Key Points
Lenalidomide treatment has variable transient effects on the clonal architecture of myelodysplastic syndromes without 5q deletion.
Lenalidomide is unlikely to eradicate myelodysplastic clones characterized by combinations of SF3B1, TET2, DNMT3A, and ASXL1 mutations.
Abstract
Non-del(5q) transfusion-dependent low/intermediate-1 myelodysplastic syndrome (MDS) patients achieve an erythroid response with lenalidomide in 25% of cases. Addition of an erythropoiesis-stimulating agent could improve response rate. The impact of recurrent somatic mutations identified in the diseased clone in response to lenalidomide and the drug’s effects on clonal evolution remain unknown. We investigated recurrent mutations by next-generation sequencing in 94 non-del(5q) MDS patients randomized in the GFM-Len-Epo-08 clinical trial to lenalidomide or lenalidomide plus epoetin β. Clonal evolution was analyzed after 4 cycles of treatment in 42 cases and reanalyzed at later time points in 18 cases. The fate of clonal architecture of single CD34+CD38− hematopoietic stem cells was also determined in 5 cases. Mutation frequency was >10%: SF3B1 (74.5%), TET2 (45.7%), DNMT3A (20.2%), and ASXL1 (19.1%). Analysis of variant allele frequencies indicated a decrease of major mutations in 15 of 20 responders compared with 10 of 22 nonresponders after 4 cycles. The decrease in the variant allele frequency of major mutations was more significant in responders than in nonresponders (P < .001). Genotyping of single CD34+CD38− cell–derived colonies showed that the decrease in the size of dominant subclones could be associated with the rise of founding clones or of hematopoietic stem cells devoid of recurrent mutations. These effects remained transient, and disease escape was associated with the re-emergence of the dominant subclones. In conclusion, we show that, although the drug initially modulates the distribution of subclones, loss of treatment efficacy coincides with the re-expansion of the dominant subclone. This trial was registered at www.clinicaltrials.gov as #NCT01718379.
Introduction
Myelodysplastic syndromes (MDSs) are acquired, neoplastic disorders of hematopoietic stem cells (HSCs) characterized by ineffective and dysplastic myeloid cell differentiation and a high rate of progression to acute myeloid leukemia (AML). In low- or intermediate-risk MDS, treatment with erythropoiesis-stimulating agents (ESAs) aims at correcting anemia. Lenalidomide is another compound approved for treating anemia in lower-risk MDS patients, with deletion of the long arm of chromosome 5 (del[5q]), where it yields red blood cell (RBC) transfusion independence in 60% to 75% of cases and a complete or partial cytogenetic response in 40% to 60%.1-5 In this setting, the molecular target of lenalidomide was shown to be cereblon, a protein that is part of an E3 ubiquitin ligase complex, cereblon-CUL4-ROC1. Lenalidomide activates this E3 ligase that, in turn, ubiquitinates and degrades casein kinase 1A, encoded by the CSNK1A1 gene located in the 5q commonly deleted region, which may cause cell death.6
Lenalidomide also yields an erythroid response (hematologic improvement of the erythroid lineage [HI-E]) and RBC transfusion independence in 20% to 30% of MDS patients without del(5q) who are resistant to ESAs.7,8 In this context, the addition of high doses of epoetin was suggested to increase the hematologic response to lenalidomide.9,10 Predictive biomarkers of response are needed to guide the use of lenalidomide (with or without ESAs) in these patients, and recurrent somatic mutations could be such molecular predictors. The present study investigated mutations in 26 selected genes11-13 in a cohort of 94 non-del(5q) International Prognostic Scoring System low and intermediate-1 MDS patients who received lenalidomide with or without epoetin β (EPO) as second-line treatment of their ESA-resistant anemia. Deep sequencing of bulk bone marrow (BM) mononuclear cells and single CD34+CD38− genotyping were used to assess the evolution of the clonal architecture under therapy.
Patients and methods
Patient characteristics
Low and intermediate-1 non-del(5q) MDS patients (n = 131) were enrolled in the GFM-Len-Epo-08 clinical trial of the Groupe Francophone des Myélodysplasies (registered at clinicaltrials.gov as #NCT01718379) after specific informed consent according to the recommendations of the local ethics committee. All the studied patients had failed to respond to ESAs (used for at least 12 consecutive weeks at a minimum dosage of 60 000 IU of epoetin or 250 μg of darbepoetin per week) or had relapsed after initial response to ESAs and were RBC-transfusion dependent of at least 4 RBC units over the last 8 weeks. These patients received lenalidomide at a dose of 10 mg/d for 21 days every 28 days, either alone or combined with 60 000 IU of EPO per week. Transfusion dependency and HI-E, defined according to International Working Group 2006 criteria, were evaluated after the fourth cycle in 99 patients.14 Treatments were continued in responders until relapse. BM aspirates were collected from 94 patients (47 patients in each treatment arm) at inclusion, from 42 of 94 patients after 4 cycles of treatment, and from 18 patients during later follow-up (between 12 and 26 months) and processed for biological studies. No patient was excluded on the basis of initial clinical and biological characteristics (Table 1 and supplemental Table 1, available on the Blood Web site).
Clinical characteristics of patients
| Characteristics . | Whole cohort, N = 94 . | Responders, n = 40 . | Nonresponders, n = 54 . | P . |
|---|---|---|---|---|
| Age | ||||
| Median, y (IQR) | 73 (67–76) | 73 (69–77) | 73 (66–76) | .519 |
| Gender | ||||
| Male, n (%) | 67 (71.3) | 30 (73.2) | 37 (63.8) | .645 |
| WHO subtype, n (%) | ||||
| RA | 3 (3.2) | 2 (5.0) | 1 (1.8) | .386 |
| RARS | 46 (48.9) | 17 (42.5) | 29 (53.7) | |
| RCMD-RS | 15 (16.0) | 5 (12.5) | 10 (25.0) | |
| RCMD | 12 (12.8) | 9 (22.5) | 3 (7.5) | |
| RAEB-1 | 16 (17.0) | 7 (17.5) | 9 (22.5) | |
| MDS-U | 2 (2.1) | 0 (0.0) | 2 (5.0) | |
| Hemoglobin, g/L | ||||
| Median (IQR) | 8.1 (7.6–8.7) | 8.1 (7.6–8.6) | 8.1 (7.6–8.7) | .756 |
| Neutrophil count, g/L | ||||
| Median (IQR) | 2.2 (1.4–3.4) | 1.6 (1.1–2.8) | 2.4 (1.7–3.5) | .119 |
| Platelet count, g/L | ||||
| Median (IQR) | 231 (170–325) | 241 (159–304) | 230 (170–344) | .443 |
| BM blasts, % | ||||
| Median (IQR) | 2 (1–4) | 2 (1.9–4) | 2 (1–3.7) | .118 |
| BM erythroblast, % | ||||
| Median (IQR) | 30 (19–45) | 39 (20–48) | 29 (18–43) | .210 |
| BM ring sideroblasts | ||||
| >15%, n (%) | 63/69 (91) | 24/25 (96) | 39/44 (89) | .406 |
| Karyotype, n (%) | ||||
