Key Points
Agents that inhibit both complexes containing the mammalian target of rapamycin are particularly toxic to acute lymphocytic leukemia cells.
This killing reflects engagement of a 4EBP1/c-MYC/PUMA axis downstream of mTORC1 and an NF-κB/EGR1/BIM axis downstream of mTORC2.
Abstract
The mammalian target of rapamycin (mTOR), a kinase that regulates proliferation and apoptosis, has been extensively evaluated as a therapeutic target in multiple malignancies. Rapamycin analogs, which partially inhibit mTOR complex 1 (mTORC1), exhibit immunosuppressive and limited antitumor activity, but sometimes activate survival pathways through feedback mechanisms involving mTORC2. Thus, attention has turned to agents targeting both mTOR complexes by binding the mTOR active site. Here we show that disruption of either mTOR-containing complex is toxic to acute lymphocytic leukemia (ALL) cells and identify 2 previously unrecognized pathways leading to this cell death. Inhibition of mTORC1-mediated 4EBP1 phosphorylation leads to decreased expression of c-MYC and subsequent upregulation of the proapoptotic BCL2 family member PUMA, whereas inhibition of mTORC2 results in nuclear factor-κB–mediated expression of the Early Growth Response 1 (EGR1) gene, which encodes a transcription factor that binds and transactivates the proapoptotic BCL2L11 locus encoding BIM. Importantly, 1 or both pathways contribute to death of malignant lymphoid cells after treatment with dual mTORC1/mTORC2 inhibitors. Collectively, these observations not only provide new insight into the survival roles of mTOR in lymphoid malignancies, but also identify alterations that potentially modulate the action of mTOR dual inhibitors in ALL.
Introduction
The mammalian target of rapamycin (mTOR) is a serine/threonine kinase implicated in cell growth, actin cytoskeleton modulation, and inhibition of apoptosis.1-4 The observation that mTOR is aberrantly activated in a variety of malignancies has generated intense interest in this kinase as a target for antineoplastic therapy, particularly for lymphoid malignancies.1,3,5-11 Over the last decade, rapamycin-based mTOR inhibitors have proven effective in certain lymphomas.7,9,10 However, their efficacy is limited by incomplete inhibition of mTOR complex 1 (mTORC1) and by activation of AKT and downstream prosurvival pathways through a variety of feedback mechanisms.6,11-15 To overcome this limitation, inhibitors targeting the kinase activities of both mTORC1 and mTORC2 have been developed.6,9,11,16-21 Because these agents also more effectively inhibit mTORC1,16-18,21,22 it has been unclear whether inhibition of mTORC1 or mTORC2 is responsible for the cytotoxic effects. Moreover, the specific mechanisms underlying killing by these agents remain incompletely understood.
We previously showed that mTOR dual inhibitors induce apoptosis in a variety of malignant lymphoid cell lines and clinical samples of certain lymphoid neoplasms, with some cases of acute lymphocytic leukemia (ALL) being particularly sensitive.21 Further investigation indicated that this killing involves upregulation of the proapoptotic BCL2 family members BIM and PUMA.21 The present study was performed to better understand this response, which is not observed in other cell types.23 Genes encoding both BIM and PUMA are known to be transcriptionally activated by FOXO3A when phosphorylation of this transcription factor by AKT is inhibited24,25 or by a c-Jun N-terminal kinase (JNK)/cJUN axis after mTORC1 inhibition in other cell types.26,27 Surprisingly, however, we demonstrate here that upregulation of PUMA and BIM by mTOR dual inhibitors appears to occur independent of these pathways. Instead, mTOR dual inhibitors induce PUMA by inhibiting mTORC1-mediated phosphorylation of 4EBP1, thereby stabilizing its interaction with EIF4E to inhibit translation, downregulate c-MYC (abbreviated MYC throughout this work), and derepress PUMA mRNA. Simultaneously, mTOR dual inhibitors activate nuclear factor (NF)-κB, leading to transactivation of EGR1, which encodes a transcription factor for BIM. These observations provide new insight into the antineoplastic activity of mTOR inhibitors in lymphoid neoplasms.
Methods
Reagents
Reagents were purchased as follows: OSI-027 and MLN0128 from ChemieTek (Indianapolis, IN), allophycocyanin-conjugated annexin V from BD Biosciences (San Jose, CA); propidium iodide (PI) and rapamycin from Sigma-Aldrich (St Louis, MO); 7-methyl-guanosine triphosphate-Sepharose (7Me-GTP-Sepharose) from GE Healthcare (Pittsburgh, PA); the broad spectrum caspase inhibitor Q-VD-OPh from SM Biochemicals (Anaheim, CA); and 4EGI-1 from EMD (Billerica, MA). Antibodies were obtained as follows: β-ACTIN, BAX, PUMA (SC-28226 and SC-374223), LAMIN B1 and cRAF from Santa Cruz Biotechnology (Santa Cruz, CA), cleaved poly(ADP-ribose) polymerase 1 from Promega (Madison, WI), heat shock protein 90β (HSP90β) from D. Toft (Mayo Clinic, Rochester, MN), and MYC from Chi Dang (University of Pennsylvania School of Medicine, Philadelphia, PA). A second antibody to MYC and antibodies to all other proteins, including phosphorylated epitopes, were from Cell Signaling Technology (Beverly, MA).
Tissue culture
P388 cells were from Y. Pommier (National Cancer Institute, Bethesda, MD). All other cell lines were obtained as previously described.21 Cell lines were propagated at densities of <1 × 106 cells/mL in RPMI 1640 medium containing 10% heat-inactivated fetal bovine serum, 100 U/mL penicillin G, 100 μg/mL streptomycin, and 2 mM glutamine (medium A) except for SeAx, which received medium A with 15% fetal bovine serum.
Assessment of cell killing
Lentiviral transduction
For hairpin-mediated knockdown, lentivirus encoding 4EBP1 short hairpin RNAs (shRNAs) #31 (AGGATCATCTATGACCGGAAA) and #35 (GCCAGGCCTTATGAAAGTGAT) (R. Bram, Mayo Clinic, MN) or control shRNAs A8 and A9 in pLKO1 (Addgene, Cambridge, MA) were packaged in HEK293T cells as previously described.29 Cells were exposed to virus for 48 hours, washed, and selected with 2 µg/mL puromycin. Knockdown was verified by immunoblotting.
Transient transfections
For transient knockdown or overexpression, 5 to 10 × 106 cells were electroporated with 40 µg DNA consisting of plasmid encoding shRNAs targeting RAPTOR or RICTOR (Addgene), myristoylated AKT (myrAKT), or AKT S473D (F. Sinicrope, Mayo Clinic, Rochester, MN) at 240 V with a single 10-ms pulse using a BTX830 square-wave electroporator (Harvard Apparatus, Holliston, MA). Cells were incubated for 12 hours before drug treatment.
