Key Points
Runx1 is a key determinant of megakaryocyte cell-fate decisions in multipotent progenitors.
Runx1 downregulates cell-adhesion factors that promote residency of stem cells and megakaryocytes in their bone marrow niche.
Abstract
Disrupting mutations of the RUNX1 gene are found in 10% of patients with myelodysplasia (MDS) and 30% of patients with acute myeloid leukemia (AML). Previous studies have revealed an increase in hematopoietic stem cells (HSCs) and multipotent progenitor (MPP) cells in conditional Runx1-knockout (KO) mice, but the molecular mechanism is unresolved. We investigated the myeloid progenitor (MP) compartment in KO mice, arguing that disruptions at the HSC/MPP level may be amplified in downstream cells. We demonstrate that the MP compartment is increased by more than fivefold in Runx1 KO mice, with a prominent skewing toward megakaryocyte (Meg) progenitors. Runx1-deficient granulocyte-macrophage progenitors are characterized by increased cloning capacity, impaired development into mature cells, and HSC and Meg transcription signatures. An HSC/MPP subpopulation expressing Meg markers was also increased in Runx1-deficient mice. Rescue experiments coupled with transcriptome analysis and Runx1 DNA-binding assays demonstrated that granulocytic/monocytic (G/M) commitment is marked by Runx1 suppression of genes encoding adherence and motility proteins (Tek, Jam3, Plxnc1, Pcdh7, and Selp) that support HSC–Meg interactions with the BM niche. In vitro assays confirmed that enforced Tek expression in HSCs/MPPs increases Meg output. Interestingly, besides this key repressor function of Runx1 to control lineage decisions and cell numbers in progenitors, our study also revealed a critical activating function in erythroblast differentiation, in addition to its known importance in Meg and G/M maturation. Thus both repressor and activator functions of Runx1 at multiple hematopoietic stages and lineages likely contribute to the tumor suppressor activity in MDS and AML.
Introduction
The RUNX1 transcription factor (TF) is encoded by one of the most frequently mutated genes in myeloid malignancies, including de novo and secondary acute myeloid leukemia (AML),1-5 myelodysplasia (MDS),6-11 and chronic myelomonocytic leukemia.12,13 RUNX1 mutations lead to the expression of no protein, a crippled protein, or a dominant-negative fusion protein (in the case of the (8;21) translocation). Somatic alterations of the second RUNX1 allele, found at a high percentage in AML,1,5 suggest a classical tumor suppressor. Runx1 has been implicated in mechanisms controlling apoptosis, cell-cycle control, ribosome biogenesis, and decisions involving self-renewal and differentiation.14-18 Although the disruption of any one of these processes likely contributes to neoplastic transformation, its pivotal tumor-protective function is likely regulating myeloid differentiation.
Current models of hematopoietic differentiation posit that lineage decisions between multiple cell fates occur within a small number of phenotypically distinct cells within the bone marrow (BM), referred to as LSK (Lin−Sca1+Kit+) cells in mice. A small fraction of LSK cells are true long-term hematopoietic stem cells (HSCs) with high regenerative and proliferation capacity (ie, self-renewal), but the majority are multipotent progenitors (MPPs) that lack extended self-renewal capacity and demonstrate a continuum of restricted differentiation potential.19-22 Recent results suggest that megakaryocyte (Meg) potential can be closely linked to self-renewal capacity,23,24 and an MPP subset with pronounced Meg but no lymphoid potential has been identified.22,25,26 MPPs give rise to lineage-committed myeloid progenitors (MPs), which have lost self-renewal capacity and multipotency but whose numbers are expanded through proliferation.
The extra- and intracellular mechanisms that regulate lineage-restriction and self-renewal capacity during this differentiation process remain poorly understood. Lineage-specific cis-regulatory elements that are gained or lost during commitment have been identified and TF complexes that bind these elements delineated.27,28 Runx1 colocalizes with TFs specific for the HSC/MPP compartment29 but is also implicated in TF complexes associated with Meg and erythroid (Ery) progenitors,30,31 granulocyte (G) and/or monocyte (M) progenitors,32 and cells of the lymphoid compartment.33,34 To decipher the function and importance of Runx1 within these TF complexes, the consequence of its inactivation has been studied in mouse models. An important finding is the observed three- to eightfold expansion of the LSK population in the absence of Runx1,33,35-37 suggesting a key function in self-renewal and differentiation decisions. The MP compartment is also expanded, whereas lymphoid progenitors are nearly absent in Runx1-deficient BM. This latter trait can be traced to crucial functions in initiating the B-cell program33 and during early stages of T-cell development,34 but a pivotal function of Runx1 in myeloid-cell fate has not been discerned. Indeed the sole, characterized myeloid defect in Runx1-deficient mice is impaired Meg maturation.35,38
We reasoned that defects in cell fate decision in the early HSC/MPP compartment in Runx1-deficient mice should read out in the downstream MP compartment, which has only been superficially examined in previous studies. This compartment may also be the target of leukemic transformation, as the transcriptome of the leukemic stem cell (LSC) can share high homology to granulocyte-macrophage progenitors (GMPs).39
Methods
Mouse strains used for this study and the protocols used for fluorescence-activated cell sorting (FACS), gene expression analysis, and chromatin immunoprecipitation have been reported previously33 and are detailed in the supplemental Methods and supplemental Tables 1 and 2 (available on the Blood Web site).
Transplantation experiments
For rescue experiments, BM cells were isolated from Runx1fl/fl-Tg(vav-Cre) mice on day 3 postinjection of 150 mg/kg 5-fluorouracil (Medac), infected twice with ecotropic pseudotypes of MIG retroviral vectors expressing GFP with RUNX1-ERt2 or ERt2, and transplanted into irradiated recipient mice.40 After 6 weeks, RUNX1-ERt2 was induced in half of the mice by i.p. tamoxifen injection (49.5 µg/g body weight; Sigma T5648) on 3 consecutive days. BM cells were harvested 24 hours later, and viable GFP+ GMP cells were sorted.
In vitro functional assays
Colony-forming units (CFUs) were determined by plating single-cell suspensions of sorted cells in methylcellulose (MethCult GF M3434; StemCell Technologies) and scoring 7 days after plating. Cell-adhesion assays41 were performed on progenitor-enriched populations seeded onto MS-5 stroma cells. To evaluated candidate genes, complementary DNAs were generated by reverse-transcription polymerase chain reaction and incorporated into MYs vectors42 coexpressing BFP, which were used to transduce sorted LSK cells. Subsequently, cells were cultured on OP9 cells43 in StemPro-34 Media (Thermo Fisher Scientific) supplemented with stem cell factor (50 ng/mL), interleukin-3 (10 ng/mL), and thrombopoietin (Thpo) (50 ng/mL) for 6 days.
