Key Points
New genome-wide maps for 17 TFs, 3 histone modifications, DNase I sites, Hi-C, and Promoter Capture Hi-C in a stem/progenitor model.
Integrated analysis shows that chromatin loops in a stem/progenitor model are characterized by specific TF occupancy patterns.
Abstract
Comprehensive study of transcriptional control processes will be required to enhance our understanding of both normal and malignant hematopoiesis. Modern sequencing technologies have revolutionized our ability to generate genome-scale expression and histone modification profiles, transcription factor (TF)-binding maps, and also comprehensive chromatin-looping information. Many of these technologies, however, require large numbers of cells, and therefore cannot be applied to rare hematopoietic stem/progenitor cell (HSPC) populations. The stem cell factor–dependent multipotent progenitor cell line HPC-7 represents a well-recognized cell line model for HSPCs. Here we report genome-wide maps for 17 TFs, 3 histone modifications, DNase I hypersensitive sites, and high-resolution promoter-enhancer interactomes in HPC-7 cells. Integrated analysis of these complementary data sets revealed TF occupancy patterns of genomic regions involved in promoter-anchored loops. Moreover, preferential associations between pairs of TFs bound at either ends of chromatin loops led to the identification of 4 previously unrecognized protein-protein interactions between key blood stem cell regulators. All HPC-7 data sets are freely available both through standard repositories and a user-friendly Web interface. Together with previously generated genome-wide data sets, this study integrates HPC-7 data into a genomic resource on par with ENCODE tier 1 cell lines and, importantly, is the only current model with comprehensive genome-scale data that is relevant to HSPC biology.
Introduction
Modern DNA-sequencing technologies have revolutionized our ability to generate genome-wide data sets that capture a wide range of processes involved in the transcriptional control of gene expression. In addition to gene expression profiling, these range from genome-wide maps of histone modification status and open chromatin to comprehensive information on transcription factor (TF) binding, and, more recently, the genome-wide analysis of the 3-dimensional architecture of chromosomes that mediate the interactions between gene promoters and distal regulatory elements. When interrogated in isolation, however, it has become increasingly recognized that only limited new biological insights can be extracted from individual genome-scale data sets. Large consortia efforts have therefore been assembled to generate integrated multiomics data sets that cover multiple levels of the transcriptional control process.1-7
Hematopoietic stem/progenitor cells (HSPCs) ensure the lifelong supply of mature blood cells, and their dysregulation forms the basis for a wide range of hematopoietic diseases. HSPC function critically depends on finely tuned transcriptional control processes, a fact highlighted by the common occurrence of leukemogenic driver mutations in transcriptional and epigenetic regulators.8-10 HSPCs represent exceedingly rare cell populations in both human and mouse, with <1:20 000 bone marrow cells estimated to possess stem cell activity. Although gene expression profiles have been reported for highly purified single HSPCs11,12 and histone modifications have been mapped in purified bone marrow HSPC populations,13 no protocols exist for the application of other genome-wide mapping techniques for highly purified stem and/or progenitor cells. Researchers have therefore relied on the use of either heterogeneous primary cell sources such as human CD34+ cells,14,15 or the use of cytokine-dependent model cell lines such as the multipotent stem cell factor (SCF)-dependent HPC-7 cell line.16 Importantly, however, none of these studies, nor any of the large consortia efforts have so far reported the whole range of complementary genome-scale data sets for a single HSPC model.
We previously reported genome-wide TF-binding maps as well as RNA-Seq expression and histone H3 lysine 27 acetylation (H3K27ac) profiles in HPC-7 cells.17,18 Here we report binding maps for an additional 17 TFs, 3 histone marks, genome-wide DNase I hypersensitive sites, genome-wide chromosomal contacts maps generated by Hi-C,19 and high-resolution genome-wide promoter–distal element interactions mapped by the recently reported Promoter Capture Hi-C method,20,21 all generated within uniformly cultured HPC-7 cells. Integrated analysis of these complementary data sets demonstrated that (1) active looping of distal TF-bound regions provides a powerful way to identify new enhancers that are active in vivo in transgenic mice in blood-forming tissues, (2) TF colocalization analysis identifies distinct transcriptional programs operating within a single-cell type with a program driven by 13 TFs being specifically associated with HSPC identity, (3) individual TFs differ in their preference for promoter or enhancer binding within genomic regions that are involved in promoter-distal interactions, and (4) computational analysis of preferential pairwise interactions of TFs involved in promoter-distal looping can correlate with their ability for direct protein-protein interactions. All data sets are freely accessible through an intuitive Web browser interface (CODEX),22 thus providing the hematopoietic research community, for the first time, with comprehensive genome-scale data that cover the whole range of the transcriptional control processes within a single model for HSPCs.
Materials and methods
For more detailed protocols, see supplemental Materials and methods (available on the Blood Web site).
Hi-C with sequence capture enrichment
Hi-C raw data processing
Four replicates of CHi-C paired-end sequencing data (2 technical replicates per each biological replicate) were quality controlled, aligned to mm9, and filtered with HiCUP (http://www.bioinformatics.babraham.ac.uk/projects/hicup/). Technical replicates were then merged and de-duplicated. Signal detection on the resulting 2 aligned, pooled biological replicates was then jointly performed using CHiCAGO24 and the associated chicagoTools suite; a score threshold of 5 was used to define significant interactions. Promoter-promoter interactions and known promoter elements (taken from MPromDB promoters) which had not been included in the custom-designed capture bait library were also removed using in-house scripts. Further analysis was performed using SeqMonk (http://www.bioinformatics.babraham.ac.uk/projects/seqmonk/) and the data were visualized using the WashU Epigenome browser.25
ChIP-Seq similarity analysis
Chromatin immunoprecipitation sequencing (ChIP-Seq) data were processed as previously described22 ; peaks were called using MACS226 and lifted over to mm9. The peaks were remapped to restriction fragment regions and used to generate a binary binding matrix. Similarity analysis was performed using normalized pointwise mutual information (NPMI).27,28 After normalization, NPMI ranged from 1 for complete co-occurrence (correlation limit), 0 for independent peaks profiles, and −1 when peaks did not occur together (anticorrelation limit). NPMI values were clustered using Euclidean distance and Ward linkage in R.