| Favorable | 79 (84.1) | 32 (80.0) | 47 (87.0) | .462 |
| Intermediate | 13 (13.8) | 7 (17.5) | 6 (11.1) | |
| Unavailable | 2 (2.1) | 1 (2.5) | 1 (1.9) | |
| IPSS, n (%) | ||||
| Low risk | 44 (46.8) | 14 (35.0) | 30 (55.5) | .499 |
| Int-1 | 48 (51.1) | 25 (62.5) | 23 (42.6) | |
| Unavailable | 2 (2.1) | 1 (2.5) | 1 (1.9) | |
| Serum erythropoietin level, U/L | ||||
| Median (IQR) | 190 (76–407) | 108 (59–228) | 269 (102–689) | .306 |
| Characteristics . | Whole cohort, N = 94 . | Responders, n = 40 . | Nonresponders, n = 54 . | P . |
|---|---|---|---|---|
| Age | ||||
| Median, y (IQR) | 73 (67–76) | 73 (69–77) | 73 (66–76) | .519 |
| Gender | ||||
| Male, n (%) | 67 (71.3) | 30 (73.2) | 37 (63.8) | .645 |
| WHO subtype, n (%) | ||||
| RA | 3 (3.2) | 2 (5.0) | 1 (1.8) | .386 |
| RARS | 46 (48.9) | 17 (42.5) | 29 (53.7) | |
| RCMD-RS | 15 (16.0) | 5 (12.5) | 10 (25.0) | |
| RCMD | 12 (12.8) | 9 (22.5) | 3 (7.5) | |
| RAEB-1 | 16 (17.0) | 7 (17.5) | 9 (22.5) | |
| MDS-U | 2 (2.1) | 0 (0.0) | 2 (5.0) | |
| Hemoglobin, g/L | ||||
| Median (IQR) | 8.1 (7.6–8.7) | 8.1 (7.6–8.6) | 8.1 (7.6–8.7) | .756 |
| Neutrophil count, g/L | ||||
| Median (IQR) | 2.2 (1.4–3.4) | 1.6 (1.1–2.8) | 2.4 (1.7–3.5) | .119 |
| Platelet count, g/L | ||||
| Median (IQR) | 231 (170–325) | 241 (159–304) | 230 (170–344) | .443 |
| BM blasts, % | ||||
| Median (IQR) | 2 (1–4) | 2 (1.9–4) | 2 (1–3.7) | .118 |
| BM erythroblast, % | ||||
| Median (IQR) | 30 (19–45) | 39 (20–48) | 29 (18–43) | .210 |
| BM ring sideroblasts | ||||
| >15%, n (%) | 63/69 (91) | 24/25 (96) | 39/44 (89) | .406 |
| Karyotype, n (%) | ||||
| Favorable | 79 (84.1) | 32 (80.0) | 47 (87.0) | .462 |
| Intermediate | 13 (13.8) | 7 (17.5) | 6 (11.1) | |
| Unavailable | 2 (2.1) | 1 (2.5) | 1 (1.9) | |
| IPSS, n (%) | ||||
| Low risk | 44 (46.8) | 14 (35.0) | 30 (55.5) | .499 |
| Int-1 | 48 (51.1) | 25 (62.5) | 23 (42.6) | |
| Unavailable | 2 (2.1) | 1 (2.5) | 1 (1.9) | |
| Serum erythropoietin level, U/L | ||||
| Median (IQR) | 190 (76–407) | 108 (59–228) | 269 (102–689) | .306 |
Clinical and biological characteristics were collected at the time of inclusion and compared between responders and nonresponders to treatment by lenalidomide or lenalidomide plus EPO. Perls staining was available in 69 of 94 (73%) patients.
Int-1, intermediate-1; IPSS, International Prognostic Scoring System; IQR, interquartile range; MDS-U, undefined MDS; RA, refractory anemia; RAEB1, refractory anemia with excess of blasts type 1; RARS, refractory anemia with ring sideroblasts; RCMD, refractory cytopenia with multilineage dysplasia; RCMD-RS, RCMD with ring sideroblasts; WHO, World Health Organization.
Genomic studies
BM mononuclear cells were purified on Ficoll gradient and processed for DNA extraction using the DNA/RNA Kit (Qiagen, Hilden, Germany). Mutations in a selected panel of 26 genes (ASXL1, CBL, DNMT3A, ETV6, EZH2, FLT3-TKD, IDH1, IDH2, JAK2, KIT, KRAS, NRAS, MPL, NPM1, PHF6, PTPN11, RIT1, RUNX1, SETBP1, SF3B1, SRSF2, TET2, TP53, U2AF1, WT1, and ZRSR2) (supplemental Table 2) were screened by a next-generation sequencing (NGS) assay using the Ion AmpliSeq Library Kit 2.0 (384 reactions; Thermo Fisher Scientific, Carlsbad, CA). Multiplex PCR amplifications (233 primer pairs) were performed from 2 × 10 ng of genomic DNA. After amplification, barcodes and adaptors were added to amplicons by ligation. Products were subjected to a selective purification on AMPure beads (Beckman Coulter, Brea, CA). Emulsion polymerase chain reaction (PCR) was performed using the OneTouch 2 instrument (Thermo Fisher Scientific). Sequencing was performed with the Ion PGM system (Thermo Fisher Scientific) onto the 318 V2 chip (15 samples per chip). All the samples were also screened for ASXL1 (including c.1934dupG; p.G646WfsX12) and SRSF2 mutations by Sanger sequencing. JAK2, NPM1, and FLT3–internal tandem duplication mutations were also investigated by qualitative PCR and fluorescent PCR.
Bioinformatics analysis
Base calls were generated by the Torrent Browser software using the included variant caller with an additional plug-in (Thermo Fisher Scientific). The .bam and .vcf files were used for further analysis. The .vcf files were annotated with the Ion reporter software (Thermo Fisher Scientific) and processed for a second analysis of the indexed files using the NextGENe software (Softgenetics, State College, PA). Results were compared with select abnormalities that will be further considered. For each mutation, depth at the variant position (number of mutated reads and unmutated reads) was considered to calculate variant allele frequency, which is the proportion of mutated reads among total reads and its 95% confidence interval (CI). For each patient, pairwise comparisons between the variant allele frequencies of mutations were performed using Fisher’s exact test, and mutations with the significantly highest variant allele frequencies were considered major mutations. For patients with 1 unique genetic event, the isolated mutation was considered a major mutation. To assess clonal evolution after treatment, for each patient, variant allele frequencies of a given mutation were compared between 2 consecutive time points of the follow-up period using Fisher’s exact test. P values <.05 were considered significant, and analyses were performed using the Statsmodels package (Python Software Foundation, Beaverton, OR).
Single-cell cloning and genotyping
Cryopreserved BM mononuclear cells were thawed at 37°C in RPMI 1640 medium and treated with 150 IU of RNase-free DNAse (Qiagen) for 1 hour at 37°C. CD34+ cells were enriched on the MidiMacs system (Miltenyi Biotech, Bergisch Badgach, Germany). CD34+CD38− HSCs were isolated by cell sorting on a BD FACS Aria Cell Sorting System (Becton Dickinson, Franklin Lakes, NJ) and then distributed at 1 cell per well in 96-well plates coated with murine MS-5 stromal cell line.
Long-term expansion of single CD34+CD38− HSCs was performed in H5100 MyeloCult medium complemented with 10−6 M hydrocortisone (StemCell Technologies, Vancouver, Canada) and 1 mM penicillin/streptomycin in the presence of 20 ng/μL stem cell factor, 0.2 ng/μL interleukin-3, 10 ng/μL thrombopoietin, and 10 ng/μL FLT3 ligand (PeproTech, Rocky Hill, NJ). Half of the medium was changed weekly. After 6 weeks, colonies were harvested, lysed in water containing 1× Tween-20 and 10 mg/mL proteinase K at 95°C for 1 hour. Specific primers were designed for each PCR, and single-cell–derived colonies were sequenced using the Sanger method. The architecture of the CD34+CD38− compartment was represented as the proportion of mutated colonies of each genotype among all sequenced colonies.