7Me-GTP pulldown assay
2 × 107 cells were treated with OSI-027, MLN0128, rapamycin, or 4EGI-1 for 48 hours in the presence of 5 µM Q-VD-OPh. All further steps were performed at 4°C. Cells were washed in PBS and lysed in Nonidet P-40 (NP-40) lysis buffer (150 mM NaCl, 1.0% [w/v] NP-40, 50 mM Tris-HCl, pH 8.0). After sedimentation at 14 000g for 10 minutes to remove insoluble material, lysates were incubated with 7Me-GTP-Sepharose beads overnight. Bound protein was washed 5 times with NP-40 lysis buffer, released in 2× sodium dodecyl sulfate sample buffer, and subjected to immunoblotting.
Luciferase assays and chromatin immunoprecipitation
RNA sequencing analysis
Jurkat cells were treated with diluent or 10 μM OSI-027 for 48 hours in 5 µM Q-VD-OPh. Total RNA was extracted using a Qiagen RNA extraction kit. After RNA sample quality was assessed by RNA integration number, an Illumina TruSeq mRNA kit was used to generate cDNA for next-generation sequencing. RNAs were poly-A selected and fragmented, then subjected to reverse transcription with random primers and second-strand synthesis to generate double-stranded cDNA. Ends were repaired and poly(adenyl)ated, followed by adaptor and index ligation. The cDNAs were then denatured and polymerase chain reaction (PCR) enriched to generate the final genomic library, which was analyzed on an Illumina HiSeq 2000. Each mRNA count number was normalized to counts per million.
Human primary ALL cells
After pretreatment bone marrow aspirates from newly diagnosed ALL patients (supplemental Table 1) were obtained with institutional review board approval, cells were isolated on Histopaque-1077 (Sigma-Aldrich) step gradients, washed with serum-free RPMI 1640 medium, cultured for 48 hours in medium A with 5 µM Q-VD-OPh and the indicated concentrations of OSI-027 or MLN0128, sedimented at 100g, washed once with ice cold RPMI containing 10 mM 4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid (pH 7.4 at 4°C), lysed in buffered 6 M guanidine hydrochloride, and prepared for sodium dodecyl sulfate-polyacrylamide gel electrophoresis and immunoblotting as described.31 Alternatively, samples cultured for 5 days in the presence of 10 nM rapamycin or the indicated concentrations of OSI-027 and MLN0128 (in the absence of Q-VD-OPh) were assayed for viable cell mass by 3-(4,5 dimethylthiazol-2-yl)-5-(3-carboxymethoxyphenyl-2-(4-sulfophenyl)-2H-tetrazolium (MTS) assay21 or induction of DNA fragmentation by flow cytometry after propidium iodide staining.28
Statistical methods
Unless otherwise indicated, all error bars represent mean ± standard error of the mean of 3 independent experiments. Effects of OSI-027 or MLN0128 vs rapamycin on the same samples were compared using paired Student t tests for all ALL samples treated with dual mTORC1/mTORC2 inhibitor vs rapamycin. All other comparisons involved unpaired Student t tests with P values that are corrected for the effects of multiple comparisons.32
Results
Effects of dual mTORC1/mTORC2 inhibitors on human ALL ex vivo
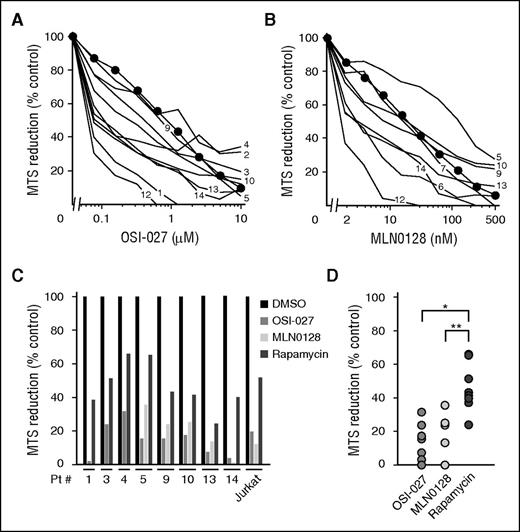
Building on previous work, which found that ALL cells are more sensitive to dual mTORC1/mTORC2 inhibition than other lymphoid neoplasms,21 we assessed the sensitivity of ALL samples (supplemental Table 1) to the mTORC1/mTORC2 dual inhibitors OSI-027 and MLN0128 using tetrazolium dye reduction assays and flow cytometry. The dual inhibitor-sensitive human T cell ALL cell line Jurkat21 served as a control. As indicated in Figure 1A-B, clinical ALL specimens exhibited IC50 values of 80 to 2000 nM for OSI-027 and 2 to 100 nM for MLN0128 in MTS assays but were generally as sensitive as Jurkat cells. Further experiments demonstrated DNA fragmentation (subdiploid cells), a hallmark of apoptosis, in clinical ALL specimens after treatment with either agent (supplemental Figure 1), extending earlier observations that OSI-027 induces phosphatidylserine externalization and poly(ADP-ribose) polymerase cleavage in clinical lymphoma isolates.21 Both dual inhibitors were also more cytotoxic to ALL isolates than the maximum clinically achievable rapamycin concentration (Figure 1C-D), providing the impetus for trying to better understand the unique action of mTORC1/mTORC2 dual inhibitors in malignant lymphoid cells.
OSI-027 and MLN0128 diminish viable cell mass in ALL cultures ex vivo. After samples from patients with newly diagnosed ALL were treated for 5 days with (A-B) OSI-027 or MLN0128 as indicated or (C-D) with 5 µM OSI-027, 250 nM MLN0128, or 10 nM rapamycin, MTS reduction was assayed. Results in cells treated with diluent (0.1% dimethylsulfoxide) were set at 100%. Numbers next to lines in panels A and B refer to patient numbers in supplemental Table 1. Mean values for Jurkat cells (included in each assay) are indicated by circles. (C-D) The same data separated by (C) patient or (D) treatment. In D, *P < .001 and **P = .011.
OSI-027 and MLN0128 diminish viable cell mass in ALL cultures ex vivo. After samples from patients with newly diagnosed ALL were treated for 5 days with (A-B) OSI-027 or MLN0128 as indicated or (C-D) with 5 µM OSI-027, 250 nM MLN0128, or 10 nM rapamycin, MTS reduction was assayed. Results in cells treated with diluent (0.1% dimethylsulfoxide) were set at 100%. Numbers next to lines in panels A and B refer to patient numbers in supplemental Table 1. Mean values for Jurkat cells (included in each assay) are indicated by circles. (C-D) The same data separated by (C) patient or (D) treatment. In D, *P < .001 and **P = .011.
mTORC1 and mTORC2 inhibition both contribute to lymphoid cell death
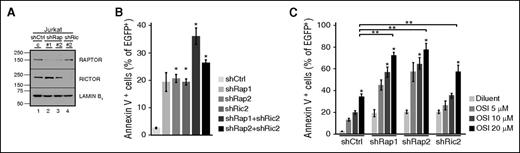
To confirm that the effects of these inhibitors reflect mTOR inhibition, we examined the effect of shRNA-mediated downregulation of RAPTOR and RICTOR (Figure 2; supplemental Figure 2), critical components of mTORC1 and mTORC2, respectively.33,34 Knockdown of either RAPTOR or RICTOR induced apoptosis in multiple malignant human lymphoid lines (Figure 2B; supplemental Figure 2B,E), with a trend toward a further increase in apoptosis when both were knocked down (Figure 2B; supplemental Figure 2E). Moreover, knockdown of either RAPTOR or RICTOR sensitized cells to OSI-027 (Figure 2C; supplemental Figure 2C,F). Collectively, these results suggest that inhibition of both mTORC1 and mTORC2 can contribute to apoptosis.