Results and discussion
Phenotypically abnormal MP compartment in Runx1-deficient mice
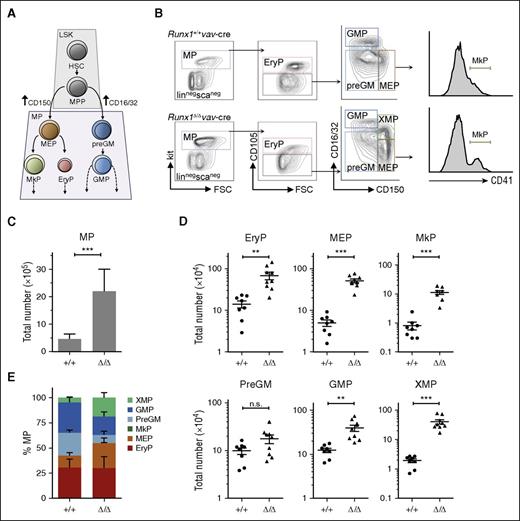
In a first step to characterize the MP compartment of Runx1-deficient BM, we performed FACS analysis that permits separation of G/M and Meg/Ery bipotent progenitors (GMP and MEP), as well as Meg or Ery monopotent progenitors (MkP and EryP)44 (Figure 1A-B). A fivefold increase in absolute MP numbers was observed in Runx1Δ/Δ as compared with Runx1+/+ mice (Figure 1C). This number is higher than that observed in interferon-inducible Runx1-deficient models,38,45 in which excision was induced in newborns as opposed to fetal liver and may have been incomplete. A striking difference in the distribution of the various BM progenitors in knockout (KO) and wild-type (WT) mice was also found (Figure 1D-E). Although the relative proportion of EryP was similar in mice of both genotypes (with an overall fivefold increase in KOs), relatively higher proportions of MEP and MkP were observed in the KOs (with a 10-fold increase for each population) and a two- to threefold increase in actual numbers in G/M progenitors. Several cell-surface markers showed altered expression levels on Runx1-deficient progenitors; whereas the CD105/Endoglin marker on EryP was diminished, the CD150/Slamf1 marker on MEP was increased (Figure 1B), as previously reported for HSCs/MPPs.36
Runx1 deletion by Tg(vav-Cre) expression profoundly impacts the size and distribution of the MP compartment. (A) Schematic of the bi- and monopotent MPs that differentiate from the HSC/MPP compartment and can be distinguished by antigen markers.44 (B) Representative FACS analysis to detect G/M and Meg/Ery progenitors performed on BM cells isolated from mice with indicated genotype. The genotype of the isolated fractions was confirmed by polymerase chain reaction (supplemental Figure 1A). (C) Bar graph showing the average number of MPs per mouse (2 femora, 2 tibia); n = 9 per cohort. Error bars, ± standard deviation (SD). (D) Total number of MP subpopulation per mouse is depicted by a dot, n ≥ 8 per cohort; horizontal line, median; error bars, ± standard error (SE). Total absolute BM cell counts were similar (1.03 × 108 ± 0.20 × 108 vs 1.06 × 108 ± 0.20 × 108 cells/mouse for Runx1Δ/Δ versus Runx1+/+ (n = 13 per genotype). (E) Relative distribution of progenitor types for each genotype based on a minimum of 8 mice per cohort. P values for panels D and E were calculated by a unpaired Student t test; n.s., not significant; **P < .05; ***P < .01. FSC, forward scatter.
Runx1 deletion by Tg(vav-Cre) expression profoundly impacts the size and distribution of the MP compartment. (A) Schematic of the bi- and monopotent MPs that differentiate from the HSC/MPP compartment and can be distinguished by antigen markers.44 (B) Representative FACS analysis to detect G/M and Meg/Ery progenitors performed on BM cells isolated from mice with indicated genotype. The genotype of the isolated fractions was confirmed by polymerase chain reaction (supplemental Figure 1A). (C) Bar graph showing the average number of MPs per mouse (2 femora, 2 tibia); n = 9 per cohort. Error bars, ± standard deviation (SD). (D) Total number of MP subpopulation per mouse is depicted by a dot, n ≥ 8 per cohort; horizontal line, median; error bars, ± standard error (SE). Total absolute BM cell counts were similar (1.03 × 108 ± 0.20 × 108 vs 1.06 × 108 ± 0.20 × 108 cells/mouse for Runx1Δ/Δ versus Runx1+/+ (n = 13 per genotype). (E) Relative distribution of progenitor types for each genotype based on a minimum of 8 mice per cohort. P values for panels D and E were calculated by a unpaired Student t test; n.s., not significant; **P < .05; ***P < .01. FSC, forward scatter.
Importantly, Runx1-deficient BM also contained an undefined myeloid progenitor (XMP) that was positive for both the MEP marker CD150 and the myeloid markers CD16/32 (FcγRII/III) (Figure 1B,D). Previous reports have used the CD34 myeloid marker to distinguish between MEP and GMP and define a common myeloid progenitor (CMP),38 which is now known to be composed of G/M and Meg/Ery bi- and monopotent progenitors.44 Using this older FACS strategy, the XMP population appears as a CD34lo “CMP” (supplemental Figure 1B). However, staining with the MkP CD41 marker showed high positivity on XMP (supplemental Figure 1C), supporting the hypothesis that XMP may be an aberrant Meg progenitor. Of note, high CD41 expression on an undefined MP cell has also been previously reported.35
Functional assays of MP progenitors reveal differentiation defects in G/M and Ery lineages
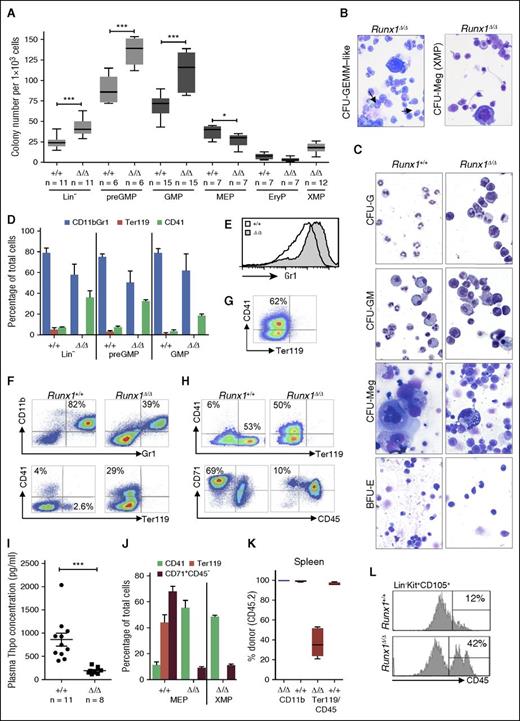
To further assess the impact of Runx1 deficiency on the MP compartment, cloning assays were performed on various progenitors (Figure 2A; supplemental Figure 2). Four conclusions can be surmised from this analysis. Firstly, and in accordance with previous results,35,38 in the absence of Runx1, an absolute increase in the clonogenic potential of the MP was observed. However, our analysis demonstrated that this was specific for G/M-committed progenitors, which gave rise to ∼50% more colonies than controls, in contrast to MEPs or EryPs with similar or slightly reduced colony numbers (Figure 2A). G/M colonies were also larger, as reflected in their morphology and cell counts (supplemental Figure 2B-C). These results are consistent with the presence of more primitive cells exhibiting an increased regenerative capacity, perhaps reflecting a skewed differentiation program that favors self-renewal to differentiation/maturation.