Binding site and looping region overlap densities
R was used to generate a histogram showing the number of ChIP-Seq peaks which were overlapping with either mate in an interacting region when compared with an equal number of arbitrary regions randomly chosen from the University of California, Santa Cruz (UCSC) repeat masker table file (this represents the mouse genome with all annotated repeats removed, to ensure that no repeat regions are considered within the background calculations due to the problems of mapping ChIP-Seq peaks reliably to repeats).
Enhancer and promoter ChIP-Seq overlaps
The R statistical environment was used to generate a bar chart counting TF-binding sites overlaps with baits/promoters vs distal regions (promoter-interacting regions).
Enhancer and promoter loops
Using in-house scripts, a matrix was generated by counting the number of either promoter or distal element regions from the CHi-C data that overlap with the ChIP-Seq peaks. Simulated matrices were generated using arbitrary peak regions (as described previously), and used to normalize the observed matrix. A P value was assigned to each element of the matrix, calculated using the number of times that the value was greater in the simulated matrices than in the observed matrix (B) plus 1, divided by the number of simulations (M) plus 1; pval = (B + 1)/(M + 1).29 A heatmap was generated in R using the ggplots library. The resulting heatmap reveals significant TF-binding patterns at interacting regions.
In vivo validation of potential regulatory elements
Identified genomic regions were polymerase chain reaction amplified from mouse genomic DNA and inserted in lacZ reporter plasmids. F0 transgenic mouse embryos were generated by Cyagen Biosciences. Expression of the transgene in the fetal liver and the dorsal aorta was confirmed in selected embryos by performing histologic sections, as described previously.30 All animal studies were performed according to United Kingdom Home Office guidelines with Home Office approval.
Chromatin immunoprecipitation
HPC-7 cells16 were grown in SCF, ChIP assays were performed as previously described,18 and all samples were crosslinked using 1% formaldehyde unless otherwise stated. For a list of antibodies used, see supplemental Materials and methods. Each sample was amplified and sequenced using the Illumina HiSeq 2500 following the manufacturer’s instructions. Sequencing reads were mapped to the mouse reference genome (GRCm38/mm10) using bowtie2, lifted over to mm9, converted to a density plot, and displayed as UCSC genome browser custom tracks.
DNase I hypersensitive site mapping
DNase I treatment was performed on permeabilized cells as described previously.31,32 HPC-7 cells were harvested and enriched for live cells, and 6 × 106 cells were incubated with 20 U of DNase I for 3 minutes. DNA was purified by phenol/chloroform extraction. DNase I treated DNA was size-selected and sequencing libraries were prepared using the Illumina Truseq ChIP kit according to the manufacturer’s instructions. Peaks were called with F-Seq33 using a standard deviation threshold of 14.
Coimmunoprecipitation
293T cells were transiently transfected with expression plasmids using the Protransfection Mammalian Transfection System (Promega) and incubated 48 hours before analysis. Cells were lysed and supernatants were precleared. Relevant antibodies were added. The immune complexes were washed, boiled in sample buffer, and analyzed by western blot.
Results
Genome-wide capture Hi-C data for HPC-7 reveals promoter contacts for known distal regulators
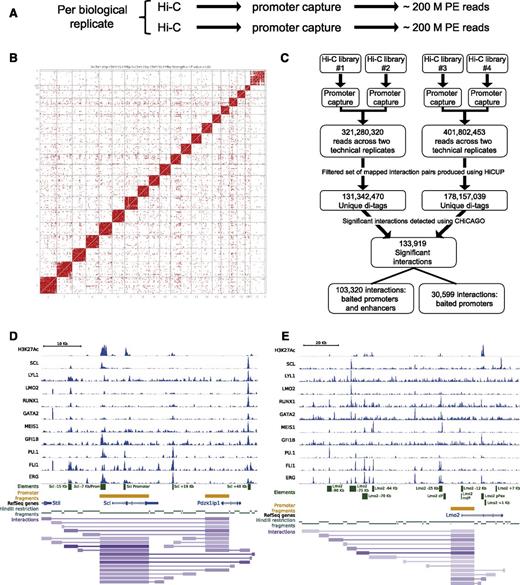
Comprehensive knowledge of distal interactions is vital to understanding gene-regulatory programs at genome scale, yet traditional Hi-C methods suffer from lack of coverage due to the highly complex nature of genomic interactions. Several laboratories have developed adaptations of genome-wide capture protocols, where interactions involving promoters are enriched by sequence homology-based capture and thus gain sufficient sequencing depth for this subset of all possible interactions.34,35 To generate such a genome-wide data set for the HPC-7 cells, we followed the Promoter Capture Hi-C protocol (CHi-C) from Schoenfelder et al,21 enriching the Hi-C material for 22 225 annotated promoters using sequence capture with a library of custom-synthesized biotinylated RNAs.
