Key Points
Wild-type AML1 and AML1/ETO form a complex on chromatin via binding to adjacent different motifs and interacting through the runt homology domain.
The relative binding signals of AML1/ETO and AML1 and AP-1 recruitment determine whether AML1/ETO activates or represses its targets.
Abstract
The AML1/ETO fusion protein is essential to the development of t(8;21) acute myeloid leukemia (AML) and is well recognized for its dominant-negative effect on the coexisting wild-type protein AML1. However, the genome-wide interplay between AML1/ETO and wild-type AML1 remains elusive in the leukemogenesis of t(8;21) AML. Through chromatin immunoprecipitation sequencing and computational analysis, followed by a series of experimental validations, we report here that wild-type AML1 is able to orchestrate the expression of AML1/ETO targets regardless of being activated or repressed; this is achieved via forming a complex with AML1/ETO and via recruiting the cofactor AP-1 on chromatin. On chromatin occupancy, AML1/ETO and wild-type AML1 largely overlap and preferentially bind to adjacent and distinct short and long AML1 motifs on the colocalized regions, respectively. On physical interaction, AML1/ETO can form a complex with wild-type AML1 on chromatin, and the runt homology domain of both proteins are responsible for their interactions. More importantly, the relative binding signals of AML1 and AML1/ETO on chromatin determine which genes are repressed or activated by AML1/ETO. Further analysis of coregulators indicates that AML1/ETO transactivates gene expression through recruiting AP-1 to the AML1/ETO-AML1 complex. These findings enrich our knowledge of understanding the significance of the interplay between the wild-type protein and the oncogenic fusion protein in the development of leukemia.
Introduction
Many oncogenic fusion proteins generated by chromosomal translocations play a causal role in the development of leukemia. The oncogenic lesion almost exclusively occurs in a single allele within an individual leukemia, whereas the wild-type protein produced from the nontranslocated allele generally still exists.1,2 The coexistence of the wild-type protein with the oncogenic fusion protein raises the question about their significance in leukemogenesis. To address this question, acute myeloid leukemia (AML) with the t(8;21) translocation and resultant AML1/ETO fusion gene is an ideal model, particularly through understanding the interplay between the AML1/ETO fusion protein and the AML1 wild-type protein.
AML1 (also known as RUNX1 or core-binding factor α [CBFα]), a critical regulator of normal hematopoiesis,3 is widely expressed in multiple hematopoietic lineages and regulates the expression of a variety of hematopoietic genes through recognizing the motif TGTGGT.4 AML1 is frequently involved in chromosomal abnormalities in AML,5-7 among which AML1/ETO is the most common fusion protein resulted from the t(8;21) translocation.7 Structurally, AML1/ETO consists of the N-terminal portion of AML1 and the majority of the ETO protein.7 The AML1 portion includes the runt homology domain (RHD) that is responsible for the DNA binding and the interaction with its heterodimerization partner core-binding factor β (CBFβ) and other transcription regulators.8 AML1/ETO functions as a dominant inhibitor of wild-type AML1 by recruiting corepressor complexes (such as NCoR/SMRT,9 mSin3a, and HDACs10 ) through the ETO moiety to repress genes that are primarily transactivated by AML1.11 On the other hand, AML1/ETO can also recruit coactivators (such as p300 and PRMT1) to activate genes, especially those involved in stem cell self-renewal, such as ID1, CDKN1A, and EGR1.12,13 In addition to dynamic interactions with various regulatory (co)factors, a recent study has reported that AML1/ETO resides in and functions through a stable multiprotein complex that at least contains HEB, LYL1, LMO2, and CBFβ,14 providing another layer of regulatory complexity for AML1/ETO in regulating its target genes.
Although it is commonly accepted that AML1/ETO exerts a dominant-negative effect on the function of wild-type AML1 during leukemia development,7 increasing evidence suggests that the real relationship between AML1/ETO and AML1 may be substantially more complex. Clinical data have shown that inactivating mutations of AML1 are frequently identified in patients without the AML1/ETO fusion protein but not in those with this fusion protein,8,15 implicating that wild-type AML1 may function in AML1/ETO-induced leukemogenesis. This implication is reinforced by recent findings that wild-type AML1 is required for the survival of t(8;21) and other CBF leukemic cells.16,17 Furthermore, AML1/ETO9a, an alternatively spliced isoform of AML1/ETO that has a lower capacity to inhibit wild-type AML1 activity,18 shows a stronger leukemogenic potential in mice.19 In addition to the functional importance of wild-type AML1 in t(8;21) AML, evidence from several high-throughput binding site studies raises the possibility that wild-type AML1 is colocalized with AML1/ETO on chromatin in t(8;21) AML,16,20-22 although the degree of the overlap varies due to the antibodies used in the immunoprecipitation. All these studies pointed out a potential role of wild-type AML1 in AML1/ETO-induced leukemogenesis.
In an attempt to clarify the functional and physical interplay between AML1/ETO and AML1, it is essential to uncover the binding feature of their colocalized regions and the associated mechanisms. In this study, we demonstrate an indispensable role of wild-type AML1 in the AML1/ETO-mediated transcriptional regulation in t(8;21) AML.
Methods
Chromatin immunoprecipitation sequencing
Chromatin immunoprecipitation (ChIP) was performed in Kasumi-1 and U937 cells according to the manufacturer’s protocol (Active Motif, Carlsbad, CA). The total input was used as control. Libraries were prepared according to the Illumina genomic prep kit (Illumina, San Diego, CA) and then sequenced by Illumina Genome Analyzer II platform (Illumina) to generate 35-bp single-end reads. The detailed analysis of ChIP sequencing (ChIP-seq) data, including our data and published ChIP-seq data,14,16,20,23-25 is available in supplemental Methods available on the Blood Web site.
DNA pull-down, coimmunoprecipitation, GST pull-down, and immunofluorescence microscopy
DNA pull-down assays were performed as described previously with several modifications.26 Coimmunoprecipitation (Co-IP) assays in Kasumi-1 and AML1/ETO-induced U937-A/E/9/14/18 cells were performed strictly according to the manufacturer’s protocol (Active Motif). Glutathione S-transferase (GST) fusion proteins were generated according to the GST Gene Fusion System Handbook (GE Healthcare, Piscataway, NJ). Immunofluorescence microscopy was performed in Kasumi-1 and HEK-293T cells. Details are available in supplemental Methods.
Motif analysis
DNA sequences for motif analysis were extracted from the repeat-masked human reference genome sequence hg19 using BEDtools getfasta (version 2.17.0). Details are available in supplemental Methods.
Gene expression analysis
RNA-seq raw data (GSE43834)14 of Kasumi-1 cells before and after AML1/ETO knockdown were aligned to the human reference genome sequence hg19 using TopHat (version 2.17.0).27 Differentially expressed genes before and after AML1/ETO knockdown were detected by the GFOLD (generalized fold change) algorithm28 (Version 1.0.9). The detailed analysis of the expression data of Kasumi-1 (GSE43834),14 SKNO-1 (GSE34594),20 and AML patients (GSE14468)29 is available in supplemental Methods.