Runx1-deficient progenitors are impaired in their in vitro differentiation potential. (A) Two-dimensional box plots showing mean colony numbers obtained per 103 sorted progenitors. Each progenitor type was evaluated in at least 2 biological replicates (independent sorts) performed in duplicate. Horizontal line, median; box, range; whiskers, ±SD. (B-C) Cytospins of dispersed cells from the indicated colonies and genotypes. Giemsa staining; original magnification ×20. Arrow in panel B indicates immature Ery progenitors and granulocytic forms. (D) Bar graph representing FACS data for expression of lineage markers on dispersed cells derived from colony assays of indicated progenitors and genotypes. Horizontal line, median; error bars, ±SD. (E-H) FACS analysis of dispersed cells from (E) GMP, (F) pre-GM, (G) XMP, and (H) MEP methylcellulose colonies confirming impaired myeloid differentiation and Meg-skewing of Runx1Δ/Δ progenitors. (I) Thpo plasma level determined per mouse is depicted by a dot. Horizontal line, median; error bars, ±SE. (J) Bar graph representing FACS data for expression of lineage markers on dispersed cells derived from colony assays of indicated progenitors and genotypes. Horizontal line, median; error bars, ±SD. (K) Donor contribution in spleen in competitive transplantation experiment shows no impact of Runx1-deletion on myeloid contribution but a significant reduction in the contribution to Ery CD45+Ter119+ progenitors. (L) Representative FACS analysis for CD45 expression on cells within the Ery progenitor population (CD105+) in BM isolated from mice of the indicated genotype (n = 4 per cohort). P values for panels A, D, I, J, and K were calculated by an unpaired Student t test; **P < .05; ***P < .01.
Runx1-deficient progenitors are impaired in their in vitro differentiation potential. (A) Two-dimensional box plots showing mean colony numbers obtained per 103 sorted progenitors. Each progenitor type was evaluated in at least 2 biological replicates (independent sorts) performed in duplicate. Horizontal line, median; box, range; whiskers, ±SD. (B-C) Cytospins of dispersed cells from the indicated colonies and genotypes. Giemsa staining; original magnification ×20. Arrow in panel B indicates immature Ery progenitors and granulocytic forms. (D) Bar graph representing FACS data for expression of lineage markers on dispersed cells derived from colony assays of indicated progenitors and genotypes. Horizontal line, median; error bars, ±SD. (E-H) FACS analysis of dispersed cells from (E) GMP, (F) pre-GM, (G) XMP, and (H) MEP methylcellulose colonies confirming impaired myeloid differentiation and Meg-skewing of Runx1Δ/Δ progenitors. (I) Thpo plasma level determined per mouse is depicted by a dot. Horizontal line, median; error bars, ±SE. (J) Bar graph representing FACS data for expression of lineage markers on dispersed cells derived from colony assays of indicated progenitors and genotypes. Horizontal line, median; error bars, ±SD. (K) Donor contribution in spleen in competitive transplantation experiment shows no impact of Runx1-deletion on myeloid contribution but a significant reduction in the contribution to Ery CD45+Ter119+ progenitors. (L) Representative FACS analysis for CD45 expression on cells within the Ery progenitor population (CD105+) in BM isolated from mice of the indicated genotype (n = 4 per cohort). P values for panels A, D, I, J, and K were calculated by an unpaired Student t test; **P < .05; ***P < .01.
The second novel observation was the impact of Runx1-deficiency on G and M maturation. Atypical clone morphology made classification difficult (supplemental Figure 2B), and could be attributed to the presence of immature G/M cells (Figure 2B-C). This was confirmed by FACS analysis, which showed an increase in cells with reduced Gr1 expression levels on CD11b+ cells, an indicator of immature granulocytes46 (Figure 2E-F). A fivefold increase in the relative levels of immature (Gr1med) to mature (Gr1hi) cells was also observed in the BM of Runx1-deficient mice (supplemental Figure 3A-B). Low Gr1-antigen levels on Runx1-deficient cells in vitro was also reported in a previous study, but this was attributed to a switch from granulopoiesis to monopoiesis.47 Using specific antibodies for markers highly expressed on monocytes (Ly6c) and granulocytes (Ly6g), rather than the Gr1 antibody that recognizes both antigens,48 our analysis demonstrated that the M to G ratio in BM and peripheral blood was unchanged in Runx1-deficient mice, but the expression levels of both Ly6c and Ly6g on granulocytic cells varied (supplemental Figure 3C-E). Furthermore, blood values for monocytes and neutrophils were comparable to controls (supplemental Figure 3F). Consistent with the observed immature morphology in colony assays, we conclude that in the absence of Runx1, granulocytic and monocytic maturation is impaired.
Thirdly, our analysis confirmed an increased propensity toward Meg differentiation in Runx1-deficient progenitors. This was most striking in colony assays from early pre-GMP, but also observed using total progenitors (Lin−). Low levels of Ery and Meg differentiation have been reported for pre-GMP,44 but we observed a large proportion of granulocyte-erythrocyte-monocyte-megakaryocyte (GEMM)–like colonies that contained high levels of Meg precursors, in addition to G/M progenitors (Figure 2B; supplemental Figure 2A-B). Using FACS analysis as an unbiased tool to quantitate the lineage affiliation of progeny cells, a fivefold increase in CD41+ cells was confirmed (Figure 2D,F). We also confirmed that the unique XMP were Meg progenitors, as they gave rise to predominantly aberrant CFU-Meg colonies containing CD41+ cells (Figure 2B,G). Runx1 deficiency is known to impede Meg maturation and production of platelets,35,38,49 which normally bind Thpo to control their own production.50 Thus aberrantly high Thpo levels may lead to the increase in Meg progenitors in Runx1Δ/Δ mice. However, Thpo plasma levels were significantly reduced in Runx1-deficient mice (Figure 2I), likely due to the ability of Meg precursors to also “sink” Thpo.50
Finally, our results unveiled a striking defect in erythropoiesis. This was most evident in MEP-derived colonies, in which Runx1+/+ cultures contained colonies typical of burst-forming units erythroid (BFU-E) and CFU-Meg/Ery; in contrast, only few and very small BFU-E colonies were found in Runx1Δ/Δ cultures, with the majority of colonies having a CFU-Meg morphology (supplemental Figure 2A-B). No Ter119+ erythrocytes were observed in any Runx1Δ/Δ cultures (Figure 2H,J). We could exclude the possibility that aberrant downregulation of the gene encoding the Ter119 antigen prevented detection of Ery progenitors, as a profound absence of CD71+CD45− cells (proerythroblasts) was also evident in KO compared with WT cultures (Figure 2H,J). Consistently, although we observed increased numbers of CD105+ EryPs, which are reported to give rise exclusively to CFU-E,44 Runx1Δ/Δ EryPs gave rise to a few colonies that did not contain Ter119+ cells and thus are likely contaminating CFU-Meg or CFU-GM. Because the CD105+ EryP population is primarily composed of proerythroblasts,44 we investigated the possibility that the low expression of CD105+ on the Runx1Δ/Δ EryP population may actually reflect overall differentiation impairment. Strikingly, 49% ± 8.9% (n = 4) cells within the CD105+ population still expressed CD45, suggesting differentiation impairment at the erythroblast stage (Figure 2L).