Two Hi-C libraries were generated per biological replicate (4 in total) and of these, 2 were analyzed by Illumina sequencing to ensure high complexity of the generated libraries at this initial stage in the protocol (1 per biological replicate). Promoter capture was then performed on each of the Hi-C libraries, resulting in 4 CHi-C libraries (Figure 1A). High-throughput sequencing generated a total of over 400 million paired-end reads, which were aligned (see “Materials and methods”) to generate a contact map showing both intra- and interchromosomal ligation products (Figure 1B). To identify significant interactions, we took advantage of a newly developed statistical method, CHiCAGO,24 whose background model accounts for both technical noise and the distance-dependent random collisions between DNA fragments (Figure 1C). This analysis identified over 133 000 significant interactions, of which >100 000 were specific interactions between promoters and nonpromoter distal elements. Of note, the promoter regions/baits are contained within a restriction fragment which commonly encompasses a larger fragment of the genome than the specific promoter region. On average, the promoter fragments/baits are 6880 bp. Visualization of the interaction files together with our previously published 10 TF ChIP-Seq demonstrated specific interactions of the Scl (also known as Tal1) and Lmo2 promoters with the previously characterized enhancer regions at Scl −15 kb, +19 kb, and +40 kb, as well as Lmo2 −75 kb, −70 kb, −64 kb, and the proximal promoter (pPex) (Figure 1D-E). Of interest, the characterized Scl enhancer elements also interact with the promoter of the neighboring Pdzk1ip1 gene, consistent with previous reports suggesting that Scl and Pdzk1ip1 form a single transcriptional domain.36 Analysis of well-characterized gene loci encoding key HSPC regulators therefore suggests that the newly generated CHi-C data set represents a valuable resource to advance our understanding of transcriptional control mechanisms in HSPCs.
Genome-wide Promoter Capture Hi-C data reiterate previously known promoter-distal element interactions. (A) Brief schematic of CHi-C experimental pipeline. (B) Contact map generated in Seqmonk using the 2 biological replicates of Hi-C data. (C) Schematic of the CHi-C processing pipeline. Significant interactions as identified by the CHiCAGO pipeline24 were loaded into the WashU browser as a custom track (interactions) along with the bigwig tracks of the previously published HPC-7 ChIP-Seq data.17,18 Only interactions where both interacting fragments are within the genomic window are shown. For visualization purposes, the promoters used as “bait” in the CHi-C protocol are also shown (promoter fragments), the individual restriction enzyme fragments (HindIII restriction fragments) and a track showing RefSeq genes. All tracks shown are in mouse genome build mm9. (D) Scl locus; shown for verification are the previously identified regulatory elements of the Scl locus. (E) Lmo2 locus; shown for verification are the previously identified regulatory elements of the Lmo2 locus. PE, paired end.
Genome-wide Promoter Capture Hi-C data reiterate previously known promoter-distal element interactions. (A) Brief schematic of CHi-C experimental pipeline. (B) Contact map generated in Seqmonk using the 2 biological replicates of Hi-C data. (C) Schematic of the CHi-C processing pipeline. Significant interactions as identified by the CHiCAGO pipeline24 were loaded into the WashU browser as a custom track (interactions) along with the bigwig tracks of the previously published HPC-7 ChIP-Seq data.17,18 Only interactions where both interacting fragments are within the genomic window are shown. For visualization purposes, the promoters used as “bait” in the CHi-C protocol are also shown (promoter fragments), the individual restriction enzyme fragments (HindIII restriction fragments) and a track showing RefSeq genes. All tracks shown are in mouse genome build mm9. (D) Scl locus; shown for verification are the previously identified regulatory elements of the Scl locus. (E) Lmo2 locus; shown for verification are the previously identified regulatory elements of the Lmo2 locus. PE, paired end.
Colocalized TF binding coupled with genome-wide Promoter Capture Hi-C identifies previously unknown hematopoietic enhancers
We had shown previously that HSPC enhancer elements can be identified successfully from SCL ChIP-Seq data in HPC-7 cells.37 To extend this approach, we searched for regions in the genome which were bound by at least 7 of the 10 TFs previously mapped,18 and also showed elevated levels of the histone-modification H3K27ac which is known to be associated with active enhancer regions.38 Identification of putative enhancers based on ChIP-Seq data alone cannot assign distal regions to specific genes with confidence because enhancers are known to have the ability to act over large distances, and may loop over intervening genes.39 To overcome this limitation, we made use of our CHi-C interaction list, and filtered our list of putative enhancers to only retain those that looped to the promoter regions of known regulators of HSPC function.