Statistical analysis
For continuous variables, values were expressed as median and interquartile range and compared using Mann Whitney or Student t tests. Categorical variables are reported as count and percentage and were compared using Fisher’s exact or χ2 tests. Evaluation of the erythroid response (according to International Working Group 2006 criteria) after 4 cycles of treatment (the primary efficacy end point) was considered a binary variable: responders vs nonresponders. The search for prognostic factors of response was performed by logistic regression, and results were presented by the odds ratio (OR) and its 95% CI. The 4 most common genes mutated in >10% patients were included in a multivariate logistic model, adjusting for treatment arm to assess independent prognostic factors. Interactions between each of the 4 mutations and the treatment arm were assessed using the Gail-Simon heterogeneity test.15 All statistical tests were considered significant for P values <.05. Analyses were performed using the SAS 9.3 software (SAS, Cary, NC), R version 3.0.2 (The R Foundation for Statistical Computing), and Python Statsmodels package (Python Software Foundation).
Results
Baseline characteristics including mutational profile and response to treatment
Twenty-six genes recurrently mutated in MDS patients were sequenced in the 94 BM samples obtained before treatment (supplemental Table 2). According to World Health Organization 2008 classification, 3 patients had refractory anemia, 46 had RARS, 12 had RCMD, 15 had RCMD-RS, 16 had RAEB1, and 2 had undefined MDS (Table 1; Figure 1A). Two patients with an RAEB1 classification had >15% ring sideroblasts. Conventional karyotype, available in 92 of 94 patients, was normal in 73 cases, and showed loss of chromosome Y in 5 cases, del(20q) in 5 cases, trisomy 8 in 10 cases, and del11q in 4 cases (Table 1).
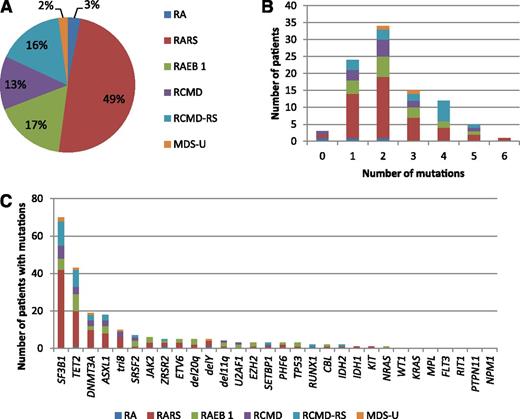
Genotyping of 94 low/int-1 MDS patients using a panel of 26 genes. (A) Repartition of WHO subtypes. (B) Number of patients of each WHO subtype having 0 to 6 mutations. (C) Number of patients of each WHO subtype exhibiting genetic or cytogenetic lesions. SF3B1, TET2, DNMT3A, and ASXL1 mutations were the most frequent genetic events in the cohort.
Genotyping of 94 low/int-1 MDS patients using a panel of 26 genes. (A) Repartition of WHO subtypes. (B) Number of patients of each WHO subtype having 0 to 6 mutations. (C) Number of patients of each WHO subtype exhibiting genetic or cytogenetic lesions. SF3B1, TET2, DNMT3A, and ASXL1 mutations were the most frequent genetic events in the cohort.
More than one-third of patients (33/94) displayed at least 2 different gene mutations, whereas only 4 patients had no mutations of the studied genes (Figure 1B). Detailed annotation of the mutations is provided in supplemental Table 3. In accordance with the high number of RARS/RCMD-RS (n = 61) or RAEB1 with ring sideroblasts classifications, and considering that MDS patients with ≤15% of ring sideroblasts may also have an SF3B1 mutation at lower frequency,16 the SF3B1 gene was not surprisingly the most frequently mutated gene (74.5%). Three other genes were mutated in >10% of patients, namely, TET2 (45.7%), DNMT3A (20.2%), and ASXL1 (19.1%). Fifteen genes (SRSF2, ZRSR2, JAK2, RUNX1, ETV6, TP53, U2AF1, EZH2, IDH1, IDH2, SETBP1, CBL, PHF6, KIT, and NRAS) were mutated in 1% to 9% of patients, and 7 genes (WT1, KRAS, MPL, PTPN11, RIT1, FLT3, and NPM1) were never found mutated in this cohort (Figure 1C). To assess the genetic heterogeneity of each patient sample containing at least 1 mutation among the SF3B1, TET2, ASXL1, and DNMT3A genes, pairwise comparisons between the variant allele frequencies of mutations were performed (Figure 2A). Among 72 SF3B1 and 20 DNMT3A mutations, 67 (93%) and 15 (75%) were expressed at the highest variant allele frequencies, meaning that mutations in SF3B1 or DNMT3A genes were mainly major mutations. In contrast, only 31 of 63 (49%) TET2 mutations and 11 of 20 (55%) ASXL1 mutations were considered major mutations. Loss of heterozygosity in the TET2 gene was identified in 2 cases. We observed a significant association between SF3B1 and DNMT3A mutations (OR, 7.96 [95% CI, 1.01–63.29]; P = .035), but not between SF3B1 and TET2 (OR, 1.25 [95% CI, 0.49–3.19]; P = .812) or between SF3B1 and ASXL1 (OR, 0.45 [95% CI, 0.15–1.35]; P = .227) mutations (Figure 2B).
Landscape of mutations. (A) Distribution of variant allele frequencies of SF3B1, TET2, ASXL1, and DNMT3A gene mutations at inclusion. For each mutation, depth at the variant position was considered to calculate variant allele frequency; that is, the proportion of mutated reads among total reads and its 95% CI. For each patient having at least 1 of the 4 SF3B1, TET2, ASXL1, or DNMT3A mutations, pairwise comparisons between the variant allele frequencies of mutations were performed using Fisher’s exact test. Mutations with the significantly highest variant allele frequencies were considered major mutations and are indicated by closed symbols; minor mutations are indicated by open symbols. (B) Barcode representation of genetic lesions, cytogenetic abnormalities, and response status of 94 patients. Two patients had no mutation and a normal karyotype. Each column represents an individual sample, and each row represents a gene. Black boxes represent abnormal karyotypes and responders to treatment. Gray boxes represent unavailable karyotype.
Landscape of mutations. (A) Distribution of variant allele frequencies of SF3B1, TET2, ASXL1, and DNMT3A gene mutations at inclusion. For each mutation, depth at the variant position was considered to calculate variant allele frequency; that is, the proportion of mutated reads among total reads and its 95% CI. For each patient having at least 1 of the 4 SF3B1, TET2, ASXL1, or DNMT3A mutations, pairwise comparisons between the variant allele frequencies of mutations were performed using Fisher’s exact test. Mutations with the significantly highest variant allele frequencies were considered major mutations and are indicated by closed symbols; minor mutations are indicated by open symbols. (B) Barcode representation of genetic lesions, cytogenetic abnormalities, and response status of 94 patients. Two patients had no mutation and a normal karyotype. Each column represents an individual sample, and each row represents a gene. Black boxes represent abnormal karyotypes and responders to treatment. Gray boxes represent unavailable karyotype.
DNMT3A mutations predict better response
Of the 94 patients, 47 were randomized to the lenalidomide treatment arm and 47 to the lenalidomide plus EPO arm. Forty patients achieved HI-E after 4 cycles of treatment and 54 patients were nonresponders. None of the pretreatment clinical parameters, including gender, age, World Health Organization classification, hemoglobin level, absolute neutrophil or platelet counts, BM blast percentage, BM erythroblast percentage, karyotype, International Prognostic Scoring System, or serum erythropoietin level, was predictive of HI-E in the whole cohort whatever the treatment (Table 1) or in each arm of treatment (supplemental Table 1). The mean number of mutations per patient was not different between responders (2.4 [95% CI, 2.0–2.9]) and nonresponders (2.2 [95% CI, 1.9–2.5]; Student t test, P = .306). Presence of a DNMT3A mutation was predictive of HI-E after 4 cycles of treatment (OR, 2.88 [95% CI, 1.01–8.17]; χ2 test, P = .043), whereas other frequent mutations of SF3B1 (OR, 0.84 [95% CI, 0.33−2.13]; P = .706), TET2 (OR, 1.92 [95% CI, 0.84–4.40]; P = .121), and ASXL1 genes (OR, 0.45 [95% CI, 0.15–1.39]; P = .158) had no predictive value. In a multivariate analysis including the 4 most frequent mutations, and adjusting for treatment arm (supplemental Table 4), DNMT3A mutation was independently associated with HI-E (OR, 3.51 [95% CI, 1.10–11.16], P = .029). Of note, no significant interaction was found between DNMT3A mutation and treatment arm.