Effects of RAPTOR and RICTOR knockdown. (A-B) Seventy-two hours after transfection with shRNAs targeting RAPTOR (shRap) or RICTOR (shRic) along with plasmid encoding EGFP-histone H2B (to mark transfected cells), Jurkat cells were harvested for immunoblotting or stained with allophycocyanin-annexin V and analyzed by flow cytometry for annexin V binding to EGFP-histone H2B+ cells. (C) Cells were transfected with shRNAs, incubated for 12 hours, and treated with diluent or OSI-027 for 72 hours before annexin V staining. Error bars in panels B, C, and all subsequent figures indicate mean ± standard error of the mean of 3 independent assays unless otherwise stated. In panels B and C, *P < .05 relative to diluent-treated cells. In panel C, **P < .05 compared with cells treated with control shRNA and 20 µM OSI-027. All P values in this and subsequent figures are corrected for multiple comparisons.
Effects of RAPTOR and RICTOR knockdown. (A-B) Seventy-two hours after transfection with shRNAs targeting RAPTOR (shRap) or RICTOR (shRic) along with plasmid encoding EGFP-histone H2B (to mark transfected cells), Jurkat cells were harvested for immunoblotting or stained with allophycocyanin-annexin V and analyzed by flow cytometry for annexin V binding to EGFP-histone H2B+ cells. (C) Cells were transfected with shRNAs, incubated for 12 hours, and treated with diluent or OSI-027 for 72 hours before annexin V staining. Error bars in panels B, C, and all subsequent figures indicate mean ± standard error of the mean of 3 independent assays unless otherwise stated. In panels B and C, *P < .05 relative to diluent-treated cells. In panel C, **P < .05 compared with cells treated with control shRNA and 20 µM OSI-027. All P values in this and subsequent figures are corrected for multiple comparisons.
Further experiments examined the possibility that mTORC1/mTORC2 dual inhibition induces apoptosis in ALL through previously described pathways. In view of earlier suggestions that mTORC1/mTORC2 dual inhibitors are more cytotoxic because they inhibit mTORC2-mediated AKT activation, we assessed the impact of constitutively active AKT constructs on dual inhibitor-induced apoptosis. Although myr-AKT and AKT S473D preserved AKT activity during dual inhibitor treatment, as indicated by PRAS40 Thr246 phosphorylation, neither prevented dual inhibitor-induced apoptosis (supplemental Figure 3). Because upregulation of BIM and PUMA contributes to dual inhibitor-induced apoptosis,21 we also examined involvement FOXO3A, which transactivates the genes for both BH3-only family members after AKT inhibition.24,25,35 FOXO3A depletion did not affect OSI-027–induced cell death (supplemental Figure 4A-B). Moreover, luciferase assays showed undiminished dual inhibitor-induced activation of the BIM promoter when the FOXO3A binding sites were mutated (supplemental Figure 4C). We similarly investigated whether an ASK1/JNK/cJUN pathway implicated in rapamycin-induced death in osteosarcoma cell lines26,27 plays a major role in mTOR dual inhibitor-induced lymphoid cell death. In Jurkat cells, OSI-027 increased cJUN phosphorylation (supplemental Figure 5A) and activation of an AP1 reporter construct (supplemental Figure 5B). Importantly, however, dominant-negative (dn) cJUN or JNK inhibited apoptosis induction by cytarabine (another JNK-dependent apoptotic stimulus36 ) but did not alter dual inhibitor-induced BIM promoter activation or apoptosis (supplemental Figure 5C-E). These observations prompted us to explore alternative explanations for OSI-027- and MLN0128-induced killing of ALL cells.
4EBP1 dephosphorylation induces apoptosis through PUMA upregulation
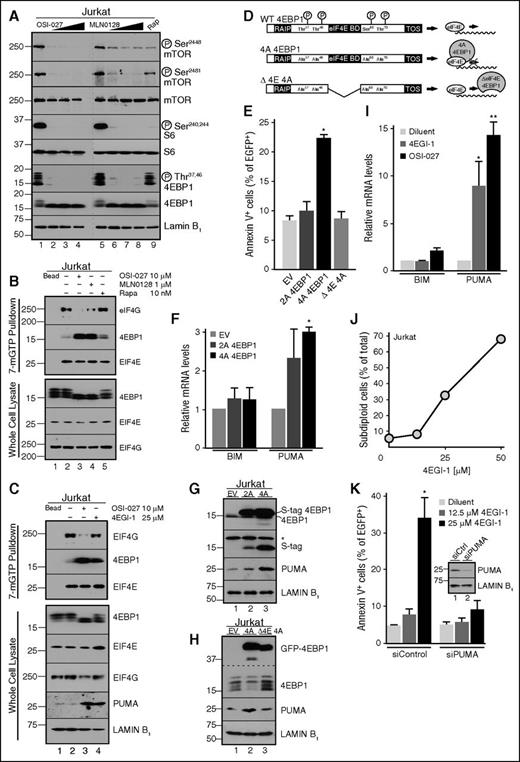
The observation that 4EBP1 phosphorylation is inhibited by mTOR dual inhibitors, but not rapamycin (Figure 3A; supplemental Figure 6),21,22,37,38 led us to hypothesize that 4EBP1 dephosphorylation might contribute to mTOR dual inhibitor-induced apoptosis, perhaps through 4EBP1-mediated disruption of EIF4E/EIF4G interactions and inhibition of translation.39 Consistent with this possibility, 7Me-GTP pulldown assays showed that mTOR dual inhibitors, but not rapamycin, cause dissociation of the EIF4E/EIF4G translational complex and enhanced 4EBP1/EIF4E binding (Figure 3B-C). Moreover, expression of 4EBP1 T37A/T46A/S65A/T70A (4A), which mimics dephosphorylated 4EBP1 (Figure 3D), increased PUMA mRNA and protein levels and apoptosis (Figure 3E-G). In contrast, a 4EBP1 4A construct lacking the EIF4E binding domain (Δ4E 4A; Figure 3D) failed to increase PUMA protein or apoptosis (Figure 3H and E, respectively).