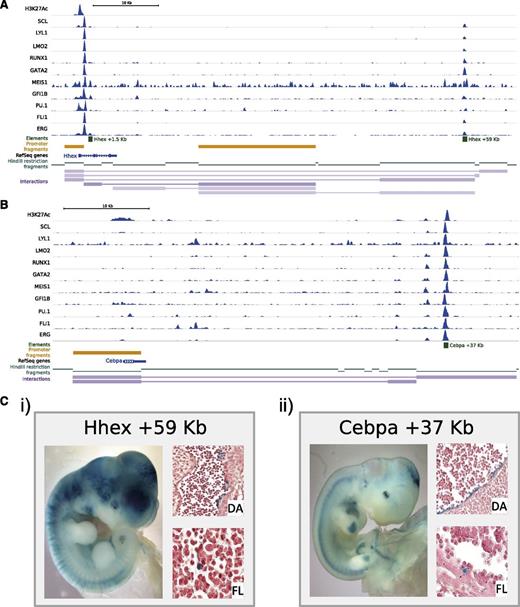
Of the specific regions that were identified, we focused on Hhex +59 kb and the Cebpα +37 kb40,41 distal elements (Figure 2A-B). Whereas previously enhancer elements have been linked to genes based on proximity or because the element could recapitulate the endogenous expression pattern of the gene, the CHi-C data allowed us to convincingly associate distal regions with specific gene promoters. Because the classic method to test the in vivo activity of a potential element is to perform F0 transgenic assays,42 we next generated lacZ reporter constructs containing a basal promoter element with the Hhex +59 kb and Cebpα +37 kb elements, respectively. Consistent tissue-specific lacZ expression in multiple independent embryos can confirm the true in vivo activity of potential regulatory elements. Importantly, analysis of midgestation mouse embryos can capture activity of key anatomic sites of HSPC location including the fetal liver (FL) and aorta-gonad-mesonephros region. The Hhex +59 kb element showed consistent staining of the vessels (3 of 3), FL (3 of 3), heart (2 of 3), and yolk sac (3 of 3), whereas the Cebpα +37 kb element showed staining of the central nervous system (5 of 8), somites (4 of 8), FL (4 of 8), and yolk sac (5 of 8) (Figure 2Ci-ii), thus validating both regions as novel transcriptional enhancers active in relevant expression domains for these 2 key regulatory genes. Further in-depth investigation into the staining pattern by histologic sectioning of the embryos showed specific localized lacZ staining of the FL, heart, and dorsal aorta (DA) (Figure 2Ci-ii and data not shown). Taken together, our approaches demonstrate that integrated analyses of ChIP-Seq data sets with genome-wide Promoter Capture Hi-C information streamlines the identification of regulatory elements, and thus integrates key regulatory genes into wider transcriptional networks.
A combination of colocalization of TF binding and genome-wide interaction data identifies previously unknown hematopoietic enhancer elements. Significant interactions as identified by the CHiCAGO pipeline were loaded into the WashU browser as a custom track (interactions) along with the bigwig tracks of the previously published HPC-7 ChIP-Seq data.17,18 Only interactions where both interacting fragments are within the genomic window are shown. For visualization purposes, the promoters used as “bait” in the CHi-C protocol are also shown (promoter fragments), the individual restriction enzyme fragments (HindIII restriction fragments) and a track showing RefSeq genes. All tracks shown are in mouse genome build mm9. (A) Hhex locus; shown for verification are the previously identified regulatory elements and the newly identified Hhex +59 kb enhancer. (B) Cebpα locus; shown for verification is the Cebpα +37 kb enhancer. (C) Transgenic analysis of E11.5 X-Gal (5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside)-stained whole-mount embryos and paraffin sections of dorsal aorta (DA) and fetal liver (FL). (i) Hhex +59 kb. (ii) Cebpα +37 kb.
A combination of colocalization of TF binding and genome-wide interaction data identifies previously unknown hematopoietic enhancer elements. Significant interactions as identified by the CHiCAGO pipeline were loaded into the WashU browser as a custom track (interactions) along with the bigwig tracks of the previously published HPC-7 ChIP-Seq data.17,18 Only interactions where both interacting fragments are within the genomic window are shown. For visualization purposes, the promoters used as “bait” in the CHi-C protocol are also shown (promoter fragments), the individual restriction enzyme fragments (HindIII restriction fragments) and a track showing RefSeq genes. All tracks shown are in mouse genome build mm9. (A) Hhex locus; shown for verification are the previously identified regulatory elements and the newly identified Hhex +59 kb enhancer. (B) Cebpα locus; shown for verification is the Cebpα +37 kb enhancer. (C) Transgenic analysis of E11.5 X-Gal (5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside)-stained whole-mount embryos and paraffin sections of dorsal aorta (DA) and fetal liver (FL). (i) Hhex +59 kb. (ii) Cebpα +37 kb.
Seventeen new genome-wide TF-binding profiles and DNase I hypersensitive site mapping enrich the combinatorial binding information of the HSPC cell model, HPC-7
Large consortia efforts have highlighted the benefits of generating large numbers of genome scale data for individual cell types, such as the tier 1 ENCODE cell lines.6,7,43 Given that HPC-7 represents one of the best in vitro models for HSPCs, we wanted to bring genomic information for these cells up to a similar level of completeness, and therefore performed ChIP-Seq experiments for a further 17 TFs (CEBPα, CEBPβ, cFOS, cMYC, E2F4, EGR1, ELF1, ETO2, c-JUN, LDB1, MAX, MYB, NFE2, p53, RAD21, pSTAT1, and STAT3) as well as genome-wide DNase I hypersensitive mapping and 3 additional histone marks (H2AK5ac, H3K4me3, and H3K36me3) (Figure 3). The additional histone marks included in this study all mark regions of active chromatin. H2AK5ac specifically marks expressed gene loci and is complementary to the repressive H3K27me3.44,45 Visual inspection of the genome-wide binding profiles for the new total of 29 TFs showed a wide variety of binding patterns with hematopoietic TFs commonly colocalized whereas additional factors such as cFOS, cMYC, E2F4, and STAT3 exhibit independent binding profiles. Of interest, while the binding patterns of RAD21 and CTCF appear to be very similar, many of these genomic locations do not exhibit particularly prominent DNase I hypersensitive sites (Figure 3; supplemental Figure 1). We investigated this phenomenon across our entire data sets, which demonstrated that CTCF/RAD21 peaks which were not called as DNase I peaks (15 038 peak regions) had a much lower signal for DNase I than those CTCF/RAD21 peaks that were also called as DNase I peaks (11 521 peak regions). Strikingly, a subset of CTCF/RAD21 peaks displayed a complete absence of DNase I signal (supplemental Figure 1).