The impact on Ery differentiation was unexpected, as Runx1-deficient mice exhibit normal red blood cell parameters (supplemental Figure 3F) and previous studies have reported normal numbers of BFU-Es within Runx1-deficient BM progenitors based on clone morphology.35,38 We postulate that this latter discrepancy may reflect either mistaken scoring of the morphologically similar CFU-GEMM–like colonies as BFU-E (supplemental Figure 2B) or incomplete excision of the Runx1 gene. As compensation mechanisms may obscure ineffective erythropoiesis in vivo, we performed competitive transplantation experiments. Analysis of BM and blood showed the expected loss of donor-derived B cells (supplemental Figure 3G). Notably, despite the observed maturation defect in vitro, CD11b+ G/M cells were almost exclusively derived from Runx1Δ/Δ-donor cells in recipient mice, likely reflecting the proliferation advantage of their progenitors (supplemental Figure 3G). In contrast, a small but significant reduction in the contribution of donor-derived Ery CD45+Ter119+ progenitors was observed. This effect was most pronounced in the spleens, in which only 40% of the early Ery progenitors were derived from Runx1Δ/Δ donor cells in recipient mice (Figure 2K). These results confirm that Runx1-deficiency impacts the early stages of erythropoiesis, although this effect is greatly mitigated in vivo. This conclusion is also supported by the observation that 20% of erythrocytes in Runx1-deficient mice show Howell-Jolly bodies,51 a characteristic of dysplastic erythropoiesis observed in MDS. Furthermore, defects in primitive erythropoiesis have been observed in Runx1-deficient embryos, as well as in cultures from Runx1-mutated induced pluripotent stem cells.52,53
Transcriptome analyses of MP populations confirm a deregulated lineage-commitment program skewed toward Meg lineage
To further assess the MP components of Runx1Δ/Δ mice, microarray and RNA-sequencing (RNA-seq) analyses were performed on sorted progenitors (Figure 3A). Principal-component and sample-to-sample distance analyses of expressed genes confirmed that both MEP and GMP populations from Runx1Δ/Δ mice expressed transcriptomes that were distinct from but related to their normal counterparts (Figure 3B; supplemental Figure 4A). A comparison of the deregulated genes in Runx1Δ/Δ vs Runx1+/+ MEP or GMP populations showed a high number of mutually downregulated or upregulated genes, suggesting that common transcriptome pathways are deregulated in both populations (Figure 3C).
Trancriptome analysis of XMP and MEP populations in Runx1-deficient mice. (A) Sorting strategy for analysis of bipotent MPs in Runx1+/+ and Runx1Δ/Δ mice. Shown are cells that have been gated for Lin−Sca1−Kit+CD41−CD105−. (B) Principle-component analysis was performed using DESeq282 and demonstrates the relatedness of the transcriptomes of the indicated populations. (C) Venn diagram of stringently filtered (at least threefold deregulated, false discovery rate [FDR] P ≤ .05, base mean > 100) genes whose expression is either up or down in Δ/Δ vs +/+ GMPs or MEPs. Bold numbers refer to the number of shared genes that are deregulated in GMPs and MEPs. (D) Venn diagram of stringently filtered (at least threefold deregulated, P ≤ .05) genes whose expression is either up or down in Δ/Δ XMPs vs +/+ MEPs or Δ/Δ MEPs. Bold numbers refer to the number of shared genes that are deregulated in XMP in both comparisons. (E) Bar graph showing the relative gene expression (log2) of the shared genes that are either downregulated (left) or upregulated (right) in XMPs as compared with MEPs of either genotype. Each pair of bars corresponds to the gene either to the far left (downregulated) or far right (upregulated). Heat maps show the relative expression levels of these genes in monopotent Ery or MkP progenitors or bipotent GMPS-based on normalized values in Gene Expression Commons (GEXC).83 Genes not annotated in GEXC or showing spurious expression levels are not shown. (F) Relative expression levels determined from reads per kilobase of transcripts per million mapped reads (RPKM) values of genes encoding key Ery or Meg TFs are shown. Expression levels in WT MEP (+/+) were set as 1. Shown are the mean values of 2 independent experiments. Error bars, ±SD. Original P values for the comparison relative to +/+ MEP values are indicated as *P < .05, **P < .0, and ***P < .001, the latter of which have FDR P values ≤ .05. A list of all TF genes used for this analysis and gene expression levels is found in supplemental Table 2.
Trancriptome analysis of XMP and MEP populations in Runx1-deficient mice. (A) Sorting strategy for analysis of bipotent MPs in Runx1+/+ and Runx1Δ/Δ mice. Shown are cells that have been gated for Lin−Sca1−Kit+CD41−CD105−. (B) Principle-component analysis was performed using DESeq282 and demonstrates the relatedness of the transcriptomes of the indicated populations. (C) Venn diagram of stringently filtered (at least threefold deregulated, false discovery rate [FDR] P ≤ .05, base mean > 100) genes whose expression is either up or down in Δ/Δ vs +/+ GMPs or MEPs. Bold numbers refer to the number of shared genes that are deregulated in GMPs and MEPs. (D) Venn diagram of stringently filtered (at least threefold deregulated, P ≤ .05) genes whose expression is either up or down in Δ/Δ XMPs vs +/+ MEPs or Δ/Δ MEPs. Bold numbers refer to the number of shared genes that are deregulated in XMP in both comparisons. (E) Bar graph showing the relative gene expression (log2) of the shared genes that are either downregulated (left) or upregulated (right) in XMPs as compared with MEPs of either genotype. Each pair of bars corresponds to the gene either to the far left (downregulated) or far right (upregulated). Heat maps show the relative expression levels of these genes in monopotent Ery or MkP progenitors or bipotent GMPS-based on normalized values in Gene Expression Commons (GEXC).83 Genes not annotated in GEXC or showing spurious expression levels are not shown. (F) Relative expression levels determined from reads per kilobase of transcripts per million mapped reads (RPKM) values of genes encoding key Ery or Meg TFs are shown. Expression levels in WT MEP (+/+) were set as 1. Shown are the mean values of 2 independent experiments. Error bars, ±SD. Original P values for the comparison relative to +/+ MEP values are indicated as *P < .05, **P < .0, and ***P < .001, the latter of which have FDR P values ≤ .05. A list of all TF genes used for this analysis and gene expression levels is found in supplemental Table 2.
The XMP transcriptome was most closely related to that of Runx1-deficient MEP cells, consistent with its strong Meg cloning potential. Comparison of the gene expression patterns in XMP as compared with both KO MEP and WT MEP revealed that genes downregulated in XMP as compared with MEP are normally highly expressed in Ery progenitors; conversely, the upregulated genes are normally expressed in either MkP or GMPs (Figure 3D-E). We next assessed the expression levels of 1745 genes encoding TF54 and found 8 genes significantly differentially expressed in XMPs, including high levels of Fli1 and low levels of Klf1 transcripts (Figure 3F; supplemental Table 2). These TF are reciprocal determinants of Meg versus Ery differentiation,30,55 supporting the hypothesis that XMP is committed to the Meg lineage. Furthermore, several transcripts for TF expressed in the HSC/MPP compartment and associated with Meg, but not Ery, differentiation (eg, Erg, Meis1, and Pbx1) were significantly upregulated in XMP as compared with MEP populations (Figure 3F). No obvious upregulation of G/M-associated TFs was observed in the XMP population, except for significantly higher levels of Cebpa transcripts in XMPs as compared with those in Runx1Δ/Δ MEPs (Figure 3F). We cannot exclude that low C/ebpα levels are sufficient to activate G/M marker genes in synergy with abnormal expression of other deregulated TFs in the XMP population.