Effects of mutant 4EBP1 constructs and 4EGI-1 on cell survival. (A) After Jurkat cells were treated with diluent (lanes 1 and 5); OSI-027 at 5, 10, or 20 µM (lanes 2-4); MLN0128 at 0.25, 0.,5 or 1 µM (lanes 6-8); or 10 nM rapamycin (lane 9), whole cell lysates were subjected to immunoblotting. Dashed line indicates juxtaposition of nonadjacent lanes from the same X-ray films. (B-C) Cells treated for 48 hours with diluent (lane 2), 10 µM OSI-027 (lane 3), 1 µM MLN0128 (lane 4, B), 10 nM rapamycin (lane 5, B), or 25 µM 4EGI-1 (lane 4, C) were lysed with NP-40 buffer. After clarification, lysates were incubated with sepharose (lane 1) or 7Me-GTP-sepharose beads overnight and washed 5 times with NP-40 buffer before immunoblotting. (D) 4EBP1 constructs used in panels E-H. In the remainder of this figure, 4EBP1 T37A/T46A is designated 2A and 4EBP1 T37A/T46A/S65A/T70A is 4A. (E) Cells transfected with the indicated 4EBP1 construct were incubated for 72 hours and analyzed for annexin V binding. In panel E, *P = .03 relative to empty vector. (F) Seventy-two hours after transfection with S peptide-tagged 4EBP1 2A or 4A, qRT-PCR was performed to quantify BIM and PUMA messages, which were normalized to GAPDH mRNA. In panel F, *P = .006 relative to empty vector. (G-H) Cells transfected with the indicated 4EBP1 construct tagged with (G) S peptide or (H) EGFP were incubated for 72 hours and harvested for immunoblotting. In panel G, *nonspecific band. (I) qRT-PCR for BIM and PUMA mRNA after treatment with 25 µM 4EGI-1 or 10 µM OSI-027. In panel I, *P = .05 and **P = .002, relative to diluent. (J) Subdiploid cells, another indicator of apoptosis,21,28 after treatment for 48 hours with 4EGI-1. Error bars in this panel (n = 3 independent experiments) are smaller than symbols. (K) Cells transfected with PUMA siRNA or nontargeting control were treated for 48 hours with diluent or 4EGI-1 and stained with annexin V. In panel K, *P = .036 relative to control. (K, inset) Immunoblot of whole cell lysates.
Effects of mutant 4EBP1 constructs and 4EGI-1 on cell survival. (A) After Jurkat cells were treated with diluent (lanes 1 and 5); OSI-027 at 5, 10, or 20 µM (lanes 2-4); MLN0128 at 0.25, 0.,5 or 1 µM (lanes 6-8); or 10 nM rapamycin (lane 9), whole cell lysates were subjected to immunoblotting. Dashed line indicates juxtaposition of nonadjacent lanes from the same X-ray films. (B-C) Cells treated for 48 hours with diluent (lane 2), 10 µM OSI-027 (lane 3), 1 µM MLN0128 (lane 4, B), 10 nM rapamycin (lane 5, B), or 25 µM 4EGI-1 (lane 4, C) were lysed with NP-40 buffer. After clarification, lysates were incubated with sepharose (lane 1) or 7Me-GTP-sepharose beads overnight and washed 5 times with NP-40 buffer before immunoblotting. (D) 4EBP1 constructs used in panels E-H. In the remainder of this figure, 4EBP1 T37A/T46A is designated 2A and 4EBP1 T37A/T46A/S65A/T70A is 4A. (E) Cells transfected with the indicated 4EBP1 construct were incubated for 72 hours and analyzed for annexin V binding. In panel E, *P = .03 relative to empty vector. (F) Seventy-two hours after transfection with S peptide-tagged 4EBP1 2A or 4A, qRT-PCR was performed to quantify BIM and PUMA messages, which were normalized to GAPDH mRNA. In panel F, *P = .006 relative to empty vector. (G-H) Cells transfected with the indicated 4EBP1 construct tagged with (G) S peptide or (H) EGFP were incubated for 72 hours and harvested for immunoblotting. In panel G, *nonspecific band. (I) qRT-PCR for BIM and PUMA mRNA after treatment with 25 µM 4EGI-1 or 10 µM OSI-027. In panel I, *P = .05 and **P = .002, relative to diluent. (J) Subdiploid cells, another indicator of apoptosis,21,28 after treatment for 48 hours with 4EGI-1. Error bars in this panel (n = 3 independent experiments) are smaller than symbols. (K) Cells transfected with PUMA siRNA or nontargeting control were treated for 48 hours with diluent or 4EGI-1 and stained with annexin V. In panel K, *P = .036 relative to control. (K, inset) Immunoblot of whole cell lysates.
If 4EBP1-mediated disruption of EIF4E/EIF4G interactions contributes to apoptosis, the effect should be phenocopied by 4EGI-1, a small molecule that disrupts EIF4E/EIF4G interactions,40-44 and diminished by 4EBP1 knockdown. In agreement with these predictions, 4EGI-1 not only decreased EIF4G binding to EIF4E (Figure 3C), but also increased PUMA expression (Figure 3C,I) and induced apoptosis (Figure 3J) that was PUMA dependent (Figure 3K). Conversely, 4EBP1 knockdown (Figure 4A) diminished the ability of mTOR dual inhibitors to induce PUMA mRNA (Figure 4B), PUMA protein (Figure 4C-D), and apoptosis (Figure 4E-F); this phenotype was reversed by expression of shRNA-resistant 4EBP1 (Figure 4G-H). Collectively, these observations indicate that mTORC1 inhibition, acting through 4EBP1 to disrupt EIF4E/EIF4G interactions, leads to PUMA upregulation.
Effect of 4EBP1 knockdown on PUMA expression and dual inhibitor-induced apoptosis. (A) Immunoblot of Jurkat cells transfected with different control (lanes 1 and 2) or 4EBP1 knockdown constructs (lanes 3 and 4). (B) qRT-PCR for BIM and PUMA mRNA in control or 4EBP1 knockdown cells after 48-hour treatment with diluent or 10 µM OSI-027. In panel B, *P < .001 compared with diluent control. (C-D) Immunoblot of cells transfected with empty vector or 4EBP1 shRNA and treated for 48 hours with (C) OSI-027 or (D) MLN0128. (E-F) Cells were stained with PI and subjected to flow microfluorimetry after treatment with (E) OSI-027 or (F) MLN0128. (G-H) Cells expressing control plasmid or 4EBP1 shRNA were transfected with pSPN empty vector or plasmid expressing shRNA-resistant S peptide-tagged 4EBP1, incubated for 48 hours (G) with or (H) without OSI-027, and harvested for annexin V (G) binding or (H) blotting. In panels E-G, *P < .05 relative to A8 control.
Effect of 4EBP1 knockdown on PUMA expression and dual inhibitor-induced apoptosis. (A) Immunoblot of Jurkat cells transfected with different control (lanes 1 and 2) or 4EBP1 knockdown constructs (lanes 3 and 4). (B) qRT-PCR for BIM and PUMA mRNA in control or 4EBP1 knockdown cells after 48-hour treatment with diluent or 10 µM OSI-027. In panel B, *P < .001 compared with diluent control. (C-D) Immunoblot of cells transfected with empty vector or 4EBP1 shRNA and treated for 48 hours with (C) OSI-027 or (D) MLN0128. (E-F) Cells were stained with PI and subjected to flow microfluorimetry after treatment with (E) OSI-027 or (F) MLN0128. (G-H) Cells expressing control plasmid or 4EBP1 shRNA were transfected with pSPN empty vector or plasmid expressing shRNA-resistant S peptide-tagged 4EBP1, incubated for 48 hours (G) with or (H) without OSI-027, and harvested for annexin V (G) binding or (H) blotting. In panels E-G, *P < .05 relative to A8 control.