Lenalidomide often decreases the size of the dominant subclone
Using NGS, we sequentially analyzed the clonal architecture of 42 BM samples. Before treatment, all these samples harbored a dominant subclone containing 1 to 4 different mutations, with variation in mutated genes between patients. We also identified minor subclones in 29 cases. After 4 cycles of treatment, 20 of these patients were classified as responders and 22 as nonresponders. In 25 cases, we observed a decrease in the size of the dominant subclone, indicated by a decrease in SF3B1, DNMT3A, or TET2 mutated allele burden as compared with the pretreatment sample. Fifteen of 25 (60%) patients were responders. In the other 17 cases, the variant allele frequency of major mutations remained stable or increased, and only 5 of 17 (29%) were responders (supplemental Table 5; Figure 3; supplemental Figure 2). Based on these numbers, there was a trend to associate treatment response with decrease in the size of the dominant clone (χ2 test, P = .066). These results are summarized in Figure 4A and show the evolution of variant allele frequencies before and after 4 cycles of treatment. Furthermore, treatment-induced decrease of variant allele frequencies was more significant in responders compared with nonresponders (Mann Whitney U test, P < .001; Figure 4B), which suggests that response to treatment is associated with changes of dominant clone size. Strikingly, in 9 cases (UPNs 17, 24, 29, 40, 61, 69, 91, 96, and 110), major DNMT3A, SF3B1, TET2, or ASXL1 mutations remained stable, whereas other mutations of the dominant clone decreased. Only 2 of the 9 cases were responders. By contrast, 12 of 16 patients with a significant decrease of all the mutations that define the dominant clone were responders (Figure 3; supplemental Figure 2; supplemental Table 5). The evolution of minor subclones was also examined. In 15 cases, at least 1 of the identified minor mutations decreased, whereas in 14 cases, the variant allele frequencies of these minor mutations increased or remained stable. There was no correlation between response to treatment and evolution of minor subclones (Figure 3; supplemental Figure 2; supplemental Table 5). We also evaluated the impact of EPO on clonal evolution in 22 patients treated by lenalidomide plus EPO vs 20 patients treated by lenalidomide alone. The addition of EPO did not change the proportion of patients demonstrating a decrease of major or minor mutations compared to lenalidomide alone.
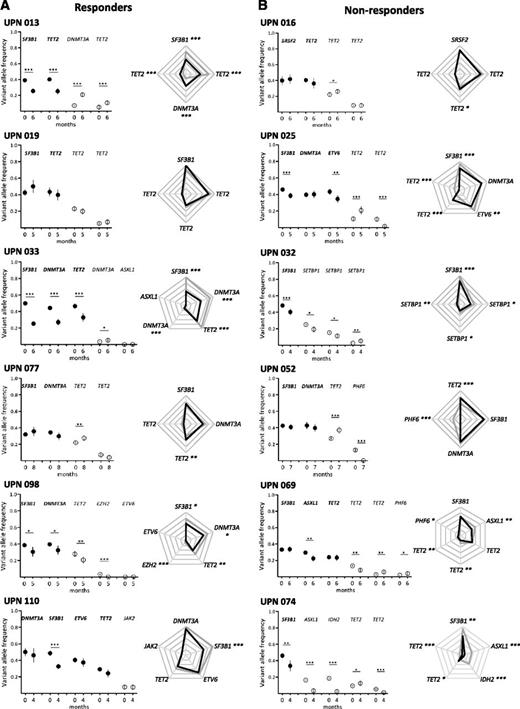
Clonal evolution after short-term treatment with lenalidomide. Results from 6 responders (A) and 6 nonresponders (B) are shown. Other results from 30 additional patients are available in supplemental Figure 2. Paired BM mononuclear cell samples (n = 42) were collected for genotyping before and after 4 cycles of treatment expressed in months of follow-up. Depth at the variant position was considered to calculate variant allele frequency and its 95% CI. For each patient, variant allele frequencies of a given mutation were compared between 2 consecutive time points of the follow-up period using Fisher’s exact test. Left: dot representation; closed symbols represent major mutations and open symbols represent minor mutations for each patient. Vertical bars represent 95% CIs. Right: radar plots of the variant allele frequencies at inclusion and after 4 cycles of treatment. Maximum scale value is 50%, with intervals of 10%. Each extremity of the spider web represents 1 mutation. Gray lines indicate the variant allele frequencies at inclusion and black lines indicate the variant allele frequencies after treatment. In both panels, significant differences between the 2 time points are indicated. *P < .05; **P < .01; ***P < .001. UPN, unique patient number.
Clonal evolution after short-term treatment with lenalidomide. Results from 6 responders (A) and 6 nonresponders (B) are shown. Other results from 30 additional patients are available in supplemental Figure 2. Paired BM mononuclear cell samples (n = 42) were collected for genotyping before and after 4 cycles of treatment expressed in months of follow-up. Depth at the variant position was considered to calculate variant allele frequency and its 95% CI. For each patient, variant allele frequencies of a given mutation were compared between 2 consecutive time points of the follow-up period using Fisher’s exact test. Left: dot representation; closed symbols represent major mutations and open symbols represent minor mutations for each patient. Vertical bars represent 95% CIs. Right: radar plots of the variant allele frequencies at inclusion and after 4 cycles of treatment. Maximum scale value is 50%, with intervals of 10%. Each extremity of the spider web represents 1 mutation. Gray lines indicate the variant allele frequencies at inclusion and black lines indicate the variant allele frequencies after treatment. In both panels, significant differences between the 2 time points are indicated. *P < .05; **P < .01; ***P < .001. UPN, unique patient number.
Evolution of variant allele frequency of dominant mutations in responders and nonresponders to lenalidomide after short-term treatment. (A) Variant allele frequency of each major mutation was shown before (0 cycles) and after 4 cycles of treatment in responders (n = 20; left) and nonresponders (n = 22; right). (B) Variation of variant allele frequency was calculated as follow-up minus screening for each responder and each nonresponder. Box plots represent medians and IQRs [25%–75%], and whiskers extend to 1.5 times the IQR. Variations are compared between responders and nonresponders using the Mann Whitney U test.
Evolution of variant allele frequency of dominant mutations in responders and nonresponders to lenalidomide after short-term treatment. (A) Variant allele frequency of each major mutation was shown before (0 cycles) and after 4 cycles of treatment in responders (n = 20; left) and nonresponders (n = 22; right). (B) Variation of variant allele frequency was calculated as follow-up minus screening for each responder and each nonresponder. Box plots represent medians and IQRs [25%–75%], and whiskers extend to 1.5 times the IQR. Variations are compared between responders and nonresponders using the Mann Whitney U test.
Together, these results show that, after 4 cycles of treatment by lenalidomide with or without EPO, the size of the dominant clone decreased more importantly in responders than in nonresponders, and that clonal dynamics was not influenced by the addition of EPO.