Additional genome-wide TF-binding profiles and DNase I hypersensitive site mapping enrich the combination binding information of the HSPC cell model, HPC-7. Raw ChIP-Seq read data were transformed into a density plot for each TF and loaded into the UCSC genome browser as custom tracks above the UCSC tracks for gene structure; all tracks are shown in mouse genome build mm10. (A) Gfi1 locus. (B) Atp6v1c1 locus. (C) Ralgps2t and Tex35 loci.
Additional genome-wide TF-binding profiles and DNase I hypersensitive site mapping enrich the combination binding information of the HSPC cell model, HPC-7. Raw ChIP-Seq read data were transformed into a density plot for each TF and loaded into the UCSC genome browser as custom tracks above the UCSC tracks for gene structure; all tracks are shown in mouse genome build mm10. (A) Gfi1 locus. (B) Atp6v1c1 locus. (C) Ralgps2t and Tex35 loci.
Having 29 TF-binding profiles from the same HSPC model allowed us to perform correlation analysis of global binding profiles (Figure 4). Using NPMI,27,28 we observed association between the so-called HSPC TFs (ERG, FLI1, MEIS1, GFI1B, pSTAT1, MYB, GATA2, LYL1, LMO2, RUNX1, E2A, LDB1, and SCL) and, furthermore, within this cluster there was even stronger correlation between a subset of these TFs (GATA2, LYL1, LMO2, RUNX1, E2A, LDB1, and SCL). A separate cluster was formed which was composed largely of more widely expressed TFs such as cMYC and E2F4, but also contained some myeloid TFs including SPI1/PU.1. A third completely independent cluster is made up of CTCF and RAD21, which, due to their known involvement in chromatin structure, can be considered as “structural” factors.46 Of interest, these “structural” factors appear to negatively correlate with the HSPC TFs, which could also be seen by visual inspection of binding profiles (Figure 3). Taken together, the new data sets generated here provide deep genomic characterization of a valuable HSPC cell model. To facilitate access for the wider community, we have made all data available on the CODEX Web browser and a stable Web link (http://tinyurl.com/E-MTAB-3954), in addition to the standard submission to DNA sequence archives. As a comparison, we also analyzed published data for a tier 1 cell line from ENCODE, and therefore performed NPMI on TF ChIP-Seq data sets for the K562 cell line (supplemental Figure 2). K562 ChIP-Seq data sets separated into 3 clusters, with 1 cluster including TFs which play roles in cell cycle and proliferation (MAX, cMYC, E2F4, E2F6, ETS1, ELF1, and EGR1) whereas the second cluster contained many of the myeloid TFs such as SCL, GATA2, and GATA1. The final cluster contained only CTCF and RAD21 as seen in the HPC-7 data. A similar number of TFs were covered for HPC-7 and K562, but because several of the “HSPC” TFs were not studied within K562, the HSPC TF cluster could not be observed in this cell line.
TF correlation analysis highlights combinatorial binding patterns in an HSPC cell model. Correlation analysis was performed using NPMI. The heatmap separates the 29 factors into “structural,” myeloid/generic TFs, and HSPC TFs.
TF correlation analysis highlights combinatorial binding patterns in an HSPC cell model. Correlation analysis was performed using NPMI. The heatmap separates the 29 factors into “structural,” myeloid/generic TFs, and HSPC TFs.
Combinatorial TF binding characterizes genomic regions interacting with promoters
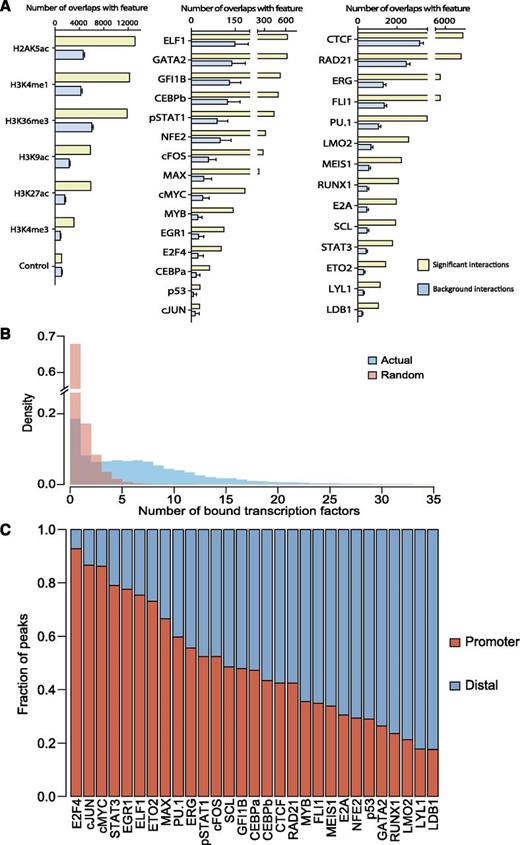
Having multi-TF binding and CHi-C data for the same cell type allowed us to investigate patterns of TF binding associated with promoter-distal element interactions. We first assessed the enrichment of individual TFs and histone modifications at promoter-interacting fragments. To do this, we calculated the number of promoter-interacting fragments that overlap with a given TF/histone mark, and compared this to distance-matched samples of “background noninteracting” regions (fragments for which no promoter interactions were detected as significant by the CHiCAGO pipeline) (Figure 5A). For this analysis, we used 29 TF and 6 histone modifications,17,18,37,47 all of which were found to be significantly enriched at promoter-interacting regions, in line with previous suggestions that TFs and their cofactors play critical roles in genomic looping.48,49 Having established significant binding to looping regions for all TFs when considered individually, we next investigated combinatorial binding of multiple TFs. To this end, we calculated the number of TFs bound to all promoter-interacting regions and compared this to random genomic locations (selected by taking an equal number of genomic coordinates randomly selected) (Figure 5B). This analysis clearly showed that for the control set of regions, most were bound by just 1 TF, and very few by >5. In contrast, regions involved in looping were commonly bound by multiple TFs.