Role of MYC in dual mTOR inhibitor-induced PUMA upregulation
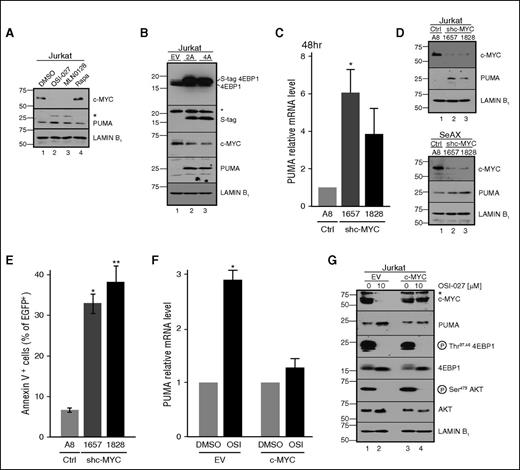
In accord with the known dependence of MYC translation on the EIF4E/EIF4G complex,45 we also observed MYC downregulation after treatment with OSI-027, MLN0128, and 4EGI-1, but not rapamycin (Figure 5A; supplemental Figure 7A-B). Importantly, MYC downregulation paralleled PUMA induction under a variety of conditions, including expression of nonphosphorylatable 4EBP1 in parental Jurkat cells (Figure 5B) or wild-type 4EBP1 in 4EBP1-deficient cells (Figure 4H). Conversely, neither MYC downregulation nor PUMA induction occurred after dual inhibitor treatment if 4EBP1 was downregulated (Figure 4C).
Role of MYC downregulation in PUMA expression. (A) Immunoblot of Jurkat cells treated with diluent (lane 1), 10 µM OSI-027 (lane 2), 1 µM MLN0128 (lane 3), or 10 nM rapamycin (lane 4) for 48 hours. (B) Forty-eight hours after transient transfection with empty vector or plasmid encoding the indicated 4EBP1 construct fused to S peptide, cells were harvested for immunoblotting. (C-E) Forty-eight hours after transient transfection with control shRNA or shRNAs targeting MYC, (C) PUMA mRNA, (D) protein, and (E) annexin V binding were assessed. In panel C, *P = .036 relative to control. In panel E, *P = .002 and **P = .008 relative to control. (F-G) Cells transfected with empty vector or MYC cDNA behind the constitutive CMV promoter were treated with diluent or 10 μM OSI-027 and harvested for (F) qRT-PCR or (G) immunoblotting. Results of drug-treated samples in panels C and F were normalized to each corresponding diluent-treated control, which was set to 1. In panel F, *P = .011 relative to control. In panels A,B,G, *nonspecific bands.
Role of MYC downregulation in PUMA expression. (A) Immunoblot of Jurkat cells treated with diluent (lane 1), 10 µM OSI-027 (lane 2), 1 µM MLN0128 (lane 3), or 10 nM rapamycin (lane 4) for 48 hours. (B) Forty-eight hours after transient transfection with empty vector or plasmid encoding the indicated 4EBP1 construct fused to S peptide, cells were harvested for immunoblotting. (C-E) Forty-eight hours after transient transfection with control shRNA or shRNAs targeting MYC, (C) PUMA mRNA, (D) protein, and (E) annexin V binding were assessed. In panel C, *P = .036 relative to control. In panel E, *P = .002 and **P = .008 relative to control. (F-G) Cells transfected with empty vector or MYC cDNA behind the constitutive CMV promoter were treated with diluent or 10 μM OSI-027 and harvested for (F) qRT-PCR or (G) immunoblotting. Results of drug-treated samples in panels C and F were normalized to each corresponding diluent-treated control, which was set to 1. In panel F, *P = .011 relative to control. In panels A,B,G, *nonspecific bands.
In further studies examining the role of MYC loss in PUMA upregulation, MYC shRNA not only induced PUMA mRNA and protein (Figure 5C-D), but also increased apoptosis (Figure 5E) in a PUMA-dependent manner (supplemental Figure 7C). Conversely, MYC expression from a construct lacking the long 5′ untranslated region (UTR; and therefore less dependent on mTORC1 for translation) suppressed OSI-027–induced PUMA mRNA and protein upregulation (Figure 5F-G). Further experiments indicated that OSI-027 failed to activate the PUMA promoter (supplemental Figure 7D), but instead stabilized constructs containing the PUMA 3′ UTR (supplemental Figure 7E), consistent with the possibility that MYC represses PUMA through its widespread effects on microRNAs.46 Similar effects of OSI-027, but not rapamycin, on 4EBP1 phosphorylation, MYC expression, and PUMA upregulation, were observed in Nalm6 and Jeko cells (supplemental Figure 8A-B), ruling out the possibility that observed effects are unique to Jurkat cells. Moreover, OSI-027 caused decreased 4EBP1 phosphorylation, MYC downregulation, and PUMA upregulation in Jeko xenografts in vivo (supplemental Figure 8C). Collectively, these results support a model in which mTORC1 inhibition, acting through 4EBP1, induces PUMA upregulation and apoptosis through MYC downregulation.
Dual inhibitors transcriptionally activate BIM through EGR1
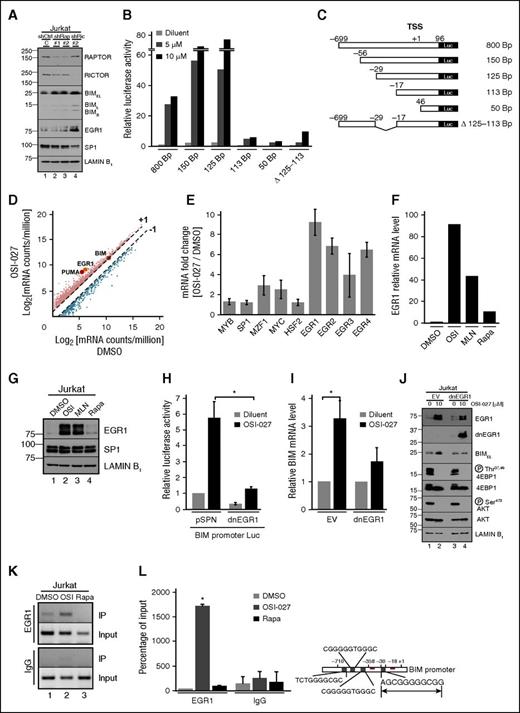
To assess the basis for BIM upregulation during dual inhibitor-induced killing,21 we used RNA interference, reporter assays, and RNAseq experiments. Initial experiments demonstrated that BIM increases after RICTOR and, to a lesser extent, RAPTOR knockdown (Figure 6A). Because this reflects BIM promoter activation (supplemental Figure 7D), we investigated dual inhibitor-induced activation of a series Bim promoter truncations in P388 mouse lymphoma cells, which have a wider dynamic range for these assays. OSI-027–induced promoter activity decreased markedly when the nucleotides −29 to −18 (relative to the transcription start site) were removed (Figure 6B-C). Similarly, the 800-bp Bim promoter lacking these 12 bp (∆113-125) was activated much less (Figure 6B), suggesting that this response element is critical for OSI-027–induced promoter activation. In silico analysis using the transcription factor databases TRANSFAC and TFSEARCH identified 8 transcription factors, including EGR1, SP1, MYB, and HSF2, that potentially bind this 12-bp region (supplemental Figure 9A).