We next examined the MEP expression data to determine if the impaired erythropoiesis could be attributed to changes at this level. Recent studies have defined separable MEP populations with either Ery or Meg-primed differentiation capacity.56,57 To exclude that Runx1 deficiency leads to the preferential generation of Meg-skewed MEP, gene expression of Runx1Δ/Δ MEPs and pro–Meg-MEPs57 were compared. Notably, over 50% of the genes deregulated in Runx1Δ/Δ vs Runx1+/+ MEPs were actually reciprocally regulated in pro–Meg-MEPs (supplemental Figure 4D). No differential expression was observed in TFs that specify Meg or Ery differentiation (eg, Gfib1, Fli1, Klf1, or Gata1); on the contrary, Runx1Δ/Δ MEPs showed significantly increased levels of Klf3, Lyl1, and Arrb1 (implicated in Ery maturation)58,59 and decreased levels of Gata2 (a known Meg factor)55 (Figure 3F; supplemental Figure 4B-C). Thus, we find it likely that the defective erythropoiesis in Runx1Δ/Δ mice is not a reflection of the skewed propensity toward Meg differentiation at the MEP level but rather a requirement for Runx1 in Ery development.
Deregulated Meg-primed lineage program identified in GMPs and confirmed in LSK
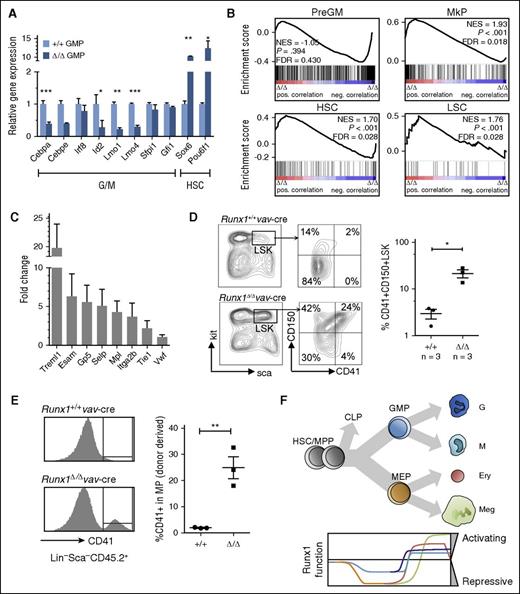
To elucidate the mechanism underlying the increased GMP clonality but impaired G/M differentiation, differentially expressed TF genes in KO vs WT GMPs were determined. The genes encoding C/ebpα and Id2, TFs involved in G/M maturation,60,61 were reduced by a factor of >2 (Figure 4A; supplemental Figure 4E-F). Lmo1 and Lmo4 were also significantly downregulated in KO GMP, and although not previously implicated in G/M development, both genes are highly expressed in mature Gs (supplemental Figure 4E). These results are consistent with the observed immature phenotype in GMP colony assays and the importance of Runx1 in G/M differentiation,18 a conclusion not reported in previous analyses of Runx1-deficient mouse models.
Runx1 is necessary to downregulate key HSC and Meg genes during transition to GMP differentiation state. (A) Relative expression levels of TF genes in Runx1Δ/Δ versus Runx1+/+ GMPs. Expression levels (RPKM) in Runx1+/+ GMPs were set as 1. Shown is the mean value of two independent experiments. Error bars, ±SD. Original P values for the comparison are indicated as *P < .05, **P < .01, ***P < .001, the latter of which have FDR P values ≤ .05. (B) GSEA plots showing top gene sets downregulated in Δ/ΔGMP compared with +/+GMP in four independently conducted microarray analyses. Signature sets for pre-GMP, MkP, HSC, and LSC were taken from published data.24,84 (C) Bar graph depicting the fold-increase in expression levels for selected genes. Shown is the mean of four independent microarray analyses. Error bars, ±SD. (D) FACS analysis of LSK population, containing all HSC/MPP subtypes for expression of CD41+ cells. Bar graph depicts the results of 3 independent experiments. Horizontal line, median; error bars, ±SE. (E) Runx1+/+ or Runx1Δ/Δ whole BM cells were transplanted into lethally irradiated recipients. CD41 expression in donor-derived progenitors was analyzed by FACS analysis 14 weeks after transplantation (left). Bar graph depicts the percentage of CD41-expressing cells in Lin−Sca1−CD45.2+ compartment. Each dot represents one recipient mouse. Horizontal line, median; error bars, ±SE. P values were calculated by a unpaired Student t test; **P < .01. (F) Schematic representation of the activating / repressing function of Runx1 in myeloid cell lineages during differentiation. CLP, common lymphoid progenitor.
Runx1 is necessary to downregulate key HSC and Meg genes during transition to GMP differentiation state. (A) Relative expression levels of TF genes in Runx1Δ/Δ versus Runx1+/+ GMPs. Expression levels (RPKM) in Runx1+/+ GMPs were set as 1. Shown is the mean value of two independent experiments. Error bars, ±SD. Original P values for the comparison are indicated as *P < .05, **P < .01, ***P < .001, the latter of which have FDR P values ≤ .05. (B) GSEA plots showing top gene sets downregulated in Δ/ΔGMP compared with +/+GMP in four independently conducted microarray analyses. Signature sets for pre-GMP, MkP, HSC, and LSC were taken from published data.24,84 (C) Bar graph depicting the fold-increase in expression levels for selected genes. Shown is the mean of four independent microarray analyses. Error bars, ±SD. (D) FACS analysis of LSK population, containing all HSC/MPP subtypes for expression of CD41+ cells. Bar graph depicts the results of 3 independent experiments. Horizontal line, median; error bars, ±SE. (E) Runx1+/+ or Runx1Δ/Δ whole BM cells were transplanted into lethally irradiated recipients. CD41 expression in donor-derived progenitors was analyzed by FACS analysis 14 weeks after transplantation (left). Bar graph depicts the percentage of CD41-expressing cells in Lin−Sca1−CD45.2+ compartment. Each dot represents one recipient mouse. Horizontal line, median; error bars, ±SE. P values were calculated by a unpaired Student t test; **P < .01. (F) Schematic representation of the activating / repressing function of Runx1 in myeloid cell lineages during differentiation. CLP, common lymphoid progenitor.
Notably, upregulation of several TF genes specifically expressed in the HSC/MPP compartment or shared by HSC and the Meg lineages (eg, Hmga2, Pouf6f1, and Sox6) was also observed in Runx1-deficient GMPs (Figure 4A; supplemental Figure 4E-F). Gene set enrichment analysis (GSEA) revealed a striking bias for HSC, leukemic stem cells (LSCs) and pre-MkP transcripts and the loss of a GMP signature (Figure 4B). These results are reminiscent of results obtained from a subset of HSCs with biased platelet production and high self-renewal activity24,25 and include Mpl, Selp, and Itga2b (encoding CD41) (Figure 4C). Expression of the latter gene has been used to select for the platelet-biased population within HSCs/MPPs.23,25
The high levels of Meg progenitors and the aberrant Meg signature in the GMP transcriptome might reflect cell-fate decisions occurring at the level of HSC/MPP. It is well-established that Runx1 deficiency leads to an increase in absolute HSC/MPP cell numbers, but aberrant expression of cell-surface markers in the absence of Runx1 make it difficult to phenotypically classify long-term HSCs or specific MPPs.33,36,45 The expression of CD41, however, has not been previously examined. Gating on the LSK population, we could confirm high levels of CD150 but also the coincident expression of CD41 (Figure 4D), a characteristic of Meg-skewed HSC/MPPs.22,23,25 Close to 20% of the LSK population showed this marker expression, compared with <3% in controls. Furthermore, 6-day liquid cultures of enriched Runx1Δ/Δ HSC/MPP yielded a 31% ± 2.9% increase in CD41+ cells as compared with controls. To confirm that the increased Meg potential arises from a long-term HSC population, we analyzed levels of MkP cells in mice >100 days after transplantation. A striking 10-fold increase in donor-derived MkP (CD41+ MPs) was observed in mice receiving Runx1Δ/Δ vs Runx1+/+ cells (Figure 4E).
