al1 provide these data in the CODEX database8 and at a stable URL for visualization in a genome browser. Thus, investigators can easily find levels of transcripts, maps of epigenomic features, and interaction frequencies in genes and loci of interest to them. These data should catalyze refinement and improve accuracy in identifying candidate enhancers and assigning them to target genes.
This improvement in the accuracy and completeness of our views of regulatory domains can also facilitate clinical research. More and more examples are being reported of the phenotypic effect of genetic variants and mutations in regulatory regions.3,9,10 For phenotypes expressed in myeloid cells, the maps and resources provided by Wilson et al1 will be particularly valuable.
These new data will serve as a strong resource for much future work, but they are not the final story. The promoter Hi-C data are valuable for the interactions they reveal, but the experimental design precludes the discovery of many interactions, and some chromatin interactions play key roles at other stages of differentiation. The binding patterns for some transcription factors are highly dynamic, and thus their occupancy needs to be mapped at multiple stages and in different lineages. The data in the article by Wilson et al1 will help guide these additional studies and provide an important point of reference for comparison with new results.
These new data, coupled with the large amount of information from many laboratories, provide a rich description of the molecular players regulating expression of each locus. The next challenge is to build on this descriptive foundation to generate predictive models of expression, in which the role of each protein complex and each cis-regulatory module is defined quantitatively as an outcome on gene expression. Such models, after extensive experimental testing, could provide a basis for consolidating information about the many regulatory complexes at genetic loci into mechanistic rules for gene regulation that apply broadly across genomes and cell types. That would be a notable achievement in our understanding of gene regulation during hematopoiesis.
Conflict-of-interest disclosure: The author declares no competing financial interests.