Clonal architecture of CD34+CD38− HSC compartment
To further assess the impact of treatment on clonal evolution, we reconstructed clonal architecture of HSC compartment at the single-cell level. We sorted BM CD34+CD38− HSCs before and after 4 cycles of treatment and cocultured them for 6 weeks on the murine MS-5 cell line. Single cell–derived colonies were harvested and genotyped using mutation-specific PCR and Sanger sequencing in a search for each mutation initially identified by NGS in BM mononuclear cells (supplemental Figure 1A). The number of colonies formed by control vs patient CD34+CD38− HSCs was not significantly different (34 [range, 15−58] vs 23 [range, 6–39], respectively; P = .187) (supplemental Figure 1B). Of importance, the percentage of colonies screened positive for a given variant was consistent with the variant allele frequency determined by NGS in BM mononuclear cells (supplemental Figure 1C), suggesting that in vitro culture might not introduce any bias in clonal representation.
Genotyping of single CD34+CD38− cells identified a dominant clone and 1 or several minor subclones generated by the occurrence of additional mutations in a cell of the dominant clone or, in 1 case, in an independent clone. In all 5 tested patients, at least 2 or 3 mutations were found in >50% of single CD34+CD38−-derived colonies, suggesting the early amplification of a dominant clone at the HSC level (Figure 5).
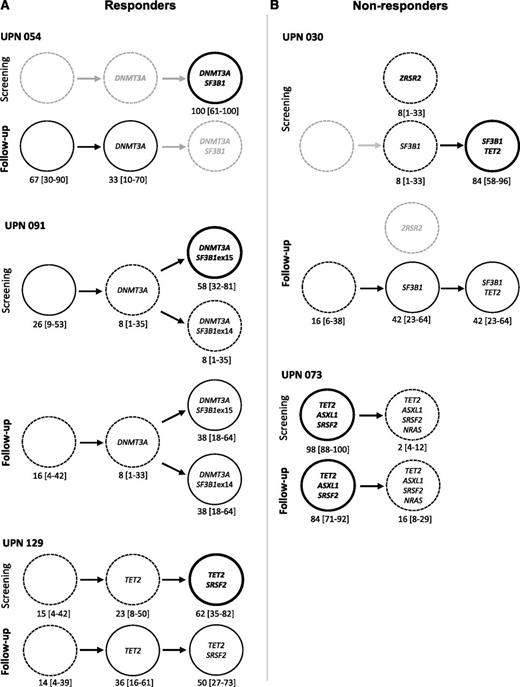
Genotyping of single HSC–derived colonies. (A) Responders. (B) Nonresponders. Graphic representation of clonal architecture of HSC compartment at the single-cell level before and after 4 cycles of treatment. Percentages of mutated colonies and 95% CIs are indicated for each patient. Bold lines are used for dominant subclones, standard and dotted lines for minor subclones, and gray lines for undetected subclones.
Genotyping of single HSC–derived colonies. (A) Responders. (B) Nonresponders. Graphic representation of clonal architecture of HSC compartment at the single-cell level before and after 4 cycles of treatment. Percentages of mutated colonies and 95% CIs are indicated for each patient. Bold lines are used for dominant subclones, standard and dotted lines for minor subclones, and gray lines for undetected subclones.
In 1 patient (UPN 054), DNMT3A and SF3B1 mutations were initially identified in all studied single cell–derived colonies. This patient responded to lenalidomide, and the response was associated with disappearance of SF3B1/DNMT3A double-mutated cells, the remaining cells being either devoid of driver mutation or mutated only for DNMT3A. In this patient, lenalidomide appeared to specifically decrease the competitiveness of the most mutated cells (Figure 5). In a second patient (UPN 091), a DNMT3A mutation was identified in 74% of the colonies, including 8% with DNMT3A alone, 58% with DNMT3A and SF3B1 exon 15 mutations, and 8% with DNMT3A and SF3B1 exon 14 mutations, suggesting that DNMT3A mutation preceded the acquisition of 2 distinct SF3B1 mutations, defining 2 branched subclones. The response to lenalidomide plus EPO observed in this patient was associated with a decrease in the size of the dominant clone (DNMT3A/SF3B1 exon 15), together with the expansion of the initially smaller clone (DNMT3A/SF3B1 exon 14) (Figure 5). In a third patient (UPN 129), 84% of the single cell–derived colonies initially contained a TET2 mutation and 23% of them contained an additional SRSF2 mutation. The response to lenalidomide plus EPO obtained in this patient was not associated with any significant change in clonal architecture (Figure 5). In a fourth patient (UPN 030), SF3B1 mutation was found alone in 8% of the colonies and combined with a TET2 mutation in 85% of them, suggesting that SF3B1 mutation preceded the occurrence of TET2 mutation. A unique ZRSR2 mutation was detected in 8% of the cells, suggesting an independent clone. Although this patient did not respond to lenalidomide plus EPO, the most mutated SF3B1/TET2 clone decreased. A subclone with only SF3B1 or cells with no mutation in these 3 genes expanded, and the isolated clone with a single ZRSR2 mutation disappeared. In a final patient (UPN 073), before treatment, 98% of the cells had mutations in TET2, ASXL1, and SRSF2 genes, and an additional NRAS mutation was detected in only 2% of the cells, defining a minor clone. This patient failed to respond to lenalidomide plus EPO, and failure was associated with the expansion of the minor, NRAS-mutated clone to 16% (Figure 5). Lastly, analysis of clonal architecture at the single-cell level helped discriminate between dominant and founding clones. In UPN 091, DNMT3A appeared as a founding mutation, preceding the onset of 2 different SF3B1 mutations in 2 distinct clones. In UPN 030 and 054, posttreatment experiments demonstrated that the double-mutated clone decreased to the benefit of a clone with a single mutation.
Together, these observations indicate that 4 cycles of lenalidomide, with or without EPO, induce changes in the size and distribution of clones in the CD34+CD38− cell compartment, but that these changes do not accurately reflect the clinical response to the drug.
Long-term follow-up
To determine whether lenalidomide with or without EPO treatment had a reproducible impact on the mutated allele burden at later time points, we repeated targeted sequencing of BM mononuclear cell samples collected from 18 patients after 5 to 28 cycles of treatment (Table 2; Figure 6; supplemental Figure 3). Four of them (UPNs 130, 124, 91, and 85) were still responding after 10, 19, 22, and 23 months of treatment, respectively; 12 had relapsed; and 2 evolved to AML at 9 and 15 months after inclusion.