Enrichment analysis
Gene set analysis on the association of targets bound by the AML1/ETO-AML1 complex with genes regulated by AML1/ETO was performed as described previously.30,31 We evaluated the association of AML1/ETO-regulated genes with the AML1/ETO and AML1 binding affinities using the gene set enrichment analysis (GSEA) algorithm,32 in which genes were ranked according to the difference in enrichment of AML1/ETO and AML1. Two online tools based on publicly available ChIP-seq data, the ENCODE ChIP-seq Significance tool33 (http://encodeqt.simple-encode.org) and the Cscan tool34 (http://159.149.160.51/cscan/), were used to identify the enriched regulators associated with AML1/ETO-activated genes. Details are available in supplemental Methods.
Data access
ChIP-seq data have been submitted to the National Center for Biotechnology Information (NCBI) Gene Expression Omnibus under accession no. GSE65427.
Results
AML1/ETO and AML1 coexist on chromatin in t(8;21) AML
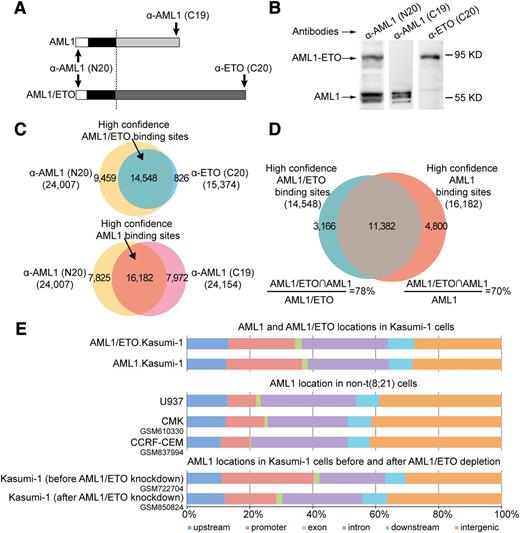
We performed ChIP-seq analysis to investigate the chromatin occupancy of AML1 and AML1/ETO in t(8;21) AML-derived Kasumi-1 cells, which express the equivalent level of AML1/ETO and AML1 but without any detectable ETO.35 We used 2 independent antibodies recognizing different parts of AML1/ETO or AML1 (supplemental Tables 1-3) to obtain the high confidence binding sites of AML1/ETO and AML1, respectively (Figure 1A-B). Accordingly, 14 548 high confidence AML1/ETO (supplemental Table 4) and 16 182 high confidence AML1 binding sites (supplemental Table 5) were identified (Figure 1C). Compared with previous genome-wide binding studies on Kasumi-1 cells14,16,20 and other AML1/ETO-positive SKNO-1 cells,25 our list of high confidence binding sites highly overlapped with those in Kasumi-1 cells (84% for AML1/ETO and 70% for AML1; supplemental Figure 1A), and with those in SKNO-1 cells (76% for AML1/ETO and 69% for AML1; supplemental Figure 1B). In addition to the binding number, the binding signals of AML1/ETO and AML1 in this study were quite similar to those previously published in these 2 cell lines (supplemental Figure 1C). Together, these results showed the accuracy of our high confidence binding sites for AML1/ETO and AML1.
AML1 coexists with AML1/ETO on chromatin in t(8;21) leukemic cells. (A) Schematic diagrams of the antibody recognition sites to wild-type AML1 and the AML1/ETO fusion protein. The anti-AML1 (N20) antibody targets the N terminus of AML1 and recognizes both AML1 and AML1/ETO; the anti-AML1 (C19) antibody targets the C terminus of AML1 and recognizes AML1 but not AML1/ETO; and the anti-ETO (C20) antibody targets the C terminus of ETO and specifically recognizes AML1/ETO. The significant ChIP regions enriched by anti-ETO (C20), anti-AML1 (C19), and anti-AML1 (N20) antibody in Kausmi-1 cells are available in supplemental Tables 1 to 3, respectively. (B) Validation of antibodies specific to AML1, AML1/ETO, and both proteins by western blotting with the Kasumi-1 cell lysates. (C) Acquisition of high confidence AML1/ETO and AML1 binding sites in Kasumi-1 cells. High confidence AML1/ETO binding sites (supplemental Table 4) were generated by overlapping ChIP regions enriched by anti-ETO (C20) (supplemental Table 1) and anti-AML1 (N20) antibodies (supplemental Table 3) (upper). High confidence AML1 binding sites (supplemental Table 5) were generated by overlapping ChIP regions enriched by anti-AML (C19) (supplemental Table 2) and anti-AML1 (N20) antibodies (supplemental Table 3) (lower). (D) Venn diagram of the overlap between high confidence AML1/ETO and AML1 binding sites in Kasumi-1 cells. AML1/ETO∩AML1, overlapped ChIP regions. The full list of the overlapped high confidence ChIP regions between AML1/ETO and AML1 in Kasumi-1 cells are available in supplemental Table 6. (E) The genomic distribution of wild-type AML1 differs between AML1/ETO-positive and -negative cells. Each region bound by AML1 or AML1/ETO was mapped to the closest Refseq gene. upstream, upstream regulatory regions between −20 and −3 kb to the transcription start site (TSS); promoter, regions between −3 and 1 kb to the TSS; downstream, 20-kb regions downstream regulatory regions of the transcription termination site (TTS); exon and intron, regions mapped to related location according to Refseq annotations; intergenic, other regions. ChIP-seq data used in this analysis were retrieved from the NCBI Gene Expression Omnibus database (GSM610330,24 GSM837994,23 GSM722704,20 and GSM85082420 ).
AML1 coexists with AML1/ETO on chromatin in t(8;21) leukemic cells. (A) Schematic diagrams of the antibody recognition sites to wild-type AML1 and the AML1/ETO fusion protein. The anti-AML1 (N20) antibody targets the N terminus of AML1 and recognizes both AML1 and AML1/ETO; the anti-AML1 (C19) antibody targets the C terminus of AML1 and recognizes AML1 but not AML1/ETO; and the anti-ETO (C20) antibody targets the C terminus of ETO and specifically recognizes AML1/ETO. The significant ChIP regions enriched by anti-ETO (C20), anti-AML1 (C19), and anti-AML1 (N20) antibody in Kausmi-1 cells are available in supplemental Tables 1 to 3, respectively. (B) Validation of antibodies specific to AML1, AML1/ETO, and both proteins by western blotting with the Kasumi-1 cell lysates. (C) Acquisition of high confidence AML1/ETO and AML1 binding sites in Kasumi-1 cells. High confidence AML1/ETO binding sites (supplemental Table 4) were generated by overlapping ChIP regions enriched by anti-ETO (C20) (supplemental Table 1) and anti-AML1 (N20) antibodies (supplemental Table 3) (upper). High confidence AML1 binding sites (supplemental Table 5) were generated by overlapping ChIP regions enriched by anti-AML (C19) (supplemental Table 2) and anti-AML1 (N20) antibodies (supplemental Table 3) (lower). (D) Venn diagram of the overlap between high confidence AML1/ETO and AML1 binding sites in Kasumi-1 cells. AML1/ETO∩AML1, overlapped ChIP regions. The full list of the overlapped high confidence ChIP regions between AML1/ETO and AML1 in Kasumi-1 cells are available in supplemental Table 6. (E) The genomic distribution of wild-type AML1 differs between AML1/ETO-positive and -negative cells. Each region bound by AML1 or AML1/ETO was mapped to the closest Refseq gene. upstream, upstream regulatory regions between −20 and −3 kb to the transcription start site (TSS); promoter, regions between −3 and 1 kb to the TSS; downstream, 20-kb regions downstream regulatory regions of the transcription termination site (TTS); exon and intron, regions mapped to related location according to Refseq annotations; intergenic, other regions. ChIP-seq data used in this analysis were retrieved from the NCBI Gene Expression Omnibus database (GSM610330,24 GSM837994,23 GSM722704,20 and GSM85082420 ).