BIM upregulation reflects increased EGR1 activity. (A) Forty-eight hours after transfection of Jurkat cells with shRNAs targeting RAPTOR or RICTOR, knockdown was assessed by immunoblotting. (B-C) Plasmids containing the 0.8-kb BIM promoter or truncation constructs in front of firefly luciferase cDNA (C) were transfected into (B) 5 × 106 log phase P388 cells. After treatment with diluent or 5 or 10 μM OSI-027, cells were assayed for firefly and Renilla luciferase activities. Panel is representative of 3 independent assays. (D-E) After Jurkat cells were treated with diluent or 10 μM OSI-027, RNAseq analysis was performed. Each mRNA count number was normalized to counts per million. Error bars in panel E, mean ± range of 2 independent assays. (F-G) After treatment with diluent, 10 μM OSI-027, 1 µM MLN0128, or 10 nM rapamycin, cells were harvested for (F) qRT-PCR and (G) immunoblotting. (H) Cells transfected with 40 µg empty vector or dnEGR1 construct along with 10 µg BIM promoter luciferase and 5 µg pTK-Renilla constructs were treated with diluent or 10 µM OSI-027 and assayed for luciferase activities. In panel H, *P = .011. (I-J) Cells were transfected with empty vector or dnEGR1 construct, treated with diluent or 10 µM OSI-027, and harvested for (I) qRT-PCR and (J) immunoblotting. Results of drug-treated samples were normalized to corresponding diluent-treated controls, which were set to 1. In panel I, *P = .02. (K) After treatment with diluent, 10 μM OSI-027, or 10 nM rapamycin, cells were harvested for ChIP assay. (L) DNA samples from ChIP assay subjected to qPCR. Results are expressed as relative % of input DNA precipitated. In panel L, *P ≤ .002 relative to dimethylsulfoxide or rapamycin. Diagram at right shows possible EGR1 binding sites in BIM promoter, with proximal site (−29 to −18) in larger font.
BIM upregulation reflects increased EGR1 activity. (A) Forty-eight hours after transfection of Jurkat cells with shRNAs targeting RAPTOR or RICTOR, knockdown was assessed by immunoblotting. (B-C) Plasmids containing the 0.8-kb BIM promoter or truncation constructs in front of firefly luciferase cDNA (C) were transfected into (B) 5 × 106 log phase P388 cells. After treatment with diluent or 5 or 10 μM OSI-027, cells were assayed for firefly and Renilla luciferase activities. Panel is representative of 3 independent assays. (D-E) After Jurkat cells were treated with diluent or 10 μM OSI-027, RNAseq analysis was performed. Each mRNA count number was normalized to counts per million. Error bars in panel E, mean ± range of 2 independent assays. (F-G) After treatment with diluent, 10 μM OSI-027, 1 µM MLN0128, or 10 nM rapamycin, cells were harvested for (F) qRT-PCR and (G) immunoblotting. (H) Cells transfected with 40 µg empty vector or dnEGR1 construct along with 10 µg BIM promoter luciferase and 5 µg pTK-Renilla constructs were treated with diluent or 10 µM OSI-027 and assayed for luciferase activities. In panel H, *P = .011. (I-J) Cells were transfected with empty vector or dnEGR1 construct, treated with diluent or 10 µM OSI-027, and harvested for (I) qRT-PCR and (J) immunoblotting. Results of drug-treated samples were normalized to corresponding diluent-treated controls, which were set to 1. In panel I, *P = .02. (K) After treatment with diluent, 10 μM OSI-027, or 10 nM rapamycin, cells were harvested for ChIP assay. (L) DNA samples from ChIP assay subjected to qPCR. Results are expressed as relative % of input DNA precipitated. In panel L, *P ≤ .002 relative to dimethylsulfoxide or rapamycin. Diagram at right shows possible EGR1 binding sites in BIM promoter, with proximal site (−29 to −18) in larger font.
In RNAseq experiments examining the impact of OSI-027 on the Jurkat cell transcriptome (supplemental Figure 9B), we detected not only 11- and 2-fold increases of PUMA and BIM mRNA (Figure 6D; supplemental Figure 9C), in good agreement with previous quantitative reverse transcriptase-PCR (qRT-PCR) data,21 but also a 9-fold increase in EGR1 mRNA and 4- to 7-fold increases in the EGR1 targets47-50 EGR2, EGR3, and EGR4 (Figure 6E). In contrast, changes in mRNAs encoding SP1 and other transcription factors predicted to bind to the same region of the BIM promoter region were minimal (Figure 6E). Dual inhibitor-induced EGR1 upregulation was confirmed in Jurkat cells by qRT-PCR (Figure 6F) and immunoblotting (Figure 6G). Moreover, RICTOR knockdown was found to induce EGR1 (Figure 6A), suggesting that mTORC2 inhibition is responsible for this effect.
Consistent with a critical role for EGR1 in BIM induction, dnEGR1 lacking the N-terminal transactivation domain51 diminished dual inhibitor-induced BIM promoter activation (Figure 6H) and BIM upregulation (Figure 6I-J). Similarly, mTORC1/mTORC2 dual inhibitors induced EGR1 and BIM in Molt4 T-cell ALL, and EGR1 knockdown diminished this BIM upregulation (supplemental Figure 9D-E).
Subsequent ChIP assays (Figure 6K-L) demonstrated that OSI-027 enhances binding of EGR1 to a region of the BIM promoter including base pairs −29 to −18, further confirming that EGR1 functions as a direct transcriptional activator for BIM during mTOR dual inhibitor treatment. Collectively, these observations support a model in which mTORC2 inhibition induces EGR1 upregulation followed by BIM transactivation.
Dual inhibitors activate NF--κB, which transactivates EGR1
To determine how EGR1 is upregulated, we examined the impact of dual inhibitors on the NF-κB pathway because (1) NF-κB is a known EGR1 transcription factor,52 (2) mTOR has been shown to regulate NF-κB,53-56 and (3) our RNAseq data demonstrated OSI-027–induced upregulation of NF-κB target genes (supplemental Figure 10A). Consistent with a role for NF-κB in dual mTOR inhibitor-induced EGR1 upregulation, we observed increased p65 in the nucleus (Figure 7A) and increased NF-κB transcriptional activity (supplemental Figure 10B) after dual inhibitor treatment. Importantly, overexpression of S32A/S36A IκB (IκB SS/AA) impaired dual inhibitor-induced NF-κB transcriptional activation (supplemental Figure 10B), EGR1 mRNA and protein upregulation (Figure 7B-C), BIM promoter activation (Figure 7D), and BIM mRNA and protein upregulation (Figure 7C,E). Likewise, pharmacologic inhibition of NF-κB activation with BAY 11-7082 diminished dual inhibitor-induced EGR1 upregulation (supplemental Figure 10C).