Taken together, our results support the model presented in Figure 4F, depicting a critical repressor role of Runx1 in inhibiting both self-renewal programs and Meg cell-fate decision in the early stage of HSC/MPP development. In addition to the well-established activating role of Runx1 in Meg maturation, our study has confirmed its critical function at late stages of G and M differentiation, consistent with early transcriptional studies,32 and has revealed a unique activating function of Runx1 during the early stages of Ery differentiation.
Rescue experiment identifies direct Runx1-target genes whose products regulate adhesion and motility
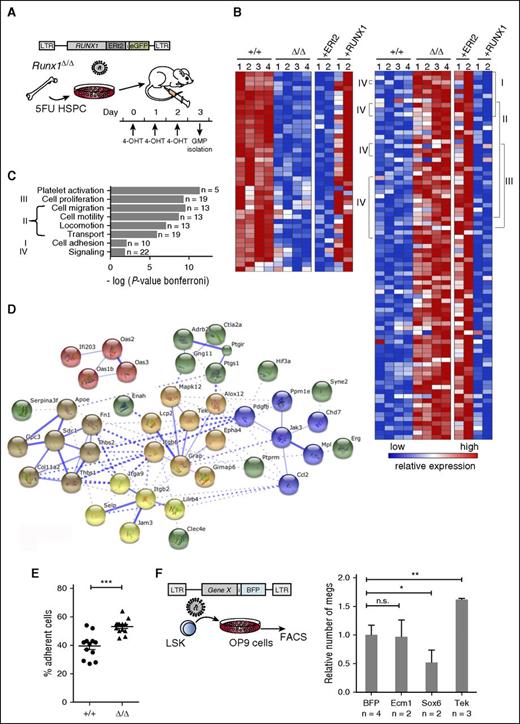
To define direct target genes of Runx1 that conferred increased regenerative capacity and Meg cell-fate decisions to G/M progenitors, we performed rescue experiments (Figure 5A). For these experiments, we used a RUNX1-ERt2 inducible system, whose ability to reverse the block to G/M development was confirmed in colony assays (supplemental Figure 5A-D). A total of 238 deregulated genes were identified whose expression pattern could be rescued (reversed) by RUNX1 reactivation (Figure 5B; supplemental Table 3). Genes upregulated in Runx1Δ/Δ GMP and repressed upon activation of RUNX1 were enriched for several Gene Ontology (GO) terms (Figure 5B-C), including regulation of localization and adhesion. Consistently, STRING analysis detected over 92 interactions (expected 2.8; p>.005) and highlighted several interactive gene clusters that connect signaling proteins (eg, Mpl, Pdfgb, Jak3, and Tek) with adhesion proteins (integrins, selectins, fibronectins, cadherins, proteoglycans, and junction adhesion molecules), which are known to support interactions between HSCs/MPPs within the BM niche and the extracellular matrix62-64 (Figure 5D). Similar enriched signatures (cell adhesion, focal adhesion, and signaling pathways) were also found by GSEA (supplemental Figure 6A). We also reassessed the deregulated genes in AML patient samples with RUNX1 inactivating mutations.65 Strikingly, GO analysis of 235 deregulated genes showed the same affected pathways found in Runx1-deficient GMPs (supplemental Figure 6B). Shared deregulated genes included ITGB2, CD302, PLXNC1, JAM3, and PYHIN1. Notably, in vitro experiments confirmed increased binding of Runx1-deficient MPs to MS-5 stromal cells (Figure 5E).
Rescue experiment identifies Runx1-target genes that mediate cell adhesion. (A) Experimental design of Runx1 rescue experiments used to define direct target genes. 4-OHT, 4-hydroxytamoxifen. (B) Heat maps depicting microarrays expression results from untreated GMP from Runx1Δ/Δ and Runx1+/+ mice, as well as rescue experiments analyzing transduced GMP. Left panel shows the relative expression levels for 42 genes downregulated in Runx1Δ/Δ GMPs and upregulated after tamoxifen treatment of RUNX1-transduced, but not control-transduced (ERt2), GMPs. Right panel shows 94 genes that are upregulated in Runx1Δ/Δ GMPs and downregulated after activation of RUNX1. Genes deregulated by a factor >2 (P ≤ .05) and that could be rescued by a mean factor of >1.5 are shown. Roman numerals indicate genes belonging to the GO Biological Processes indicated in panel C. (C) Biological processes (bar graph) that are enriched in the 94 upregulated genes were identified by GO analysis using the STRING database. The number of genes per class and the statistical significance is indicated. (D) Protein interactions clustered by k-means (n = 92; k = 5) to highlight interaction groups (http://string-db.org). (E) Adhesion assay of Runx1+/+ or Runx1Δ/Δ BM progenitors. Lin− cells were plated on MS-5 stromal cells and assessed for adherence after 48 hours. Shown are the results of 1 out of 2 independent experiments. Each dot represents an independent well. Horizontal line, median; error bars, ±SE. P values were calculated by a unpaired Student t test; ***P < .001. (F) Experimental design to test candidate genes that impact on Meg-skewing through adhesion interactions (left). The relative number of Megs was assessed by CD41 expression and confirmed microscopically (right). Expression levels in BFP expressing cultures were set as 1. Error bars, ±SE. P values were calculated by a unpaired Student t test; **P < .01. HPSC, hematopoietic stem and progenitor cells.
Rescue experiment identifies Runx1-target genes that mediate cell adhesion. (A) Experimental design of Runx1 rescue experiments used to define direct target genes. 4-OHT, 4-hydroxytamoxifen. (B) Heat maps depicting microarrays expression results from untreated GMP from Runx1Δ/Δ and Runx1+/+ mice, as well as rescue experiments analyzing transduced GMP. Left panel shows the relative expression levels for 42 genes downregulated in Runx1Δ/Δ GMPs and upregulated after tamoxifen treatment of RUNX1-transduced, but not control-transduced (ERt2), GMPs. Right panel shows 94 genes that are upregulated in Runx1Δ/Δ GMPs and downregulated after activation of RUNX1. Genes deregulated by a factor >2 (P ≤ .05) and that could be rescued by a mean factor of >1.5 are shown. Roman numerals indicate genes belonging to the GO Biological Processes indicated in panel C. (C) Biological processes (bar graph) that are enriched in the 94 upregulated genes were identified by GO analysis using the STRING database. The number of genes per class and the statistical significance is indicated. (D) Protein interactions clustered by k-means (n = 92; k = 5) to highlight interaction groups (http://string-db.org). (E) Adhesion assay of Runx1+/+ or Runx1Δ/Δ BM progenitors. Lin− cells were plated on MS-5 stromal cells and assessed for adherence after 48 hours. Shown are the results of 1 out of 2 independent experiments. Each dot represents an independent well. Horizontal line, median; error bars, ±SE. P values were calculated by a unpaired Student t test; ***P < .001. (F) Experimental design to test candidate genes that impact on Meg-skewing through adhesion interactions (left). The relative number of Megs was assessed by CD41 expression and confirmed microscopically (right). Expression levels in BFP expressing cultures were set as 1. Error bars, ±SE. P values were calculated by a unpaired Student t test; **P < .01. HPSC, hematopoietic stem and progenitor cells.