Next, we compared the chromatin occupancy between AML1/ETO and AML1 and found that 78% of AML1/ETO binding sites overlapped with 70% of AML1 binding sites (Figure 1D; supplemental Figure 2A; supplemental Table 6). Our observation was in principle consistent with previous genome-wide binding studies,16,20-22 but using 2 antibodies (thus high confidence binding sites), we were able to identify a higher fraction of chromatin co-occupancy than using only 1 antibody as in previous studies. This was also verified by ChIP-quantitative polymerse chain reaction (qPCR) using a panel of randomly selected ChIP regions (supplemental Figure 2B). The overlap of AML1/ETO and AML1 binding sites indicated that the 2 proteins were located on the same genomic regions (Figure 1E, top). However, AML1 occupancy was clustered less on promoter regions and more on the intergenic regions in AML1/ETO-negative cells (supplemental Table 7) compared with that in AML1/ETO-positive cells (Figure 1E, top and middle). This suggested that AML1/ETO might lead to the redistribution of AML1 on the genomic regions. This finding was further supported by the restoration of AML1 distribution after AML1/ETO depletion (Figure 1E, bottom). Collectively, the results suggest that the coexistence of AML1/ETO and AML1 on chromatin in t(8;21) AML cells is attributed to the formation of AML1/ETO.
AML1/ETO and AML1 preferentially bind to adjacent short and long AML1 motifs on the colocalized regions, respectively
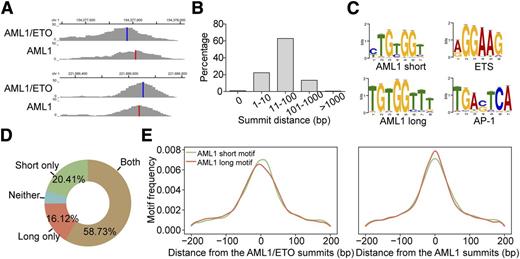
We next investigated how AML1/ETO and AML1 coexisted on chromatin. Because the peak summit is most likely the location of the actual site of transcription factor-DNA interaction,36 we first determined the exact location of AML1 and AML1/ETO by calculating the distance between their peak summits. As shown in Figure 2A-B, AML1/ETO and AML1 tended to bind to adjacent sites with a distance between 11 and 100 bp in the majority of the overlap regions (62%). We also found that 2 similar but different AML1 motifs were significantly enriched within the AML1/ETO and AML1 colocalized regions, ie, a short motif 5′-TG(T/C)GGT-3′ and a long motif 5′-TGTGGTTT-3′ (Figure 2C), respectively. The longer one contained the similar core sequence (TGTGGT) as the short one, but was appended 2 additional thymidines at the 3′ position. Moreover, the majority of the overlap peaks contained both long and short AML1 motifs (Figure 2D), implicating the coexistence of both motifs on the localized regions. Further comparison of the motif distribution on the peak summits revealed that the short one tended to be bound by AML1/ETO and the long one by AML1 (Figure 2E), which was supported by the observation that the short one was enriched in uniquely AML1/ETO-bound regions and the long one in uniquely AML1-bound regions (supplemental Figure 3). Furthermore, the ETS and AP-1 consensus sequences were also significantly enriched in the colocalized regions (Figure 2C; supplemental Table 8).
Bioinformatic analysis reveals that AML1/ETO and AML1 bind to adjacent and specified motifs on chromatin. (A) Examples of the AML1/ETO and AML1 overlap peaks. Vertical blue lines indicate the peak summits of AML1/ETO; vertical red lines indicate the peak summits of AML1. (B) The distance between the summit of AML1/ETO and AML1 binding on the overlap peaks. (C) Significant motifs identified within the AML1/ETO and AML1 overlap regions. (D) Percentages of the AML1/ETO and AML1 overlapped regions that contain the short and/or long AML1 motifs. (E) Frequency distribution of the short and long AML1 motifs relative to the AML1/ETO and AML1 peak summits on the overlap regions containing both motifs.
Bioinformatic analysis reveals that AML1/ETO and AML1 bind to adjacent and specified motifs on chromatin. (A) Examples of the AML1/ETO and AML1 overlap peaks. Vertical blue lines indicate the peak summits of AML1/ETO; vertical red lines indicate the peak summits of AML1. (B) The distance between the summit of AML1/ETO and AML1 binding on the overlap peaks. (C) Significant motifs identified within the AML1/ETO and AML1 overlap regions. (D) Percentages of the AML1/ETO and AML1 overlapped regions that contain the short and/or long AML1 motifs. (E) Frequency distribution of the short and long AML1 motifs relative to the AML1/ETO and AML1 peak summits on the overlap regions containing both motifs.
To verify the above observations, we measured the binding affinities of AML1/ETO and AML1 to the long and short motifs using biolayer interferometry. As shown in Figure 3A, AML1/ETO bound to short motifs with an ∼2000-fold higher affinity than AML1, whereas AML1 showed a 10- to 50-fold higher affinity to long motifs than AML1/ETO. Furthermore, DNA pull-down and ChIP-qPCR assays performed on multiple regions also supported this finding in vitro and in vivo, respectively (Figures 3B-C; supplemental Figure 4).
Experimental evidence shows that AML1/ETO and AML1 bind to adjacent short and long motifs. (A) Direct comparison of the kinetics of AML1 and AML1/ETO binding on short and long motifs using biolayer interferometry. (Left upper) Sequence of biotin-labeled probes used in the assays. S1 and S2 represent the short AML1 motif-containing probes, whereas L1 and L2 represent the long AML1 motif-containing probes. (Left lower) Equilibrium dissociation constants (KD). (Right) Association and the disassociation curves of each experiment group. The protein concentrations of AML1/ETO and AML1 used were 0 (light green), 14.8 (dark green), 44.6 (red), and 134 nM (blue). (B) DNA pull-down assays for AML1/ETO and AML1 with the long and short motif-containing probes. The probes used in DNA pull-down assays were the same as those used in biolayer interferometry experiments. The equal amount of AML1/ETO and AML1 was used in DNA pull-down assays (left). The protein binding was detected by western blotting with anti-AML1 (N20; Santa Cruz) antibody. Data using additional 2 short and 2 long AML1 motif-containing probes can be found in supplemental Figure 4A. (C) ChIP-qPCR analyses of AML1/ETO and AML1 recruitment on the short and long motif-containing regions in Kasumi-1 cells. Data using additional 2 short and 2 long AML1 motif-containing regions can be found in supplemental Figure 4B. Error bars represent the standard deviation (SD) of triplicate measurements.