Combinatorial TF binding is associated with promoter-distal interacting genomic regions. (A) Bar chart showing the enrichment of the individual histone modification or TF-binding peaks overlapping with the significant interaction fragments (yellow) or background interactions (blue). (B) Bar chart showing the overlapping number of bound TFs with the genomic coordinates of the CHi-C interaction data (blue) and random genomic coordinates (red). Specific enrichment for the CHi-C interaction data can be seen when 3 or more TFs are bound. (C) Individual binding peaks for each TF were separated into promoter (“baits” for CHi-C experiment) or distal (promoter-interacting regions, not considered “bait”). Distinct distributions of the 29 factors can be seen throughout the genome.
Combinatorial TF binding is associated with promoter-distal interacting genomic regions. (A) Bar chart showing the enrichment of the individual histone modification or TF-binding peaks overlapping with the significant interaction fragments (yellow) or background interactions (blue). (B) Bar chart showing the overlapping number of bound TFs with the genomic coordinates of the CHi-C interaction data (blue) and random genomic coordinates (red). Specific enrichment for the CHi-C interaction data can be seen when 3 or more TFs are bound. (C) Individual binding peaks for each TF were separated into promoter (“baits” for CHi-C experiment) or distal (promoter-interacting regions, not considered “bait”). Distinct distributions of the 29 factors can be seen throughout the genome.
We next asked whether within a looping interaction, individual TFs show a preference to be either bound to the promoter or to the distal region (only analyzing TF peaks which overlap with the looping interaction) (Figure 5C). Distinct patterns were observed for each TF, with clear trends emerging. Several TFs bind preferentially to promoter regions (E2F4, c-JUN, cMYC, STAT3, EGR1, ELF1, ETO2, and MAX), a small number bind more evenly to both promoters and promoter-interacting regions (SPI1/PU.1, ERG, pSTAT1, cFOS, SCL, GFI1B, CEBPα, CEBPβ, CTCF, and RAD21) whereas the remainder of the TFs bind preferentially to promoter-interacting regions (MYB, FLI1, MEIS1, E2A, NFE2, p53, GATA2, RUNX1, LMO2, LYL1, and LDB1). Within the last group, 3 TFs (LDB1, LMO2, and LYL1) had nearly 80% of their binding events associated with promoter-interacting elements.
Promoter-distal element loops are characterized by known and previously unknown TF associations
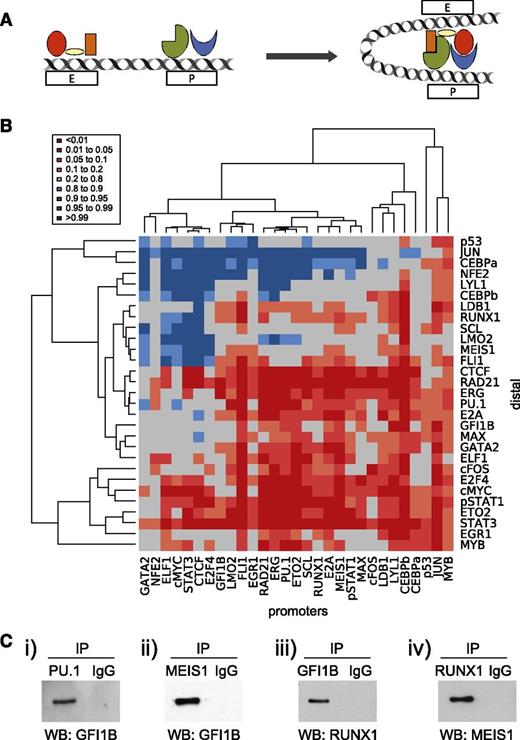
Transcriptional control of gene expression requires the complex interplay of promoter and enhancer elements, which are thought to be brought into close proximity through looping that appears to be at least in part driven by specific TF-binding events (Figure 6A). Although some factors have been associated with generic roles in the establishment of such loops,50 little is known about the specific contributions made by most TFs including the key HSPC regulators assayed in this study. So far, we have shown that looping regions are characterized by multi-TF binding and that specific TFs are associated with either promoter or promoter-interacting regions. We next asked whether binding of a given TF to either the promoters or distal component of the interaction was associated with the presence of specific partner TFs on the corresponding end of the mapped chromatin loops (Figure 6B). To interpret the results of this analysis, we curated known protein-protein interactions from the STRING database,51 which produced a list of 32 known protein-protein interactions involving the TFs analyzed here. Analysis of computationally predicted TF associations across promoter-distal loops revealed that some of the most significant pairings corresponded to known protein-protein partners, such as FLI1/GATA2 and FOS/c-JUN. Overall, this analysis showed that 24 of the 32 known protein-protein interactions corresponded to significant promoter-distal element occupancy pairings. Of note, 28% of these corresponded to modest occupancy pairings (represented by a lighter orange color, P value = .1-.2), which included known interactions between key HSPC TFs such as SCL/LDB1, LMO2/GATA2, and LMO2/LDB1. This reaffirmed that protein-protein interactions between TF pairs may play a role in the establishment of specific loops, and also that modestly significant pairings in our heatmap are potentially of importance within hematopoiesis. Of note, around 38% of TFs were significantly enriched at both ends of the interacting regions (CEBPβ, cFOS, cMYC, CTCF, E2A, ERG, ETO2, FLI1, PU.1, RAD21, and STAT3).