Long-term clonal evolution upon lenalidomide treatment in low/int-1 myelodysplastic syndromes
| Patient characteristics at baseline . | Long-term evolution . | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| UPN . | Gender/age, y . | WHO . | IPSS . | Mutated genes . | Treatment . | Last follow-up, mo . | Cycle of treatment . | Major mutations . | Minor mutation(s) . | Clinical status . |
| 13 | M/62 | RCMD | Int-1 | SF3B1, TET2, DNMT3A, TET2 | Lenalidomide | 23 | C19 | Re-increase | Increase DNMT3A | Loss of efficacy |
| 33 | M/72 | RCMD | Int-1 | SF3B1, DNMT3A, TET2, DNMT3A, ASXL1 | Lenalidomide | 12 | C10 | Re-increase | Increase | Loss of efficacy |
| 54 | M/80 | RCMD | Int-1 | DNMT3A, SF3B1 | Lenalidomide | 16 | C10 | Re-increase | NA | Loss of efficacy |
| 77 | M/73 | RARS | Int-1 | SF3B1, DNMT3A, TET2, TET2 | Lenalidomide + EPO | 26 | C21 | Increase | Decrease TET2 | Loss of efficacy |
| 85 | M/74 | RCMD | Int-1 | SF3B1, TET2, TET2, RUNX1 | Lenalidomide + EPO | 23 | C20 | Stable | Decrease RUNX1 | Still responder |
| 91 | F/84 | RCMD | Low | SF3B1, DNMT3A, SF3B1 | Lenalidomide + EPO | 22 | C20 | Decrease | Increase SF3B1 ex 14 | Still responder |
| 98 | M/70 | RAEB-1 | Int-1 | SF3B1, DNMT3A, TET2, EXH2, ETV6 | Lenalidomide + EPO | 18 | C11 | Re-increase | Decrease TET2/increase EZH2 and ETV6 | Transformation |
| 110 | M/69 | RARS | Int-1 | DMT3A, SF3B1, ETV6, TET2, JAK2 | Lenalidomide | 18 | C14 | Re-increase | Increase JAK2 | Loss of efficacy* |
| 101 | M/76 | RARS | Int-1 | SF3B1, SF3B1 | Lenalidomide + EPO | 18 | C11 | Re-increase | Decrease SF3B1 | Loss of efficacy |
| 124 | M/80 | RARS | Low | SF3B1, EZH2, TET2 | Lenalidomide + EPO | 19 | C16 | Decrease | Decrease TET2 and EZH2 | Still responder |
| 130 | M/74 | RARS | Low | SF3B1, TET2, KIT | Lenalidomide | 10 | C8 | Re-increase | Decrease TET2 and KIT | Still responder |
| 17 | M/72 | RAEB-1 | ND | DNMT3A, SRSF2, TET2 | Lenalidomide + EPO | 9 | C7 | Re-increase SRSF2 | NA | Transformation |
| 19 | M/76 | RARS | Low | SF3B1, TET2, TET2, TET2 | Lenalidomide + EPO | 11 | C8 | Stable | Stable | Loss of efficacy |
| 35 | F/46 | RARS | Int-1 | SF3B1, TET2 | Lenalidomide + EPO | 18 | C16 | Re-increase SF3B1 | NA | Loss of efficacy |
| 39 | M/63 | RARS | Int-1 | DNMT3A, SF3B1 | Lenalidomide | 23 | C28 | Re-increase | NA | Loss of efficacy |
| 60 | M/78 | RCMD | Int-1 | SF3B1, TET2 | Lenalidomide + EPO | 12 | C5 | Decrease | NA | Loss of efficacy |
| 76 | M/65 | RARS | Int-1 | SF3B1, TET2 | Lenalidomide + EPO | 20 | C18 | Re-increase | Increase TET2 | Loss of efficacy |
| 79 | F/73 | RCMD-RS | Low | SF3B1, TET2, TET2 | Lenalidomide + EPO | 15 | C12 | Decrease | Decrease TET2 | Loss of efficacy |
| Patient characteristics at baseline . | Long-term evolution . | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| UPN . | Gender/age, y . | WHO . | IPSS . | Mutated genes . | Treatment . | Last follow-up, mo . | Cycle of treatment . | Major mutations . | Minor mutation(s) . | Clinical status . |
| 13 | M/62 | RCMD | Int-1 | SF3B1, TET2, DNMT3A, TET2 | Lenalidomide | 23 | C19 | Re-increase | Increase DNMT3A | Loss of efficacy |
| 33 | M/72 | RCMD | Int-1 | SF3B1, DNMT3A, TET2, DNMT3A, ASXL1 | Lenalidomide | 12 | C10 | Re-increase | Increase | Loss of efficacy |
| 54 | M/80 | RCMD | Int-1 | DNMT3A, SF3B1 | Lenalidomide | 16 | C10 | Re-increase | NA | Loss of efficacy |
| 77 | M/73 | RARS | Int-1 | SF3B1, DNMT3A, TET2, TET2 | Lenalidomide + EPO | 26 | C21 | Increase | Decrease TET2 | Loss of efficacy |
| 85 | M/74 | RCMD | Int-1 | SF3B1, TET2, TET2, RUNX1 | Lenalidomide + EPO | 23 | C20 | Stable | Decrease RUNX1 | Still responder |
| 91 | F/84 | RCMD | Low | SF3B1, DNMT3A, SF3B1 | Lenalidomide + EPO | 22 | C20 | Decrease | Increase SF3B1 ex 14 | Still responder |
| 98 | M/70 | RAEB-1 | Int-1 | SF3B1, DNMT3A, TET2, EXH2, ETV6 | Lenalidomide + EPO | 18 | C11 | Re-increase | Decrease TET2/increase EZH2 and ETV6 | Transformation |
| 110 | M/69 | RARS | Int-1 | DMT3A, SF3B1, ETV6, TET2, JAK2 | Lenalidomide | 18 | C14 | Re-increase | Increase JAK2 | Loss of efficacy* |
| 101 | M/76 | RARS | Int-1 | SF3B1, SF3B1 | Lenalidomide + EPO | 18 | C11 | Re-increase | Decrease SF3B1 | Loss of efficacy |
| 124 | M/80 | RARS | Low | SF3B1, EZH2, TET2 | Lenalidomide + EPO | 19 | C16 | Decrease | Decrease TET2 and EZH2 | Still responder |
| 130 | M/74 | RARS | Low | SF3B1, TET2, KIT | Lenalidomide | 10 | C8 | Re-increase | Decrease TET2 and KIT | Still responder |
| 17 | M/72 | RAEB-1 | ND | DNMT3A, SRSF2, TET2 | Lenalidomide + EPO | 9 | C7 | Re-increase SRSF2 | NA | Transformation |
| 19 | M/76 | RARS | Low | SF3B1, TET2, TET2, TET2 | Lenalidomide + EPO | 11 | C8 | Stable | Stable | Loss of efficacy |
| 35 | F/46 | RARS | Int-1 | SF3B1, TET2 | Lenalidomide + EPO | 18 | C16 | Re-increase SF3B1 | NA | Loss of efficacy |
| 39 | M/63 | RARS | Int-1 | DNMT3A, SF3B1 | Lenalidomide | 23 | C28 | Re-increase | NA | Loss of efficacy |
| 60 | M/78 | RCMD | Int-1 | SF3B1, TET2 | Lenalidomide + EPO | 12 | C5 | Decrease | NA | Loss of efficacy |
| 76 | M/65 | RARS | Int-1 | SF3B1, TET2 | Lenalidomide + EPO | 20 | C18 | Re-increase | Increase TET2 | Loss of efficacy |
| 79 | F/73 | RCMD-RS | Low | SF3B1, TET2, TET2 | Lenalidomide + EPO | 15 | C12 | Decrease | Decrease TET2 | Loss of efficacy |
Targeted sequencing was performed before treatment and after >5 cycles of treatment in 18 patients. Major mutations are indicated in bold. For the last follow-up, number of treatment cycles is indicated. Based on the statistical analyses, evolution of major and minor mutations is expressed as increase, decrease, or stable. When major mutations initially decreased after 4 cycles of treatment and increased during long-term follow-up, the evolution of major mutations is indicated as re-increase.
C, treatment cycle; F, female; M, male; NA, not applicable; ND, not determined.
Severe adverse event (neuropathy).
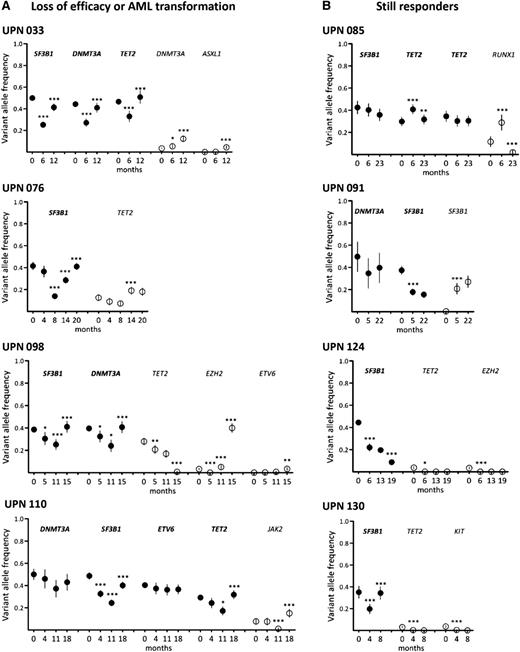
Long-term follow-up of clonal evolution under treatment by lenalidomide. Sequential samples of BM mononuclear cells were collected from 18 responder patients. Results from 4 patients with loss of efficacy or AML transformation (A) and 4 still responders (B) are shown. Other results from 10 additional patients are available in supplemental Figure 3. Depth at the variant position was considered to calculate variant allele frequency and its 95% CI. For each patient, variant allele frequencies of a given mutation were compared between 2 consecutive time points (indicated in months) of the follow-up period using Fisher’s exact test. *P < .05; **P < .01; ***P < .001. Closed symbols represent dominant mutations and open symbols represent minor mutations.