Experimental evidence shows that AML1/ETO and AML1 bind to adjacent short and long motifs. (A) Direct comparison of the kinetics of AML1 and AML1/ETO binding on short and long motifs using biolayer interferometry. (Left upper) Sequence of biotin-labeled probes used in the assays. S1 and S2 represent the short AML1 motif-containing probes, whereas L1 and L2 represent the long AML1 motif-containing probes. (Left lower) Equilibrium dissociation constants (KD). (Right) Association and the disassociation curves of each experiment group. The protein concentrations of AML1/ETO and AML1 used were 0 (light green), 14.8 (dark green), 44.6 (red), and 134 nM (blue). (B) DNA pull-down assays for AML1/ETO and AML1 with the long and short motif-containing probes. The probes used in DNA pull-down assays were the same as those used in biolayer interferometry experiments. The equal amount of AML1/ETO and AML1 was used in DNA pull-down assays (left). The protein binding was detected by western blotting with anti-AML1 (N20; Santa Cruz) antibody. Data using additional 2 short and 2 long AML1 motif-containing probes can be found in supplemental Figure 4A. (C) ChIP-qPCR analyses of AML1/ETO and AML1 recruitment on the short and long motif-containing regions in Kasumi-1 cells. Data using additional 2 short and 2 long AML1 motif-containing regions can be found in supplemental Figure 4B. Error bars represent the standard deviation (SD) of triplicate measurements.
AML1 exists in the AML1/ETO-containing transcription factor complex and directly interacts with AML1/ETO on chromatin
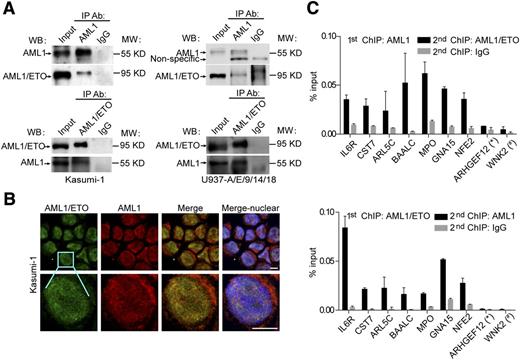
The colocalization of AML1/ETO and AML1 on chromatin might indicate a physical interaction between the 2 proteins. To show that the interaction does occur in vivo, we performed co-IP experiments and found that AML1/ETO was specifically detected in the protein complex immunoprecipitated by AML1 specific antibodies but not the control and vice versa (Figure 4A). Moreover, we performed immunofluorescence assays to globally visualize spatial relationships between AML1/ETO and AML1 in situ. As illustrated in Figure 4B, AML1/ETO and AML1 were partially colocalized with each other in Kasumi-1 cells. Furthermore, re-ChIP experiments in Kasumi-1 cells revealed that the interaction of AML1/ETO and AML1 indeed occurred on chromatin (Figure 4C). In addition, because recent studies have shown that AML1/ETO exists as a stable complex on chromatin with multiple transcription factors,14 we then assessed whether AML1 was present in this stable complex. As shown in supplemental Figures 5 and 6, the genomic occupancy of AML1 was correlated with those reported in the AML1/ETO complex, ie, E2A, HEB, and LMO2 (r = 0.492, 0.439, and 0.475, respectively; P < 2.2e−16), indicating that AML1 colocalized with the AML1/ETO stable complex on chromatin.
AML1/ETO physically interacts with AML1 on chromatin. (A) In vivo interaction between AML1/ETO and AML1 with co-IP assays in Kasumi-1 (left) and U937-A/E/9/14/18 (right) cells. The anti-AML1 (C19; Santa Cruz) and anti-AML1/ETO (fusion point; Diagenode) antibodies were used for IP. The anti-ETO (C20; Santa Cruz) and anti-AML1 (N20; Santa Cruz) antibodies were used for western blotting. Asterisks indicate nonspecific bands. The second antibody used in western blotting of AML1 immunoprecipitated with AML1/ETO in Kasumi-1 cells was horseradish peroxidase-conjugated anti-goat IgG TrueBlot (eBioscience), which could not detect the immunoglobulin heavy and light chains. The second antibody used in the remaining western blotting was horseradish peroxidase-conjugated anti-goat IgG (Santa Cruz). (B) Confocal immunofluorescence micrographs showing the distribution of AML1/ETO (green) and AML1 (red) in Kasumi-1 cells. Scale bars, 10 μm. (C) Validation for the coexistence of AML1/ETO and AML1 on chromatin through re-ChIP assays. Multiple overlapped regions were selected and validated for factor binding by a first round of ChIP followed by a second round with a different antibody or with IgG as control. The AML1 specific anti-AML1 (C19) antibody and AML1/ETO specific anti-ETO (C20) were used for re-ChIP assays. An AML1 unique region (*) and an AML1/ETO unique region (#) were included as negative controls. Error bars represent the SD of triplicate measurements.
AML1/ETO physically interacts with AML1 on chromatin. (A) In vivo interaction between AML1/ETO and AML1 with co-IP assays in Kasumi-1 (left) and U937-A/E/9/14/18 (right) cells. The anti-AML1 (C19; Santa Cruz) and anti-AML1/ETO (fusion point; Diagenode) antibodies were used for IP. The anti-ETO (C20; Santa Cruz) and anti-AML1 (N20; Santa Cruz) antibodies were used for western blotting. Asterisks indicate nonspecific bands. The second antibody used in western blotting of AML1 immunoprecipitated with AML1/ETO in Kasumi-1 cells was horseradish peroxidase-conjugated anti-goat IgG TrueBlot (eBioscience), which could not detect the immunoglobulin heavy and light chains. The second antibody used in the remaining western blotting was horseradish peroxidase-conjugated anti-goat IgG (Santa Cruz). (B) Confocal immunofluorescence micrographs showing the distribution of AML1/ETO (green) and AML1 (red) in Kasumi-1 cells. Scale bars, 10 μm. (C) Validation for the coexistence of AML1/ETO and AML1 on chromatin through re-ChIP assays. Multiple overlapped regions were selected and validated for factor binding by a first round of ChIP followed by a second round with a different antibody or with IgG as control. The AML1 specific anti-AML1 (C19) antibody and AML1/ETO specific anti-ETO (C20) were used for re-ChIP assays. An AML1 unique region (*) and an AML1/ETO unique region (#) were included as negative controls. Error bars represent the SD of triplicate measurements.