Previously unknown TF combinations are at play in promoter-distal looping interactions. (A) Schematic of promoter-distal looping interaction. TF complex(es) can be seen to be bound to both the promoter (P) and enhancer (E) elements; upon transcriptional activation, these regulatory elements are brought within close proximity allowing the interaction of these TF complexes. (B) Heatmap showing hierarchical clustering of the TFs bound at promoter and distal elements. To control for numbers of binding peaks per experiment, the data were normalized to the average of 32 iterations of a randomized selection of total ChIP-Seq peaks, weighted according to the number of peaks per TF. (C) Co-immunoprecipitation data (Co-IP) showing protein-protein interactions between PU.1/GFI1B, MEIS1/GFI1B, GFI1B/RUNX1, and RUNX1/MEIS1. Expression constructs were transfected into 293T cells and putative protein interactions assayed by Co-IP/western blot (WB) analysis. (i) Following transfection of GFI1B and SPI1/PU.1, lysates were immunoprecipitated (IP) with an anti-PU.1 antibody and immune complexes were then analyzed to detect the presence of GFI1B. (ii) After transfection of GFI1B and flag-tagged MEIS1, lysates were immunoprecipitated with an anti-FLAG antibody (MEIS1), followed by western blot using anti-GFI1B antibody. (iii) MYC-tagged RUNX1 and GFI1B were transfected, and the lysates were immunoprecipitated with an anti-GFI1B antibody, followed by western blot using anti-RUNX1 antibody. (iv) MYC-tagged RUNX1 and MEIS1 were transfected, and the lysates were immunoprecipitated with anti-MYC antibody (RUNX1) and immune complexes were analyzed by western blots using an anti-MEIS1 antibody. IgG, immunoglobulin G.
Previously unknown TF combinations are at play in promoter-distal looping interactions. (A) Schematic of promoter-distal looping interaction. TF complex(es) can be seen to be bound to both the promoter (P) and enhancer (E) elements; upon transcriptional activation, these regulatory elements are brought within close proximity allowing the interaction of these TF complexes. (B) Heatmap showing hierarchical clustering of the TFs bound at promoter and distal elements. To control for numbers of binding peaks per experiment, the data were normalized to the average of 32 iterations of a randomized selection of total ChIP-Seq peaks, weighted according to the number of peaks per TF. (C) Co-immunoprecipitation data (Co-IP) showing protein-protein interactions between PU.1/GFI1B, MEIS1/GFI1B, GFI1B/RUNX1, and RUNX1/MEIS1. Expression constructs were transfected into 293T cells and putative protein interactions assayed by Co-IP/western blot (WB) analysis. (i) Following transfection of GFI1B and SPI1/PU.1, lysates were immunoprecipitated (IP) with an anti-PU.1 antibody and immune complexes were then analyzed to detect the presence of GFI1B. (ii) After transfection of GFI1B and flag-tagged MEIS1, lysates were immunoprecipitated with an anti-FLAG antibody (MEIS1), followed by western blot using anti-GFI1B antibody. (iii) MYC-tagged RUNX1 and GFI1B were transfected, and the lysates were immunoprecipitated with an anti-GFI1B antibody, followed by western blot using anti-RUNX1 antibody. (iv) MYC-tagged RUNX1 and MEIS1 were transfected, and the lysates were immunoprecipitated with anti-MYC antibody (RUNX1) and immune complexes were analyzed by western blots using an anti-MEIS1 antibody. IgG, immunoglobulin G.
The above analysis revealed significant TF occupancy for protein pairs not known to engage in direct protein-protein interactions. To investigate this further, we focused on pairings involving the core HSPC TFs, and performed coimmunoprecipitation assays in which the relevant pairs of TFs were expressed in 293T cells (Figure 6C). Specific interactions can be seen between PU.1/GFI1B, MEIS1/GFI1B, GFI1B/RUNX1, and RUNX1/MEIS1, thus validating 4 previously unknown protein-protein interactions between key HSPC TFs. This discovery serves as an example of how the data sets presented here can be used to gain new insights into the transcriptional processes operating in HSPCs.
Discussion
Genome-wide mapping techniques based on high-throughput sequencing have revolutionized our understanding of transcriptional control processes. However, despite some progress in miniaturizing assay conditions, many of these genome-scale techniques still require the use of hundreds of thousands of cells, and are therefore not applicable to rare adult stem cell populations such as hematopoietic stem cells. International consortium efforts such as ENCODE have therefore focused on leukemic cell lines such as K562 for producing comprehensive data sets.52 Heterogeneous populations of progenitor cells such as human CD34+ cells have also been used to produce limited data sets, commonly restricted to gene expression and histone marks4 and similar histone mark data have been produced for a range of mouse stem and progenitor populations.13
We previously reported gene expression, histone acetylation, and 12 TF-binding profiles in the SCF-dependent multipotential HPC-7 cell line.17,18,37 Although these data have been validated by several groups, emphasizing the HPC-7 cell line as an authentic model for early multipotent hematopoietic cells,53-58 the HPC-7 data were limited compared with tier 1 ENCODE cell lines such as GM12878, K562, and H1 human embryonic stem cells. We have therefore now generated genome-wide maps for an additional 17 TFs, 3 histone modifications, and DNase I accessible chromatin. Because one of the most challenging processes in genome-wide experiments has been the reliable association of a TF-binding peak to a specific gene, we also generated genome-wide Hi-C and Capture Hi-C (CHi-C) data sets to complement our TF-binding data sets with information on the 3-dimensional organization of the HPC-7 genome. This means that for the first time in a HSPC cell line model, specific genes can be associated to specific TF-binding peaks and therefore transcriptional regulatory modules can be investigated on a gene-by-gene basis as well as genome-wide. For comparison, we found that there were 241 719 unique peak regions in the K562 experiments compared with 84 266 unique peak regions in the HPC-7 data set. The range of peaks per TF varies for both the HPC-7 and K562 cell lines (98-43 786 peaks can be seen in the HPC-7 experiments, whereas 1176-80 334 peaks can be seen in the K562 cell line; see supplemental Figure 2). All data are publically available both via ArrayExpress and CODEX, to ensure accessibility to the widest possible audience.