Effects of dual inhibitor-induced NF-κB activation on EGR1 expression and BIM upregulation. (A) Jurkat cells treated with diluent (lanes 1 and 2) or 10 (lanes 3 and 4) or 20 μM OSI-027 (lanes 5 and 6) were fractionated into nuclei (N) and cytoplasm (C) for immunoblotting. (B,C,E) Jurkat cells transfected with empty vector or the IκB SS/AA super-repressor were treated with diluent or 10 μM OSI-027 and harvested for (B,E) qRT-PCR or (C) immunoblotting. Results of drug-treated samples in B and E were normalized to corresponding diluent-treated controls, which were set to 1. In panel B, *P = .008 and **P = .04 relative to empty vector control. In panel E, *P = .02 and **P = .04, respectively. (D) Jurkat cells transfected with 40 µg empty vector or IκB SS/AA along with 10 µg 800-bp BIM promoter luciferase reporter and 5 µg pTK-Renilla were treated with diluent or 10 µM OSI-027 and assayed for luciferase activities. In panel D, *P < .02. (F-G) Aliquots of a (F) clinical ALL sample and (G) CML-biphenotypic blast crisis sample incubated for 48 hours with diluent (lane 1), 5 µM OSI-027 (lane 2), 10 µM OSI-027 (lane 3), 20 µM OSI-027 (lane 4), or 1 µM MLN0128 (lane 5) were harvested for immunoblotting. Patient (Pt) numbers refer to supplemental Table 1. (H) Proposed model of mTOR dual inhibitor-induced BH3-only protein upregulation. As discussed in the text, various lymphoid cell lines and ALL specimens show activation of 1 or both pathways after mTOR dual inhibitor treatment. Reagents shown in gray inhibit downstream steps.
Effects of dual inhibitor-induced NF-κB activation on EGR1 expression and BIM upregulation. (A) Jurkat cells treated with diluent (lanes 1 and 2) or 10 (lanes 3 and 4) or 20 μM OSI-027 (lanes 5 and 6) were fractionated into nuclei (N) and cytoplasm (C) for immunoblotting. (B,C,E) Jurkat cells transfected with empty vector or the IκB SS/AA super-repressor were treated with diluent or 10 μM OSI-027 and harvested for (B,E) qRT-PCR or (C) immunoblotting. Results of drug-treated samples in B and E were normalized to corresponding diluent-treated controls, which were set to 1. In panel B, *P = .008 and **P = .04 relative to empty vector control. In panel E, *P = .02 and **P = .04, respectively. (D) Jurkat cells transfected with 40 µg empty vector or IκB SS/AA along with 10 µg 800-bp BIM promoter luciferase reporter and 5 µg pTK-Renilla were treated with diluent or 10 µM OSI-027 and assayed for luciferase activities. In panel D, *P < .02. (F-G) Aliquots of a (F) clinical ALL sample and (G) CML-biphenotypic blast crisis sample incubated for 48 hours with diluent (lane 1), 5 µM OSI-027 (lane 2), 10 µM OSI-027 (lane 3), 20 µM OSI-027 (lane 4), or 1 µM MLN0128 (lane 5) were harvested for immunoblotting. Patient (Pt) numbers refer to supplemental Table 1. (H) Proposed model of mTOR dual inhibitor-induced BH3-only protein upregulation. As discussed in the text, various lymphoid cell lines and ALL specimens show activation of 1 or both pathways after mTOR dual inhibitor treatment. Reagents shown in gray inhibit downstream steps.
BIM upregulation in vivo
The engagement of the 4EBP1/MYC/PUMA and NF-κB/EGR1/BIM pathways was not limited to Jurkat cells. The 4EBP1/MYC/PUMA pathway was activated in SeAx Sézary syndrome cells (supplemental Figure 11A), which express PUMA but lack detectable BIM and EGR1, whereas the NF-κB/EGR1/BIM pathway was activated in Molt4 T-cell ALL cells (supplemental Figure 9D-E), which lack detectable PUMA. Moreover, both pathways were activated in Nalm6 B-cell ALL cells (supplemental Figures 8A and 11C) and in SuDHL-4 B-cell lymphoma cells in vitro (supplemental Figure 12A).
Curiously, after mTOR dual inhibitor administration to mice harboring Jurkat xenografts, increased BIM mRNA and protein were not detected (N.D.V. and P.A.S., unpublished observations, July 2014 and September 2015). Further investigation demonstrated that Jurkat cells contained an average of 15-fold less enhanced green fluorescent protein (EGFP)-BIM in vitro when expressing a transgene encoding this protein in the absence vs presence of caspase inhibitor (supplemental Figure 13A), suggesting high sensitivity of Jurkat cells to BIM-induced apoptosis. Moreover, mRNAs encoding BIM and other proteins decreased 4- to 32-fold when Jurkat cells underwent apoptosis (supplemental Figure 13B-C), further indicating that this line is not a good model for detecting BIM upregulation in the absence of caspase inhibition.
When the xenograft experiments were repeated using SuDHL-4 cells, which contain higher baseline BCL2, BCLXL, and MCL1 levels (supplemental Figure 12A, fourth lane), upregulation of EGR1 followed by BIM was readily detected in vivo (supplemental Figure 12B, red boxes), providing evidence for activation of the EGR1/BIM axis by mTOR dual inhibitors in vivo.
BIM and PUMA upregulation in human ALL samples
When fresh clinical ALL isolates were exposed to OSI-027 or MLN0128 ex vivo, we likewise saw activation of one or both apoptotic signaling cascades described above. In particular, some ALL samples displayed upregulation of EGR1 and BIM (Figure 7F; supplemental Figure 11D), and other samples displayed MYC downregulation accompanied by PUMA induction (supplemental Figure 11C) or activation of both pathways (Fig. 7G; supplemental Figure 11E), indicating that the pathways identified in ALL cell lines can also potentially be engaged in clinical ALLs.
Discussion
Previous studies have shown that mTORC1/mTORC2 dual inhibitors are particularly active against certain lymphoid neoplasms. Here we demonstrate the contribution of 2 separate killing mechanisms (Figure 7H): one involving mTORC1 inhibition leading to 4EBP1 dephosphorylation, MYC downregulation, and concomitant PUMA upregulation, and another involving mTORC2 inhibition leading to NF-κB activation, increased EGR1, and transcriptional activation of the BCL2L11 gene encoding BIM. These results have implications for ongoing efforts to develop these new antineoplastic agents for lymphoid malignancies.
Although rapalogs are active against a number of hematologic malignancies, responses are limited.7,9,10 The realization that mTORC2 often remains active during rapalog treatment has spurred development of small molecules targeting the mTOR kinase domain.6,9,16-18,20,21 These dual mTORC1/mTORC2 inhibitors were originally thought to be more effective because mTORC2 inhibition diminishes activation of the prosurvival kinase AKT through various mechanisms.14,57 In the present study, however, forced expression of 2 different constitutively active AKT constructs failed to rescue ALL cells from mTOR dual inhibitors (supplemental Figure 3). Moreover, downregulation of FOXO3A, an AKT-regulated transcription factor for BIM and PUMA, also failed to protect cells (supplemental Figure 4). Although AKT and its substrate FOXO3A mediate dual inhibitor effects in other cell types and might contribute to dual inhibitor-induced killing in lymphoid cells, we were unable to confirm involvement of this pathway in OSI-027– or MLN0128-induced killing of the cells studied here.