AML1/ETO interacts with AML1 through the RHD domain of both proteins
To clarify the region responsible for the formation of the AML1-AML1/ETO complex, we performed the GST pull-down assays using a series of truncated forms of AML1 and AML1/ETO. The truncated forms of AML1 that lacked the activation domain (AD) or the inhibition domain (ID) still retained the ability to interact with AML1/ETO, whereas the truncated form that lacked the RHD could not bind to AML1/ETO (Figure 5A-B; supplemental Figure 7). These results indicated that the RHD of AML1 mediated its interaction with AML1/ETO, consistent with the previous findings that the RHD is involved in the interaction with cofactors, such as PU.1, SP1, and C/EBPA.37 On the other hand, we constructed a series of AML1/ETO expression vectors, either truncated from the C-terminal of ETO or deleted the RHD of AML (Figure 5C; supplemental Figure 7), because these 2 parts are usually involved in the protein-protein interaction. As shown in Figure 5D, only AML1/ETO with the RHD could bind to AML1, indicating that the RHD of AML1/ETO was responsible for the interaction between AML1/ETO and AML1. The confocal microscopy analysis also supported the formation of the AML1/ETO-AML1 complex relied on the RHD of both proteins (Figure 5E). Furthermore, to verify the above findings, we performed ChIP-qPCR assays on a set of the overlap regions by expressing the full-length and truncated form of AML1/ETO in AML1/ETO nonexpressing U937 cells. As illustrated in Figure 5F, the association of AML1/ETO and AML1 on chromatin was indeed dependent on the RHD.
AML1/ETO and AML1 interact with each other through the RHD domain of both proteins. (A) Structures of GST-AML1 fusion proteins used in the binding assays. The GST fusion proteins containing various forms of AML1 are schematically shown. The ability of the GST-fusion proteins to bind AML1/ETO is shown at right as + or −. (B) Autoradiography showing the binding of the 35S-labeled AML1/ETO to various forms of the GST-AML1 fusion proteins. (C) Structures of the GST-AML1/ETO fusion proteins used in the binding assay. (D) Autoradiography showing the binding of the 35S-labeled AML1 to the different truncated forms of the GST-AML1/ETO fusion proteins. The expression levels of the constructs used in B and D were almost equivalent, as shown in supplemental Figure 7. (E) Fluorescence micrographs in HEK-293T cells transfected with the indicated plasmids showing the in situ localization of AML1/ETO and AML1. AML1/ETO-∆RHD, AML1/ETO construct with the RHD domain deleted; AML1-∆RHD, AML1 construct with the RHD domain deleted; AML1-∆RHD-N, AML1 construct with most of the RHD domain deleted but the nuclear localization signal (NLS) within the RHD domain intact. Scale bars, 10 μm. (F) Validation for the RHD domain responsible for the association of AML1/ETO and AML1 on chromatin. ChIP was performed with anti-AML1 (C19) and anti-ETO antibodies in U937 cells transfected with either the full length AML1/ETO or AML1/ETO with the RHD domain deleted. There might exist 2 groups of the overlap regions: (i) AML1/ETO bound to chromatin regions prebound by wild-type AML1 and (ii) wild-type AML1 was recruited by AML1/ETO after AML1/ETO expression. The left panel shows the western blot validation for the expression of AML1/ETO and AML1/ETO-∆RHD transfected in U937 cells. NC, negative control. Error bars represent the SD of triplicate measurements.
AML1/ETO and AML1 interact with each other through the RHD domain of both proteins. (A) Structures of GST-AML1 fusion proteins used in the binding assays. The GST fusion proteins containing various forms of AML1 are schematically shown. The ability of the GST-fusion proteins to bind AML1/ETO is shown at right as + or −. (B) Autoradiography showing the binding of the 35S-labeled AML1/ETO to various forms of the GST-AML1 fusion proteins. (C) Structures of the GST-AML1/ETO fusion proteins used in the binding assay. (D) Autoradiography showing the binding of the 35S-labeled AML1 to the different truncated forms of the GST-AML1/ETO fusion proteins. The expression levels of the constructs used in B and D were almost equivalent, as shown in supplemental Figure 7. (E) Fluorescence micrographs in HEK-293T cells transfected with the indicated plasmids showing the in situ localization of AML1/ETO and AML1. AML1/ETO-∆RHD, AML1/ETO construct with the RHD domain deleted; AML1-∆RHD, AML1 construct with the RHD domain deleted; AML1-∆RHD-N, AML1 construct with most of the RHD domain deleted but the nuclear localization signal (NLS) within the RHD domain intact. Scale bars, 10 μm. (F) Validation for the RHD domain responsible for the association of AML1/ETO and AML1 on chromatin. ChIP was performed with anti-AML1 (C19) and anti-ETO antibodies in U937 cells transfected with either the full length AML1/ETO or AML1/ETO with the RHD domain deleted. There might exist 2 groups of the overlap regions: (i) AML1/ETO bound to chromatin regions prebound by wild-type AML1 and (ii) wild-type AML1 was recruited by AML1/ETO after AML1/ETO expression. The left panel shows the western blot validation for the expression of AML1/ETO and AML1/ETO-∆RHD transfected in U937 cells. NC, negative control. Error bars represent the SD of triplicate measurements.
AML1/ETO-AML1 complex-targeted genes exhibit different AML1/ETO and AML1 binding patterns, which correlate with the expression of genes regulated by AML1/ETO
To investigate the biological significance of the AML1/ETO-AML1 complex, we then asked how this complex influences the expression of genes regulated by AML1/ETO. We first retrieved genes that were differentially regulated on AML1/ETO knockdown on Kasumi-1 and SKNO-1 cells14,20 and that were differentially expressed between AML1/ETO-positive and -negative AML-M2 patient samples29 (details in Methods and supplemental Methods). Based on the gene set analysis,30 we found that AML1/ETO-downregulated genes were as expected to be enriched in the genes targeted by the AML1/ETO-AML1 complex (Table 1), supporting the notion that AML1/ETO represses the AML1-regulated gene. Interestingly, the complex-targeted genes were also overrepresented in the AML1/ETO upregulated genes, although the overrepresentation was less significant.