To identify key Runx1-target genes responsible for the observed Meg-skewing and self-renewal capacity, we determined which of our identified Runx1-target genes are expressed during the transition from HSC to the pro-Meg MPP2 but repressed in MPPs that lose Meg potential26 (supplemental Figure 6C-D). Several candidate genes were examined for their ability to increase Meg-differentiation capacity in sorted LSK cells. The angiopoietin receptor Tek consistently increased Meg output (Figure 5F). Several studies have demonstrated the importance of Tek in HSC function and interaction with the BM stroma.66-68 These results suggest that although several parallel and divergent pathways mediate lineage decision, the activation of Tek by an autocrine or paracrine mechanism and consequent changes to integrin-mediated adhesion may be involved.66,69 Clearly, future studies are needed to define the functionally distinct BM niches and interacting molecules that may uniquely support the heterogenous MPP populations.70
Identification of Runx1-binding sites in GMP-like cells and verification of target genes
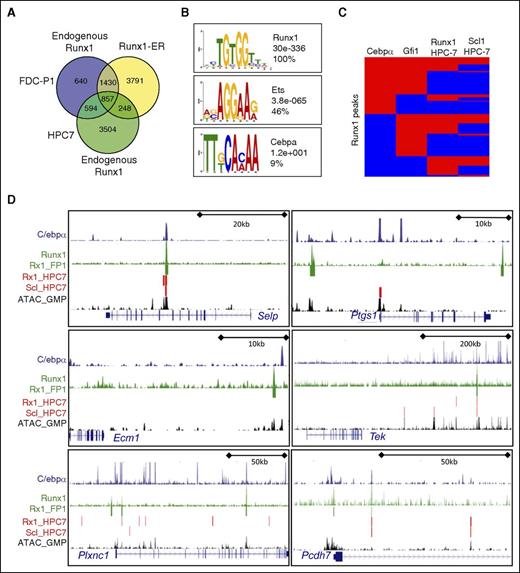
To further confirm direct Runx1 target genes, we performed ChIP-seq analysis for Runx1 binding in FDC-P1 cells, which have an immunophenotype and differentiation capacity similar to GMPs (supplemental Figure 7A). Runx1-transduced FDC-P1 revealed a larger number of Runx1-bound regions than endogenous Runx1 levels, but shared binding sites were identified (Figure 6A). Of these, 37% overlapped with Runx1-binding sites obtained with the “MPP-like” HPC7, which have maintained Meg/Ery differentiation potential.28
Runx1 binding sites in a GMP-like cell line confirm binding to Runx1 target genes that are downregulated in its absence. (A) Venn diagram showing overlap (±150 bp from peak summit) of binding sites obtained from experiments using either the GMP-like FDC-P1 cells or the HSC/MPP-like HPC7 cells.29 Differences in the binding sites between endogenous and RUNX1-ER may represent alternative binding of the fusion protein and/or higher expression levels. (B) Consensus sequences within 75 bp of the Runx1-peak summits identified by de novo motif discovery. The frequency of the observed motif in randomly selected Runx1 peaks (n = 500) and the calculated E-values are given. (C) Hierarchical clustering of the shared Runx1 peaks identified in FDC-P1 cells based on overlapping occupancy patterns with peaks identified for C/ebpα (GMPs),44 Gfi1 (M/E transformed),75 and Runx1 and Scl1 (HPC7).29 Peak regions for individual TFs were set at a standard width of 300 bp. Each line corresponds to an individual Runx1-binding region where red/blue coloring indicates the presence/absence of the given TF within 152 bp of the summit. (D) Runx1-bound regions visualized as density blots (green) and peak calling (green bar) for 6 genes, which are upregulated in KOs and downregulated after re-expression of Runx1. Density blots for C/ebpα from GMPs44 (blue) and open chromatin structure in GMPs (black),28 as determined by the ATAC methodology, are shown. Called peaks for Runx1 and Scl1 from HPC7 cells25 are depicted with red bars. Exon-intron gene structures are depicted in a 5′-3′ orientation using the UCSC Genome Browser. Asterisk by Gcnt1 denotes a packed view of 3 different transcript variants, each starting at a unique first exon but with shared second and third exons (last exons depicted).
Runx1 binding sites in a GMP-like cell line confirm binding to Runx1 target genes that are downregulated in its absence. (A) Venn diagram showing overlap (±150 bp from peak summit) of binding sites obtained from experiments using either the GMP-like FDC-P1 cells or the HSC/MPP-like HPC7 cells.29 Differences in the binding sites between endogenous and RUNX1-ER may represent alternative binding of the fusion protein and/or higher expression levels. (B) Consensus sequences within 75 bp of the Runx1-peak summits identified by de novo motif discovery. The frequency of the observed motif in randomly selected Runx1 peaks (n = 500) and the calculated E-values are given. (C) Hierarchical clustering of the shared Runx1 peaks identified in FDC-P1 cells based on overlapping occupancy patterns with peaks identified for C/ebpα (GMPs),44 Gfi1 (M/E transformed),75 and Runx1 and Scl1 (HPC7).29 Peak regions for individual TFs were set at a standard width of 300 bp. Each line corresponds to an individual Runx1-binding region where red/blue coloring indicates the presence/absence of the given TF within 152 bp of the summit. (D) Runx1-bound regions visualized as density blots (green) and peak calling (green bar) for 6 genes, which are upregulated in KOs and downregulated after re-expression of Runx1. Density blots for C/ebpα from GMPs44 (blue) and open chromatin structure in GMPs (black),28 as determined by the ATAC methodology, are shown. Called peaks for Runx1 and Scl1 from HPC7 cells25 are depicted with red bars. Exon-intron gene structures are depicted in a 5′-3′ orientation using the UCSC Genome Browser. Asterisk by Gcnt1 denotes a packed view of 3 different transcript variants, each starting at a unique first exon but with shared second and third exons (last exons depicted).