Our further studies demonstrated that downregulation of either RAPTOR or RICTOR is toxic to malignant lymphoid cells (Figure 2; supplemental Figure 2), suggesting the possibility of at least 2 distinct killing mechanisms. When we explored 4EBP1 dephosphorylation as a determinant of dual inhibitor killing, we observed that mTOR dual inhibitors facilitated binding of 4EBP1 to EIF4E, disrupting EIF4E/EIF4G interactions (Figure 3B-C) that are critical for translation of messages with 5′ terminal oligopyrimidine tracts. Some effects of dual inhibitors, notably disruption of EIF4E/EIF4G interactions, downregulation of MYC, upregulation of PUMA, and induction of apoptosis, were reproduced by nonphosphorylatable 4EBP1 mutants, the EIF4G antagonist 4EGI-1, or MYC knockdown (Figures 3 and 5). Conversely, these effects were diminished by expression of MYC constructs that are unaffected by EIF4E/EIF4G interactions or by PUMA downregulation (Figure 5F-G; supplemental Figure 7C). Collectively, these observations identify an mTORC1/4EBP1/MYC/PUMA pathway as a major determinant of dual mTOR inhibitor-induced killing in ALL cells (Figure 7H, left arm) and provide a mechanistic explanation for the recent observation that cells displaying incomplete inhibition of 4EBP1 phosphorylation or absence of 4EBP1 expression exhibit resistance to mTOR dual inhibitors.58,59 Although MYC is generally viewed as a transcription factor that mediates proliferation, its ability to induce cell death under unfavorable growth conditions has long been known60-62 ; the present studies provide an additional mechanism by which changes in MYC contribute to apoptosis under unfavorable growth conditions.
In accord with the model shown in Figure 7H (right arm), mTOR dual inhibitors also caused elevated expression of NF-κB target genes, including the transcription factor EGR1, which bound the BCL2L11 promoter to facilitate BIM upregulation and subsequent apoptosis. Some of the effects of the dual inhibitors, notably induction of BIM mRNA and protein, as well as apoptosis, were reproduced by RICTOR knockdown (Figures 2 and 6A). Conversely, proapoptotic signaling through this second pathway was inhibited by the IKK inhibitor BAY 11-7082, IκB SS/AA, dnEGR1, or EGR1 knockdown (Figure 6H-J; supplemental Figures 9E and 10). These observations identify an mTORC2/NF-κB/EGR1/BIM pathway as another major determinant of dual mTOR inhibitor-induced killing in ALL cells.
Collectively, the present studies not only identify unique pathways leading to mTOR-induced suppression of 2 different BH3-only proteins in lymphoid cells, but also raise several new questions. First, our studies indicate that PUMA upregulation is predominant in some lymphoid lines (eg, Jeko), whereas BIM and PUMA are almost equally important in other lines (eg, Jurkat).21 Second, the present results indicate that mTORC2 suppresses NF-κB/EGR1/BIM signaling in ALL cells, whereas previous studies have reported mTOR-induced NF-κB activation.53-56 Further experiments are needed to identify the factors that determine which pathway (4EBP1/MYC/PUMA or NF-κB/ERG1/BIM) predominates in various cells and to elucidate the contexts in which mTOR complexes activate versus inhibit NF-κB.
Studies in xenografts also demonstrated that each of the 2 pathways can be activated in vivo by therapeutically achievable mTOR dual inhibitor concentrations (supplemental Figures 8C and 12B). On the other hand, our studies also revealed that certain cell lines such as Jurkat are extremely sensitive to upregulation of BH3-only proteins. As a consequence, these cells are killed with relatively modest upregulation, leaving little trace of the lethal BH3-only protein upregulation at the mRNA or protein level in the absence of caspase inhibitors (supplemental Figure 13). This factor needs to be taken into account when mechanisms of cytotoxicity are evaluated.
Earlier studies also implicated diminished translation of the short-lived Bcl-2 family member MCL1 as mechanism of rapamycin-induced cytotoxicity in lymphoid cells.63,64 OSI-027 and MLN0128, which inhibit mTORC1 more effectively than rapamycin (Figure 3A), also downregulate MCL1 to a certain extent in some ALL cell lines (supplemental Figure 11A-B)21 and clinical samples (Figure 7F; supplemental Figure 11E), suggesting a third mechanism that might also promote ALL sensitivity to dual mTOR inhibitors. Although MCL1 downregulation does not always contribute to cytotoxicity of protein synthesis inhibitors,65 the loss of MCL1 might be particularly deleterious when dual mTOR inhibitors simultaneously upregulate BIM and PUMA, which are known to be neutralized by MCL1.66
Conversely, the present results also identify multiple biochemical changes (eg, variations in 4EBP1 levels or inducibility of EGR1, BIM, and PUMA) that could potentially affect sensitivity to this class of agent in lymphoid cells. Accordingly, these results not only support the notion that agents inhibiting both mTORC1 and mTORC2 may provide superior therapeutic benefit because of their ability to induce multiple distinct cell death pathways, but also identify factors that need to be examined further as preclinical and early clinical studies of mTOR dual inhibitors move forward in hematologic malignancies.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
Services of the Mayo Clinic Medical Genome Facility Gene Expression Core and kind gifts of reagents and cell lines from R. Bram, C. Dang, D. Toft, Y. Pommier, F. Sinicrope, G. Widley, P. Howe, W. Wu, and A. T. Look are gratefully acknowledged.
This work was supported in part by the National Institutes of Health (NIH), National Cancer Institute grants R01 CA166704 (S.H.K.), P30 CA06973 and UM1 CA186691 to support sample acquisition (J.E.K.), and F30 CA183507 (K.L.B.K.), NIH National Institute of General Medical Sciences grant T32 GM65841 (K.L.B.K.), a pilot grant from the Eagles Foundation (S.Y. and S.H.K.), and predoctoral fellowships from the Mayo Foundation for Education and Research (S.Y. and N.D.V.).
Authorship
Contribution: S.Y., N.D.V., K.L.B.K., M.E.F.-Z., and S.H.K. conceived and designed the study; S.H.K. and J.E.K. provided financial support; K.W.P., A.D.H., B.D.S., and J.E.K. provided study material or patients; S.Y., N.D.V., K.L.B.K., L.L.A., P.A.S., K.L.P., K.S.F., H.D., A.E.W.H., and S.H.K. collected and assembled the data; S.Y., M.E.F.-Z., and S.H.K. provided data analysis and interpretation; S.Y., N.D.V., K.L.B.K., and S.H.K. wrote the manuscript; and all authors provided final approval of manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Scott Kaufmann, Mayo Clinic, Gonda 19, 200 First St SW, Rochester, MN 55905; e-mail: kaufmann.scott@mayo.edu.
References
Author notes
S.Y. and N.D.V. contributed equally to this work.



This feature is available to Subscribers Only
Sign In or Create an Account Close Modal