Enrichment analysis of potential targets bound by the AML1/ETO-AML1 complex with gene sets associated with AML1/ETO regulation
| Gene set . | Number in the gene set . | Potential targets bound by the AML1/ETO-AML1complex . | |||
|---|---|---|---|---|---|
| Number in the gene set . | Fold enrichment . | Z-score . | P value . | ||
| Kasumi-1_AML1/ETO_down | 407 | 232 | 2.50 | 14.48 | 2.87E−35 |
| Kasumi-1_AML1/ETO_up | 359 | 166 | 2.03 | 9.32 | 4.78E−17 |
| SKNO-1_AML1/ETO_down | 660 | 255 | 2.61 | 15.92 | 1.05E−41 |
| SKNO-1_AML1/ETO_up | 700 | 200 | 1.93 | 9.45 | 6.33E−18 |
| AML-M2_ AML1/ETO_down | 544 | 267 | 2.00 | 11.59 | 1.02E−25 |
| AML-M2_AML1/ETO_up | 416 | 196 | 1.92 | 9.33 | 1.71E−17 |
| Gene set . | Number in the gene set . | Potential targets bound by the AML1/ETO-AML1complex . | |||
|---|---|---|---|---|---|
| Number in the gene set . | Fold enrichment . | Z-score . | P value . | ||
| Kasumi-1_AML1/ETO_down | 407 | 232 | 2.50 | 14.48 | 2.87E−35 |
| Kasumi-1_AML1/ETO_up | 359 | 166 | 2.03 | 9.32 | 4.78E−17 |
| SKNO-1_AML1/ETO_down | 660 | 255 | 2.61 | 15.92 | 1.05E−41 |
| SKNO-1_AML1/ETO_up | 700 | 200 | 1.93 | 9.45 | 6.33E−18 |
| AML-M2_ AML1/ETO_down | 544 | 267 | 2.00 | 11.59 | 1.02E−25 |
| AML-M2_AML1/ETO_up | 416 | 196 | 1.92 | 9.33 | 1.71E−17 |
AML-M2_AML1/ETO_down and AML-M2_AML1/ETO_up, AML1/ETO-downregulated and AML1/ETO-upregulated gene signatures in AML1/ETO-positive AML-M2 patients compared with AML1/ETO-negative AML-M2 patients, respectively; Kasumi-1_AML1/ETO_down or SKNO-1_AML1/ETO_down, AML1/ETO-repressed genes that were upregulated after AML1/ETO knockdown in Kasumi-1 or SKNO-1 cells; Kasumi-1_AML1/ETO_up or SKNO-1_AML1/ETO_up, AML1/ETO-activated genes that were downregulated after AML1/ETO knockdown in Kasumi-1 or SKNO-1 cells.
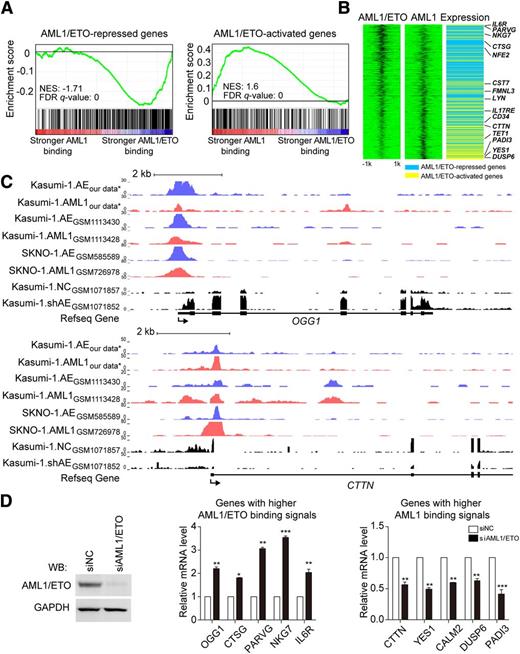
Because the AML1/ETO-AML1 complex-targeted genes have differential binding affinities to AML1/ETO and AML1, we applied GSEA to evaluate the association of AML1/ETO-regulated genes with the AML1/ETO and AML1 binding signals. As shown in Figure 6 and supplemental Figure 8, AML1/ETO-downregulated genes (supplemental Table 9) tended to show higher binding signals for AML1/ETO than for AML1. This group included many known AML1/ETO dominant-negative genes, such as OGG1, CTSG, NKG7, and IL6R.38-40 Our results suggested that AML1/ETO reduced but did not completely replace AML1 binding on the chromatin of AML1/ETO-repressed genes, which was supported by the observation that AML1/ETO depletion on chromatin increased AML1 binding signals (supplemental Figure 9).
AML1/ETO-repressed genes show more AML1/ETO binding and AML1/ETO-activated genes show more AML1 binding. (A) Correlation between the binding affinity ratio of AML1/ETO to AML1 and gene sets repressed or activated by AML1/ETO. FDR q-value, false discovery rate q value; NES, normalized enrichment score. (B) Association of the binding affinities of AML1/ETO and AML1 with the modulation of gene expression by AML1/ETO. Heat maps of AML1/ETO and AML1 ChIP-seq signals on AML1/ETO-regulated genes were sorted based on the ratio of the AML1/ETO binding affinity vs AML1 binding affinity in a 4-kb window centered on the AML1/ETO peak summit (left and center). In the gene expression panel (right), the blue indicates AML1/ETO-repressed genes and the yellow indicates AML1/ETO-activated genes. (C) Overview of the loci of representative AML1/ETO-repressed genes OGG1 and AML1/ETO-activated genes CTTN in Kasumi-1 and SKNO-1 cells. Blue, AML1/ETO ChIP-seq signals; red, AML1 ChIP-seq signals; black, RNA-seq signals before and after AML1/ETO knockdown. Refseq annotations are shown at the bottom. ChIP-seq and RNA-seq data used in this analysis were retrieved from the NCBI Gene Expression Omnibus database (ie, GSM1113430,16 GSM1113428,16 GSM585589,25 GSM726978,25 GSM1071857,14 and GSM107185214 ). *GEO accession number GSE65427. (D) Validation for the expression changes of genes with higher AML1/ETO binding signals or genes with higher AML1 binding signals by real-time RT-PCR. Western blotting in the left panel showed the AML1/ETO knockdown efficiency. On AML1/ETO knockdown, genes with higher AML1/ETO binding signals (eg, OGG1, CTSG, PARVG, NKG7, and IL6R) were significantly upregulated, whereas genes with higher AML1 binding signals (eg, CTTN, YES1, CALM2, DUSP6, and PADI3) were significantly downregulated. The overview of the loci of those genes can be seen in Figure 6C and supplemental Figures 8 and 10. Error bars represent the SD of triplicate measurements. *P < .05; **P < .01; ***P < .001.
AML1/ETO-repressed genes show more AML1/ETO binding and AML1/ETO-activated genes show more AML1 binding. (A) Correlation between the binding affinity ratio of AML1/ETO to AML1 and gene sets repressed or activated by AML1/ETO. FDR q-value, false discovery rate q value; NES, normalized enrichment score. (B) Association of the binding affinities of AML1/ETO and AML1 with the modulation of gene expression by AML1/ETO. Heat maps of AML1/ETO and AML1 ChIP-seq signals on AML1/ETO-regulated genes were sorted based on the ratio of the AML1/ETO binding affinity vs AML1 binding affinity in a 4-kb window centered on the AML1/ETO peak summit (left and center). In the gene expression panel (right), the blue indicates AML1/ETO-repressed genes and the yellow indicates AML1/ETO-activated genes. (C) Overview of the loci of representative AML1/ETO-repressed genes OGG1 and AML1/ETO-activated genes CTTN in Kasumi-1 and SKNO-1 cells. Blue, AML1/ETO ChIP-seq signals; red, AML1 ChIP-seq signals; black, RNA-seq signals before and after AML1/ETO knockdown. Refseq annotations are shown at the bottom. ChIP-seq and RNA-seq data used in this analysis were retrieved from the NCBI Gene Expression Omnibus database (ie, GSM1113430,16 GSM1113428,16 GSM585589,25 GSM726978,25 GSM1071857,14 and GSM107185214 ). *GEO accession number GSE65427. (D) Validation for the expression changes of genes with higher AML1/ETO binding signals or genes with higher AML1 binding signals by real-time RT-PCR. Western blotting in the left panel showed the AML1/ETO knockdown efficiency. On AML1/ETO knockdown, genes with higher AML1/ETO binding signals (eg, OGG1, CTSG, PARVG, NKG7, and IL6R) were significantly upregulated, whereas genes with higher AML1 binding signals (eg, CTTN, YES1, CALM2, DUSP6, and PADI3) were significantly downregulated. The overview of the loci of those genes can be seen in Figure 6C and supplemental Figures 8 and 10. Error bars represent the SD of triplicate measurements. *P < .05; **P < .01; ***P < .001.