Long-term follow-up of clonal evolution under treatment by lenalidomide. Sequential samples of BM mononuclear cells were collected from 18 responder patients. Results from 4 patients with loss of efficacy or AML transformation (A) and 4 still responders (B) are shown. Other results from 10 additional patients are available in supplemental Figure 3. Depth at the variant position was considered to calculate variant allele frequency and its 95% CI. For each patient, variant allele frequencies of a given mutation were compared between 2 consecutive time points (indicated in months) of the follow-up period using Fisher’s exact test. *P < .05; **P < .01; ***P < .001. Closed symbols represent dominant mutations and open symbols represent minor mutations.
Among the 4 still-responding patients, 1 (UPN 085) had a stable dominant clone. In UPN 091, the size of the initially dominant clone (DNMT3A/SF3B1 exon 15) had decreased at the expense of the initially smaller subclone (DNMT3A/SF3B1 exon 14), and in UPN 124, the dominant (SF3B1) and minor (TET2 and EZH2) mutations had decreased. In UPN 130, the dominant SF3B1 mutation had re-increased, whereas the 2 minor TET2 and KIT mutants remained poorly detectable (Figure 6 and supplemental Figure 3).
In 12 of 14 patients who relapsed or transformed, the dominant clone re-expanded, whereas it remained stable in UPN 077 and UPN 019 compared to levels at inclusion. In 1 case (UPN 098), AML transformation at 15 months was associated with the sudden acquisition of an EZH2 mutation in the dominant SF3B1/DNMT3A/TET2 clone whose size had significantly decreased at 11 months (Figure 6). In 4 other cases, minor subclones defined by mutations in DNMT3A (UPN 013), DNMT3A and ASXL1 (UPN 033), TET2 (UPN 076), or JAK2 (UPN 110) increased concomitantly to the loss of treatment efficacy (Figure 6; supplemental Figure 3; Table 2).
Together, these results indicate that although response to lenalidomide is often associated with a reduction in the size of the dominant subclone, it neither eradicates this clone nor prevents its re-expansion, which is sometimes associated with the emergence of minor mutations leading to loss of efficacy or AML transformation.
Discussion
The current study indicates that the presence of a DNMT3A mutation could be predictive of a better response of non-del(5q) lower-risk MDS patients to treatment. Most important, the exploration of clonal evolution under lenalidomide or lenalidomide plus EPO supports that therapeutic response is associated (in 15 of 20 patients [75%]) with a decrease in the size of the dominant clone, which re-expands upon relapse without being overtaken by an initially minor clone.
Some recurrent somatic mutations have an influence on overall survival in MDS, with ASXL1 and DNMT3A mutations being linked to a poorer prognosis, and with SF3B1 mutations linked to a better outcome.11,17-19 The prognostic value of specific mutations may vary, however, depending on the treatment administered. For example, although TET2 mutation has an overall “neutral” prognostic value in MDS, it may be associated with better response to hypomethylating agents.20,21 Our previous results also suggest that the negative prognostic value of ASXL1 in chronic myelomonocytic leukemia22,23 could be reverted by the use of demethylating agents.24 The present cohort linked to the GFM-Len-Epo-08 clinical trial prospectively enrolled ESA-resistant patients, including 61 patients with RARS or RCMD-RS, leading to an elevated percentage of SF3B1 mutations. This bias is partially explained by the fact that RCMD-RS could be frequently resistant to ESA.25 Here, we show that DNMT3A mutations could define a subgroup of non-del(5q) MDS patients with resistance to EPO that better responds to treatment. Because only 1 of 19 patients with DNMT3A mutation had no SF3B1 mutation, it is difficult to sort out the complex relationship between DNMT3A mutation or double DNMT3A/SF3B1 mutation and response to treatment. Furthermore, no interaction has been found between treatment arm and mutation. However, the interaction test may lack power; therefore, the possibility of such interaction remains. Confirmatory analyses in an independent cohort are needed.
Whole exome sequencing revealed an average of 10 somatic mutations per MDS patient, all of which were thought to arise in the same CD34+ cell.26,27 In our series, targeted sequencing before treatment identified 1 or several dominant mutations in the 42 studied patients and at least 1 minor subclone in 29 of 42 (69%) patients. Genotyping of the CD34+CD38−-derived colonies enabled us to backtrack all the genetic lesions identified by targeted sequencing. The proportion of mutant CD34+CD38− cells well reflected the variant allele frequency in the bulk of BM mononuclear cells, suggesting that the selective advantage of mutant cells occurred at the stem cell level. The fact that the dominant subclone was identified in the HSC compartment in tested patients suggested that clonal dominance might emerge in this stem cell compartment. Such an early clonal dominance of a rare initiating cell recently highlighted in MDS28 has also been observed in chronic myelomonocytic leukemia,29 and could distinguish MDS and MDS/myeloproliferative neoplasms from myeloproliferative neoplasms in which clonal dominance is amplified by differentiation.30 Our clonal analyses demonstrated not only the linear acquisition of mutations but also the possibility of branching events, as observed in post-MDS AML.26 We identified 2 cases with 2 different splice gene mutations, that is, SF3B1/ZRSR2 (UPN 030) and SF3B1/SF3B1 (UPN 091). Equivalent observations have recently been made in age-related clonal hematopoiesis.31 However, splice gene mutations have been initially described as mutually exclusive events in myeloid malignancies.12,17,32,33 Our analysis of clonal architecture at the single-cell level showed that these mutations belonged to distinct clones.
Among the 42 patients analyzed after 4 cycles of lenalidomide, a decrease in the mutant allele burden of the dominant subclone was observed in 25 patients, of whom 15 were responders. Several explanations could account for this decrease. Lenalidomide could either alter the competitiveness of the dominant subclone or improve the fitness of 1 or several minor subclones, thus decreasing the ratio between dominant and nondominant subclones. Lenalidomide could also improve the fitness of normal cells and promote the expansion of a nonclonal hematopoiesis, thus decreasing the proportion of clonal cells, as observed in UPN 054 (Figure 5). In addition, lenalidomide could theoretically promote the emergence of 1 or several subclones that carry a distinct pattern of mutations, which we did not observe in this series. In some cases in which the size of the dominant subclone had decreased, 1 major mutation could remain stable or increased. This finding suggests that lenalidomide might increase the fitness of a less-mutated subclone that, in many cases, may be the founding clone (UPNs 030 and 129). Lastly, the decrease in the size of the dominant subclone could reflect a treatment-induced change in the proportion of myeloid vs lymphoid cells in the BM mononuclear cell fraction. In a patient with 2 SF3B1 variants (UPN 101), we sorted CD3+ and CD19+ lymphoid cells and identified in the CD19+ fraction, but not in the CD3+ fraction, 1 of the SF3B1 mutations found in CD3−CD19− cells. Of interest, after treatment, the allele burden of this SF3B1 variant decreased in both CD19+ and CD3−CD19− cells. The other SF3B1 mutation was detected neither in CD3+ nor in CD19+ cells, but was present in CD3−CD19− cells and disappeared upon therapy. Therefore, we could not exclude that amplification of nonmutated lymphoid cells may lead to a relative decrement of mutant allele burden in BM mononuclear cells.