Promoter CHi-C has the advantage over other next-generation sequencing–based chromosome conformation capture-derived protocols that comprehensive coverage of all promoter-anchored genomic loops can be obtained with a realistically achievable sequencing depth, and, unlike chromatin interaction analysis by paired end tag sequencing (ChIA-PET), without the reliance on immunoprecipitation steps. Here we provide the first integrated analysis of promoter-anchored loops with genome-wide binding maps for 29 TFs which allowed us to reveal several previously unrecognized features of the transcriptional landscape in HPC-7 cells. The first observation is that there is a direct correlation between the level of TF occupancy of a distal region and the likelihood of engagement in a promoter-anchored loop. Although this might not be surprising, this observation supports mechanistic models where DNA-bound TFs directly contribute to chromatin loop formation, possibly through protein-protein interactions. Second, being able to focus analysis only on those TF-binding events that occur on actively looping regions, we were able to reexamine several aspects of TF occupancy. We show that there is a wide range of relative preference for promoter binding, from >90% for E2F4 to <20% for LDB1. This suggests that individual TFs may differ in the way they influence transcription. Of note, the most promoter-preferential TFs did not include lineage-specific factors, consistent with the notion that cell-type specific expression is largely mediated by distal elements.42,59,60
Integrated genome-wide analysis also showed that TF occupancy of promoter-distal interacting pairs is not random because we now demonstrate the presence of specific TFs at the promoter influences the likely presence of other TFs at distal regions and vice versa. This observation highlights that the data sets generated here provide much more than a catalog of genomic coordinates bound by TFs and involved in chromatin loops. Instead, our analysis demonstrates that comprehensive analysis of complementary data sets has the power to reveal potential “regulatory rules” that operate within a given cell type. To develop this argument further we investigated the potential relevance of protein-protein interactions for the observed preferential TF pairings on promoter-distal region loops. Of note, known protein-protein interactions corresponded predominantly to TF pairings that were enriched across promoter-distal region loops. These included known interactions between core HSPC TFs, which mostly occurred among moderately enriched TF pairings. This observation prompted us to investigate whether other HSPC TF pairings at a similar level of enrichment might correspond to previously unrecognized direct protein-protein interactions, which led us to experimentally validate 4 novel protein-protein interactions.
Given the dynamic nature of the hematopoietic system, transcriptional programs within multipotent progenitors must mediate both maintenance of the progenitor expression state as well as have the ability to alter expression in order to differentiate into the various mature lineages. Differentiation is known to be accompanied by widespread relocation of TFs and reorganization of promoter-enhancer chromatin loops.61 A mechanistic understanding of the underlying processes will advance our ability to design cellular programming strategies for cellular therapy and regenerative medicine, and also enhance our understanding of the perturbations of transcriptional programs associated with neoplastic disease. The data presented here may stand for many years as an important baseline comparison for such future studies.
The Hi-C, CHi-C, DNase I, and ChIP-Seq data (raw sequence data, custom track [.bigwig] files, and peak lists) have been deposited into the ArrayExpress (accession number E-MTAB-3954). The Stable UCSC Web site session can be found at: http://tinyurl.com/E-MTAB-3954.
This article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank Prof Peter Cockerill for providing protocols and advice for the DNase I experiments, and Fiona Hamey for help with P-value calculations.
Work in the B.G.'s laboratory was supported by grants from Bloodwise, the Medical Research Council (MRC), the Leukemia & Lymphoma Society, Cancer Research UK, the Biotechnology and Biological Sciences Research Council, the National Institute for Health Research Cambridge Biomedical Research Centre, and core support grants by The Wellcome Trust to the Cambridge Institute for Medical Research and The Wellcome Trust–MRC Cambridge Stem Cell Institute. For funding for the open access charge, a core support grant was provided by The Wellcome Trust–MRC Cambridge Stem Cell Institute.
Authorship
Contribution: S.S., J.S., V.L., D.K.G., F.J.C.-N., V.M., A.C.W., I.J.-M., and S.K. performed research; R.H., M.S.C., and J.M. performed bioinformatic analysis; N.K.W. designed experiments, performed research, and analyzed data; M.S. and P.F. discussed results and the manuscript; B.G. designed the study and supervised work; and B.G. and N.K.W. wrote the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Nicola K. Wilson, Department of Haematology, Wellcome Trust and MRC Cambridge Stem Cell Institute & Cambridge Institute for Medical Research, Cambridge University, Cambridge, CB2 0XY, United Kingdom; e-mail: nkw22@cam.ac.uk; or Berthold Göttgens, Department of Haematology, Wellcome Trust and MRC Cambridge Stem Cell Institute & Cambridge Institute for Medical Research, Cambridge University, Cambridge, CB2 0XY, United Kingdom; e-mail: bg200@cam.ac.uk.