When GSEA was applied to the AML1/ETO-activated genes (supplemental Table 10), we found these genes tended to exhibit higher binding signals for AML1 than for AML1/ETO (Figure 6; supplemental Figure 10). This observation probably suggested that a small amount of AML1/ETO was not capable of reducing AML1 binding on chromatin and thus provided less impact on repressing AML1-dependent transactivation. To support this assumption, we found that AML1/ETO depletion on chromatin did not alter AML1 binding on these activated genes (supplemental Figure 9), implying that AML1/ETO might serve as a platform for regulatory (co)factor binding. Genes in this category included CTTN41 and YES1 that are involved in tumor progression and DUSP642 that is the negative regulators of cell differentiation.
Finally, we performed real-time reverse transcriptase-PCR and found that the expression levels of genes with higher AML1/ETO binding signals were significantly increased, whereas those with higher AML1 binding signals were significantly decreased, when AML1/ETO was knocked down (Figure 6D).
AML1/ETO transactivates gene expression through recruiting AP-1 to the AML1/ETO-AML1 complex
Next, we addressed how the AML1/ETO-AML1 complex activated the expression of target genes. More specifically, we sought to search for regulatory (co)factors that could be recruited by the AML1/ETO-AML1 complex in AML1/ETO activation. The candidates must meet the following requirements: (1) be a transcription activator and (2) be recruited and bind to the upstream regions of activated target genes. To search for such candidate regulators, we performed 2 independent analyses, one by the ENCODE ChIP-seq Significance Tool33 and the other by Cscan,34 for possible enrichment of transcriptional regulators in the promoters of AML1/ETO-activated genes but not in the promoters of AML1/ETO-repressed genes. Both analyses showed that the AP-1 family was highly enriched among genes activated by AML1/ETO (supplemental Tables 11 and 12).
Because c-Jun is the most potent transactivator in the AP-1 complex, we next performed ChIP-qPCR assays to verify the computational predictions using c-Jun as a surrogate mark. As shown in Figure 7A and supplemental Figure 11, c-Jun binding was indeed enriched at a higher level on AML1/ETO-activated genes rather than AML1/ETO-repressed genes. Re-ChIP assays further demonstrated that AML1/ETO co-occupied with c-Jun on the promoter regions of those AML1/ETO-activated genes (Figure 7B). Furthermore, we abolished AP-1 function using SR11302, an AP-1–specific inhibitor and found the mRNA expression of AML1/ETO-activated genes was significantly decreased after the inhibition of AP-1. In contrast, AP-1 inhibition did not affect the expression level of AML1/ETO-repressed genes (Figures 7C). Finally, AML1 has been reported to interact with c-Jun through the RHD that is contained in AML1/ETO as well.43 We thus performed co-IP assays in Kasumi-1 cells and found that the protein-protein interaction also occurred between AML1/ETO and c-Jun in vivo (Figure 7D). Together, these observations suggested that AP-1 was recruited by the AML1/ETO-AML1 complex and mediated the upregulation of the complex targeted genes.
The transcriptional activation property of AML1/ETO on AML1/ETO-AML1 complex bound genes is potentiated through the recruitment of AP-1. (A) c-Jun, a surrogate mark for AP-1, binds to the AML1/ETO-activated genes, but not to the AML1/ETO-repressed genes. ChIP was performed with anti-c-Jun antibody. The detailed ChIP-qPCR values can be found in supplemental Figure 9. (B) Validation for the coexistence of AML1/ETO and c-Jun on the promoter regions of AML1/ETO-activated genes through re-ChIP assays. ChIP products of the first indicated antibodies from Kasumi-1 cells were subjected to immunoprecipitation using the second indicated antibodies. NC, negative control. (C) The expression level of AML1/ETO-activated genes (CALM2, DUSP6, and PADI3) is decreased on SR11302 (AP-1 inhibitor) treatment. AML1/ETO-repressed genes (PARVG, NKG7, and IL6R) were used as negative controls. Error bars in B and C represent the SD of triplicate measurements. (D) AML1/ETO interacts with c-Jun in Kasumi-1 cells. The c-Jun–specific antibody was used for IP and the anti-ETO antibody was used for western blotting. **P < .01.
The transcriptional activation property of AML1/ETO on AML1/ETO-AML1 complex bound genes is potentiated through the recruitment of AP-1. (A) c-Jun, a surrogate mark for AP-1, binds to the AML1/ETO-activated genes, but not to the AML1/ETO-repressed genes. ChIP was performed with anti-c-Jun antibody. The detailed ChIP-qPCR values can be found in supplemental Figure 9. (B) Validation for the coexistence of AML1/ETO and c-Jun on the promoter regions of AML1/ETO-activated genes through re-ChIP assays. ChIP products of the first indicated antibodies from Kasumi-1 cells were subjected to immunoprecipitation using the second indicated antibodies. NC, negative control. (C) The expression level of AML1/ETO-activated genes (CALM2, DUSP6, and PADI3) is decreased on SR11302 (AP-1 inhibitor) treatment. AML1/ETO-repressed genes (PARVG, NKG7, and IL6R) were used as negative controls. Error bars in B and C represent the SD of triplicate measurements. (D) AML1/ETO interacts with c-Jun in Kasumi-1 cells. The c-Jun–specific antibody was used for IP and the anti-ETO antibody was used for western blotting. **P < .01.
Discussion
The fusion protein AML1/ETO plays a major role in the pathogenesis of t(8;21) AML and is generally considered to act as a transcriptional repressor for target genes of wild-type AML1.7 In this study, we demonstrate that wild-type AML1 forms a complex with AML1/ETO on chromatin by binding to the distinct motifs and is able to orchestrate the expression of genes both repressed and activated by AML1/ETO. Our data suggest that whether genes are repressed or activated by AML1/ETO largely depends on 2 things: (1) the relative binding signals of AML1 and AML1/ETO on chromatin and (2) the recruitment of cofactors. These findings significantly enrich our understanding of the interplay between AML1 and AML1/ETO in the pathogenesis of AML1/ETO-driven AML.