Motif search analysis in the immediate vicinity of the Runx1 summit identified Runx1 consensus motifs in 100% of the assayed peaks (Figure 6B). This is in contrast to previous results using MLL-like or Meg progenitors,29,49,71 where only 40% to 50% of identified peaks contained the Runx1 consensus motif. In agreement with other studies, approximately half of the Runx1-bound regions contained binding motifs for ETS, a known RUNX1-cooperating TF.72,73 We also identified consensus sites for the G/M TF C/ebpα in 10% of the sites. To more robustly confirm that the Runx1-binding sites we identified represent the binding pattern found in the G/M versus HSC or Meg lineages, we compared our binding sites with those for the G/M regulators C/ebpα and Gfi174-76 and with Runx1- and Scl1-binding (a HSC/Meg regulator) sites identified in HPC7 cells28 (Figure 6C). Approximately 30% of the Runx1 peak summits in FDC-P1 cells were within 300 bp of both C/ebpα and Gfi1 binding sites; indeed, more than half of the Runx1 peaks overlapped with Gfi1 peaks. In contrast, Scl1 peaks showed <18% overlap with Runx1 peaks in FDC-P1 cells but ∼50% with Runx1 peaks in HPC-7 cells.28
We next mapped Runx1-binding sites to direct Runx1 target genes identified in the rescue experiment; 70% of the putative target genes were within 100 kb of a Runx1-binding site, 46% of which were either intragenic or within 10 kb of the gene body (supplemental Figure 7B). Closer examination of several target genes revealed a pattern in which de novo Runx1 binding in GMPs correlated with C/ebpα binding and open chromatin in GMP (Figure 6D). As these genes are all downregulated during the transition from MPP to GMP but upregulated in the absence of Runx1, we predict that C/ebpα-Runx1-occupied cis-regulatory elements may actively repress transcription, likely by recruiting strong repressors or corepressors, such as Gfi1, Sin3a, or protein arginine methyltransferases.77,78
In summary, we have identified a critical role of Runx1 in suppressing what may be considered the default lineage choice in HSC development, ie, the amplification and lineage skewing of progenitors destined to form Megs, essential for producing platelets that repair disrupted endothelium and inhibit excessive bleeding. We postulate that the biased differentiation toward the Meg lineage occurs at the level of the HSC/MPP but manifests itself in an amplified Meg progenitor compartment and aberrant GMPs. A large proportion of Runx1 target genes encode proteins that regulate adhesion and motility, although other mechanisms may be at play. Whereas some of the genes have been previously described to be direct Runx1 target genes (eg, Mpl, Hmga2, and Selp),71,79,80 we have identified several novel Runx1 target genes, including Jam3, Plxcn1, Tek, and Ecm1. Interestingly, several common features of HSCs and Megs are known, including shared BM niches and critical interactions with endothelial cells.81 Furthermore, many of the identified Runx1 target genes show deregulated expression within the HSC/MPP compartment.26 We speculate that by failing to repress their expression, Runx1-deficient HSCs may maintain pivotal niche interactions that lead to preferential differentiation toward the Meg lineage and increased regeneration potential.23,24 It is important to note that these regulatory factors may be distinct to those important for maintaining HSC quiescence and homing, such as CXCR4.37 Taken together, our study has revealed several novel Runx1 functions in normal hematopoiesis (Figure 4F), whose disruption contribute to the MDS and AML phenotypes associated with Runx1 mutations. The identification of downstream targets of Runx1 tumor-suppressor activity opens an avenue to bypass its mutational consequence in these myeloid neoplasms.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank M. Kauppi for providing advice for in vitro experiments and Y. Groner and D. Levanon (Weizmann Institute, Rehovot) for supplying the Runx1 antibody.
This work was funded by the Deutsche Krebshilfe e.V. and the Else Kröner-Fresenius-Stiftung, the Australian National Health and Medical Research Council, and a Victorian State Government Operational Infrastructure Support Grant. The Heinrich-Pette-Institute, Leibniz Institute for Experimental Virology is supported by the Bundesministerium für Gesundheit and the Freie und Hansestadt Hamburg.
Authorship
Contribution: K.B. designed and performed experiments, analyzed results, and wrote the manuscript; I.T., M.S., N.T., and U.M. assisted and analyzed mouse experiments; M.A. and M.S. performed bioinformatic analysis; D.I. and M.Z. performed ChIP-seq and RNA-seq experiments; W.S.A. analyzed data; and C.S. designed the research, analyzed data, and wrote the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
The current affiliation for K.B. is Cancer and Haematology Division, The Walter and Eliza Hall Institute of Medical Research, Victoria, Australia.
Correspondence: Carol Stocking, Heinrich Pette Institute, Leibniz Institute of Experimental Virology, Martinistraβe 52, 20251 Hamburg, Germany; e-mail: carol.stocking@hpi.uni-hamburg.de.

![Figure 3. Trancriptome analysis of XMP and MEP populations in Runx1-deficient mice. (A) Sorting strategy for analysis of bipotent MPs in Runx1+/+ and Runx1Δ/Δ mice. Shown are cells that have been gated for Lin−Sca1−Kit+CD41−CD105−. (B) Principle-component analysis was performed using DESeq282 and demonstrates the relatedness of the transcriptomes of the indicated populations. (C) Venn diagram of stringently filtered (at least threefold deregulated, false discovery rate [FDR] P ≤ .05, base mean > 100) genes whose expression is either up or down in Δ/Δ vs +/+ GMPs or MEPs. Bold numbers refer to the number of shared genes that are deregulated in GMPs and MEPs. (D) Venn diagram of stringently filtered (at least threefold deregulated, P ≤ .05) genes whose expression is either up or down in Δ/Δ XMPs vs +/+ MEPs or Δ/Δ MEPs. Bold numbers refer to the number of shared genes that are deregulated in XMP in both comparisons. (E) Bar graph showing the relative gene expression (log2) of the shared genes that are either downregulated (left) or upregulated (right) in XMPs as compared with MEPs of either genotype. Each pair of bars corresponds to the gene either to the far left (downregulated) or far right (upregulated). Heat maps show the relative expression levels of these genes in monopotent Ery or MkP progenitors or bipotent GMPS-based on normalized values in Gene Expression Commons (GEXC).83 Genes not annotated in GEXC or showing spurious expression levels are not shown. (F) Relative expression levels determined from reads per kilobase of transcripts per million mapped reads (RPKM) values of genes encoding key Ery or Meg TFs are shown. Expression levels in WT MEP (+/+) were set as 1. Shown are the mean values of 2 independent experiments. Error bars, ±SD. Original P values for the comparison relative to +/+ MEP values are indicated as *P < .05, **P < .0, and ***P < .001, the latter of which have FDR P values ≤ .05. A list of all TF genes used for this analysis and gene expression levels is found in supplemental Table 2.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/127/26/10.1182_blood-2015-09-668129/4/m_3369f3.jpeg?Expires=1770074283&Signature=MMvi1vaGRTKt70qh-zQrIvBWTK68pl3i59QRxhqs5~VuKU4urPyCWh3BqdNOAVzg6fpFJWCBfWUK7TQ-7V05yTbovPGMoNwWRInnO6Ph3n-rk2L28Gn61h6LpqEcWlgsK31iI2d8WveSrPdnjTxqzsmMEfAH80PnVVsEto16fYdfpHPCFM-VyeQd~3DRZ4ThXVPVuSpHEQS8SSFQRVaQgWz6IbUchrcI3ZQWkHOwP09ITJGyer75ZjkViBNDHcxVxHSK6wc~-HI9K7l8JBiGovYoglExXKm4kRvgYBU8Ps7yxR4XLwymYQ8xmuVKfgZv4klHMeMYBWOSdCq5nwcb8Q__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
This feature is available to Subscribers Only
Sign In or Create an Account Close Modal