Clonal architecture experiments at the single-cell level allowed backtracking the founding mutation. For example, in UPN 091, DNMT3A appeared as the founding mutation, which level remained stable under treatment, whereas the dominant clone containing both DNMT3A and SF3B1 exon 15 mutations decreased. In agreement with the observation that CD34+CD38−/low CD90+ stem cells with del(5q) persist under lenalidomide therapy in del(5q) MDS patients in remission,34 here, treatment did not eliminate the DNMT3A-mutated HSC population that may constitute a cellular reservoir for further clonal progression.35,36 These results suggest the possibility that a preleukemic cell with predisposing mutations (ie, TET2 or DNMT3A) appeared at a silent preclinical stage.35-37 Re-expansion of the founding clone and of hematopoiesis devoid of recurrent mutations was not sufficient to induce a definitive clinical response to treatment (UPN 030). By contrast, the evolution of minor clones was not correlated with response to treatment. Long-term follow-up showed that the loss of treatment efficacy was associated with the re-increase of the dominant clone in the majority of cases, but that limited expansion of an initially minor clone remained an option. Differences in the evolution of subclones under treatment may depend on the combination of mutations. Alternatively, these mutations could occur in subsets of HSCs with the distinct properties of aging and drug sensitivity.38,39
In conclusion, our results show that lenalidomide response is usually associated with a decrease in the size of the dominant subclone and with limited changes in the size of minor subclones. When the disease escapes lenalidomide activity or progresses to acute leukemia, the dominant subclone re-expands. Further investigation will indicate whether lenalidomide eradicates part of the cells of the dominant subclone or decreases their competitiveness, information which will be needed to understand how these cells escape lenalidomide activity and to optimize the clinical use of this drug.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank Elli Papaemmanuil for helpful discussion, Raphael Petit and Rosa Sapena for recording clinical data, and Audrey Gauthier and Marlène Dejean for technical assistance.
This study was supported by a research grant from Celgene; by Assistance Publique–Hôpitaux de Paris through the Unité de Recherche Clinique Paris Descartes Necker Cochin; by Institut National du Cancer by the Programme Hospitalier de Recherche Clinique (PHRC, MDS-04); and by fellowship grants from the Ministère de l’Enseignement supérieur et de la Recherche (France) (V.C.) and from the Site de Recherche Intégrée sur le Cancer for CARPEM (Cancer Research for PErsonalized Medicine) (O.K.).
Authorship
Contribution: M.F. and O.K. designed the study; V.C., A. Renneville, M.P., and A. Raimbault performed the experiments; M.F., O.K., A. Renneville, C.P., J. Lambert, and V.C. analyzed and interpreted the data; A.T. and F. Dreyfus designed the clinical trial; A.T., F. Dreyfus, J.D., C.R., O.B.-R., and A.S. recorded patient and clinical data; J. Lambert, J. Lejeune, S.C., F. Dumont, and V.C. performed the statistical analyses; and M.F., P.F., and E.S. wrote the paper.
Conflict-of-interest disclosure: M.F. has received research funding from Celgene. P.F. has received honoraria and research funding from Celgene. The remaining authors declare no competing financial interests.
A complete list of the members of the Groupe Francophone des Myélodysplasie appears in “Appendix.”
Correspondence: Michaela Fontenay, Service d’Hématologie Biologique, Hôpital Cochin, 27 rue du Faubourg Saint-Jacques, 75014 Paris, France; e-mail: michaela.fontenay@inserm.fr; and Olivier Kosmider, Service d’Hématologie Biologique, Hôpital Cochin, 27 rue du Faubourg Saint-Jacques, 75014 Paris, France; e-mail: olivier.kosmider@aphp.fr.
Appendix: study group members
List of the Groupe Francophone des Myélodysplasie members: Anna Banos (Centre Hospitalier Universitaire, Strasbourg, France); Agnes Guerci-Bresler (Centre Hospitalier Universitaire, Nancy, France); Stefan Wickenhauser (Centre Hospitalier Universitaire, Nimes, France); Denis Caillot (Centre Hospitalier Universitaire, Dijon, France); Kamel Laribi (Centre Hospitalier, Le Mans, France); Benoit De Renzis (Centre Hospitalier Universitaire, Clermont Ferrand, France); Dominique Bordessoule (Centre Hospitalier Universitaire, Limoges, France); Claude Gardin (Assistance Publique–Hôpitaux de Paris, Hôpital Avicenne, and Paris 13 University, Bobigny, France); Bohrane Slama (Centre Hospitalier, Avignon, France); Laurence Sanhes (Centre Hospitalier, Perpignan, France); Berangere Gruson (Centre Hospitalier Universitaire, Amiens, France); Pascal Cony-Makhoul (Centre Hospitalier, Annecy, France); Bachra Chouffi (Centre Hospitalier, Boulogne sur Mer, France); Celia Salanoubat (Centre Hospitalier, Corbeil, France); Riad Benramdane (Centre Hospitalier, Pontoise, France); Laurence Legros (Centre Hospitalier Universitaire, Nice, France); Eric Wattel (Centre Hospitalier Edouard Herriot, Lyon, France); Gerard Tertian (Assistance Publique–Hôpitaux de Paris, Kremlin Bicetre, France), Kamal Bouabdallah (Centre Hospitalier Universitaire, Bordeaux, France); François Guilhot (Centre Hospitalier Jean Bernard, Poitiers, France); Anne-Laure Taksin (Centre Hospitalier, Versailles, France); Stephane Cheze (Centre Hospitalier Universitaire, Caen, France); Karim Maloum (Assistance Publique–Hôpitaux de Paris, Hôpital Pitié-Salpetrière, and Pierre et Marie Curie University, Paris, France); Stanislas Nimuboma (Centre Hospitalier Universitaire, Rennes, France); Carole Soussain (Centre René Huguenin, Saint Cloud, France); Francoise Isnard (Assistance Publique–Hôpitaux de Paris, Hôpital Saint-Antoine, and Pierre et Marie Curie University, Paris, France); Emmanuel Gyan (Centre Hospitalier Universitaire, Tours, France).


![Figure 4. Evolution of variant allele frequency of dominant mutations in responders and nonresponders to lenalidomide after short-term treatment. (A) Variant allele frequency of each major mutation was shown before (0 cycles) and after 4 cycles of treatment in responders (n = 20; left) and nonresponders (n = 22; right). (B) Variation of variant allele frequency was calculated as follow-up minus screening for each responder and each nonresponder. Box plots represent medians and IQRs [25%–75%], and whiskers extend to 1.5 times the IQR. Variations are compared between responders and nonresponders using the Mann Whitney U test.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/127/6/10.1182_blood-2015-04-640128/4/m_749f4.jpeg?Expires=1765906036&Signature=qKdn8XHcStoiyLEawuAFoU3jLFwqGLrcfoLMjuj05IJoIYuC38KZ1jlA4PoTxc2voOb9UDiu42DA8qLp8YMHEv2K~lq6xWhiT4JF3gKMLm6wcEfWTr85e42I4Hb0BbCnYn5hKp6~pDSb3YHMR8O1iDRvus2KElinrvdlhUXR1hwcNWiwEODwXefA6k2xawyMUbb4SmAAjo~iY83RxJ3f7FUpVLaG2iV5q6LJDmFHxw32PyNH-wMNpTikoHI4uIEPAsLF5awsn8StOJno5wg7lkTFjMUAOwvkonhOCySP0dI752u08nCUThw-vIMrFaTsEA5CnydzVRZH2pnyrfUbVQ__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)