The formation of a multiprotein complex by an oncogenic fusion protein is critical for leukemogenesis, which provides a module for the fusion protein and other components to colocalize on chromatin and cooperatively regulates expression of diverse target genes. We demonstrated that wild-type AML1 formed a complex with AML1/ETO on chromatin. A recent study has shown that AML1/ETO exists as a complex with many transcription factors, including HEB, E2A, LMO2, CBFβ, and LDB1.14 It is very likely that the oligomerization of AML1/ETO might be required for its interaction with AML1 on neighboring sites of the chromatin, because the tetramerization of AML1/ETO is a prerequisite for its interaction with another cofactor HEB.14
Our observations underlie the contribution of wild-type AML1 to the pathogenesis of t(8;21) AML. AML1 has been shown to exert versatile functions in various types of leukemia. First, AML1 differs from most tumor suppressor genes that are generally inactivated through biallelic deletion or truncation mutations. Instead, AML1 genomic alterations generally only occur on 1 allele while keeping the other allele active.8,15 Second, emerging evidence has shown that wild-type AML1 is required for the leukemia development in t(8;21) AML with 1 active wild-type allele and in certain types of AML with 2 active alleles, eg, Cbfb-MYH11 and MLL-AF9 AML.16,17 Third, in addition to tumor suppression activity, AML1 can be oncogenic through amplification in certain pediatric acute lymphoblastic leukemia.44
The interaction between the fusion protein and the wild-type counterpart has also been documented in other types of leukemia, such as the interaction between PML/RARα and PML,45 and TEL/AML1 and TEL.46 In this work, we demonstrated that the RHD of AML1/ETO and AML1 mediated the complex formation. Involvement of the RHD in the interaction with AML1 (and AML1/ETO) seems to be a common feature for the complex to be formed with other proteins, such as the interactions between AML1/ETO and CEBPα,47 between AML1 and LEF-1,48 and between AML1 and MEF.49 The RHD has the ability of mediating both DNA binding and the protein-protein interaction,50 which provides a possibility for the interacting proteins to modulate the conformation of the AML1/ETO-containing complex priming for certain activities while AML1/ETO is bound to DNA.
Our genome-wide chromatin occupancy analysis of AML1 and AML1/ETO provides new clues for understanding the mechanisms on how wild-type AML1 works in cooperation with AML1/ETO to regulate gene expression. Our data demonstrated that the AML1/ETO-AML1 complex resided in target genes regardless of being repressed or activated by AML1/ETO and the ratio of AML1/ETO to AML1 bound to the targets was correlated to the transcriptional activities of target genes, ie, downregulated genes with higher AML1/ETO binding signals and upregulated genes with higher AML1 binding signals. This finding is in agreement with an earlier study showing that less amounts of AML1/ETO induces AML1 transactivation on the promoter activity of the macrophage colony-stimulating factor receptor, whereas increased AML1/ETO expression to the level higher than AML1 expression reduces AML1-dependent transactivation.51 Moreover, a recent genome-wide binding analysis has shown that the balance of MYC and MIZ1 binding on chromatin controls the direction of genes regulated by MYC in MYC-driven tumors.52
AML1/ETO used to be thought to repress the AML1-regulated gene by replacing AML1 binding through a dominant-negative effect.53 A recent study also revealed a dynamic competition between AML1/ETO and AML1 for the same binding sites.21 Our findings, nonetheless, have provided a more complex scenario. On the one hand, on AML1/ETO-AML1 colocalized regions, AML1/ETO-repressed genes might still have AML1 binding on their regulatory regions, but with lower binding signals than AML1/ETO. We speculate that on these regions, AML1/ETO may compete with AML1 in the AML1/ETO-AML1 complex through reducing AML1 binding on the adjacent sites, leading to the repression of AML1-regulated genes. This speculation is supported by our recent study on the CTSG promoter where AML1/ETO-mediated transrepression requires both AML1/ETO and AML1 binding at adjacent sites.40 On the other hand, we also observed certain chromatin regions with unique AML1 or AML1/ETO occupancy (Figure 1D). These AML1/ETO unique regions might result from a replacing of the originally bound AML1 by the fusion protein or from a totally de novo binding by AML1/ETO. Hence, the partial differences of the results from distinct studies, considering possible differences in experimental conditions, might be viewed more complementary than contradictory and probably also underscore a more complex mechanism for both AML1/ETO and AML1 to be involved in gene transrepression.
More interestingly, our findings provide a potential mechanism for AML1/ETO-mediated transactivation on the AML1/ETO-AML1 complex targeted genes. Such genes exhibited higher AML1 and lower AML1/ETO binding signals on chromatin, suggesting that a small amount of AML1/ETO might not be capable of reducing AML1 binding on chromatin as doing on AML1/ETO-repressed genes. Instead, AML1/ETO might serve as a platform for regulatory (co)factor binding as previously reported on the regulation of ID1, CDKN1A and EGR1 expression.13 We also identified AP-1 as the potential cofactor mediating such transactivation on the AML1/ETO-AML1 complex-targeted genes. AML1/ETO has recently been reported to interact with p300 or PRMT1: both are essential for AML1/ETO-mediated gene activation and leukemogenesis.12,13 Given the known modulatory effects of PRMT1 and p300 on AP-1–mediated regulation,54,55 we assume that AP-1, p300, and PRMT1 may function together in the AML1/ETO-AML1 complex.
One major contribution of this study is to update our current knowledge on the binding motif of AML1/ETO. It is generally thought that AML1/ETO recognizes the same motif as AML1 does.56 In this study, we found that AML1/ETO and AML1 tended to bind to similar yet distinct AML1 motifs on the colocalized regions. Wild-type AML1 prefers the long motif, whereas AML1/ETO is in favor of the short one. Such differential preference is probably due to the fact that, in addition to the RHD, the rest structure of AML1/ETO and AML1 differs greatly. Our finding also suggests that AML1/ETO displays a more relaxed DNA-binding specificity than wild-type AML1. This is in line with the observation that other fusion proteins, such as PML/RARα57,58 and PLZF/RARα,59 also have an extended repertoire of DNA-binding sites compared with their wild-type counterparts. Such a flexible DNA binding ability probably allows the oncogenic fusion protein to recognize more DNA sequences with a wider conformation than the wild-type protein does. Also, the binding signals of AML1 and AML1/ETO on chromatin can be determined by many factors, including the structural features of proteins in the complex (eg, the tetramerization of AML1/ETO). Further analysis will clarify the causal relationship and the nature of regulatory networks between the oncogenic fusion proteins and the wild-type counterpart and even the indirect factors in t(8;21) AML.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
This work was supported in part by Ministry of Science and Technology Grants of China (2012AA02A211), National Natural Science Foundation Grants of China (81270625, 81530003, 31171257, and 91440114), Shanghai Leading Talent Projects (2015008), the Academic Leader Program of Shanghai Science and Technology Committee (2015137), and the Samuel Waxman Cancer Research Foundation.
Authorship
Contribution: K.W. and Y.L. designed the study; Y.L., X.W., W.J., and Y.T. performed experiments; Y.L., H.W., and H.F. performed analysis; K.W. and Y.L. wrote the manuscript; K.W., Z.C., and S.C. revised the manuscript; and all authors approved the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Kankan Wang, State Key Laboratory of Medical Genomics and Shanghai Institute of Hematology, Ruijin Hospital, Shanghai Jiao Tong University of Medicine, 197 Ruijin Er Rd, Shanghai 200025, China; e-mail: kankanwang@shsmu.edu.cn.
References
Author notes
Y.L. and H.W. contributed equally to this work.