Key Points
An HA subject with a multiexon F8 deletion showed a highly clonal response to 1 FVIII epitope via an immunodominant TCR.
The same HLA-DRA*01-DRB1*01:01-restricted FVIII epitope was recognized by T cells from 3 HA subjects.
Abstract
Factor VIII (FVIII)–neutralizing antibodies (“inhibitors”) are a serious problem in hemophilia A (HA). The aim of this study was to characterize HLA-restricted T-cell responses from a severe HA subject with a persistent inhibitor and from 2 previously studied mild HA inhibitor subjects. Major histocompatibility complex II tetramers corresponding to both of the severe HA subject’s HLA-DRA-DRB1 alleles were loaded with peptides spanning FVIII-A2, C1, and C2 domains. Interestingly, only 1 epitope was identified, in peptide FVIII2194-2213, and it was identical to the HLA-DRA*01-DRB1*01:01-restricted epitope recognized by the mild HA subjects. Multiple T-cell clones and polyclonal lines having different avidities for the peptide-loaded tetramer were isolated from all subjects. Only high- and medium-avidity T cells proliferated and secreted cytokines when stimulated with FVIII2194-2213. T-cell receptor β (TCRB) gene sequencing of 15 T-cell clones from the severe HA subject revealed that all high-avidity clones expressed the same TCRB gene. High-throughput immunosequencing of high-, medium-, and low-avidity cells sorted from a severe HA polyclonal line revealed that 94% of the high-avidity cells expressed the same TCRB gene as the high-avidity clones. TCRB sequencing of clones and lines from the mild HA subjects also identified a limited TCRB gene repertoire. These results suggest a limited number of epitopes in FVIII drive inhibitor responses and that the T-cell repertoires of FVIII-responsive T cells can be quite narrow. The limited diversity of both epitopes and TCRB gene usage suggests that targeting of specific epitopes and/or T-cell clones may be a promising approach to achieve tolerance to FVIII.
Introduction
The development of factor VIII (FVIII)–neutralizing antibodies (“inhibitors”) is the most serious complication of hemophilia A (HA) treatment.1 Inhibitors occur more frequently in severe than in mild or moderate HA.2-4 Inhibitor risk is associated with genetic and nongenetic factors.5 An important predictor of inhibitor development is the F8 mutation, with large deletions and nonsense mutations associated with greater risk.6-8 F8 missense mutations are the most common cause of mild HA, and some of these carry a higher inhibitor risk.9-11
The FVIII inhibitor response is dependent on CD4 T-cell help.12-15 Protein antigens are taken up by antigen-presenting cells that process and present peptides that bind to a polymorphic groove on major histocompatibility complex class II (MHCII) proteins.16 The MHCII alleles17 carried by an individual determine which peptides can be presented to his or her immune system. The peptide-MHCII complex may (or may not) then be recognized by 1 of millions of T-cell receptors (TCRs) on T-helper (Th) cells.18 The MHCII-peptide-TCR interaction plus costimulation signals activate cytokine production promoting B-cell maturation into antibody-secreting plasma cells. Interactions between naturally processed FVIII peptides, MHCII, and TCRs are crucial in determining how a patient’s immune system will respond to FVIII replacement therapy and, subsequently, if inhibitors develop, how he or she might respond to immune tolerance induction (ITI) via intensive FVIII therapy.
FVIII consists of 2332 amino acids; thus, in principle many T-cell epitopes could contribute to inhibitor development in severe HA subjects who do not express this protein. The Conti-Fine group characterized CD4 T-cell proliferation in response to FVIII peptides spanning the A2, A3, and C2 domains.19-22 Jones et al identified a FVIII-C1 domain epitope in a severe HA subject using expanded polyclonal T-cell lines to perform comprehensive FVIII T-cell epitope mapping.23 Moise et al used computational prediction, HLA-DR peptide binding assays, and immunizations of HLA-DRA*01-DRB1*03:01 and -DRB1*04:01 transgenic mice to identify 6 immunogenic peptides in the FVIII-C2 domain.24 Van Haren et al investigated naturally processed FVIII peptides by sequencing peptides eluted from HLA-DR on dendritic cells isolated from HLA-DRB1-typed donors.25,26 These studies identified 6 to 18 FVIII core peptides per individual that could potentially be recognized by their T cells. Finally, Steinitz et al performed comprehensive T-cell epitope mapping using humanized HLA-DRA*01-DRB1*15:01 transgenic HA mice, identifying 8 HLA-DRA*01-DRB1*15:01-restricted epitopes in FVIII.27
MHCII tetramer-guided epitope mapping (TGEM) has proved useful in mapping HLA-restricted T-cell epitopes.28-30 FVIII-specific T cells have been cloned from tetramer-positive cells and by limiting dilution,31 providing definitive evidence of antigen specificity. T-cell clones have been isolated from mild HA inhibitor subjects with mutations R2150H,31 A2201P,28,29 and R593C.30 In each case, the epitope encompassed the missense mutation. FVIII-specific T-cell clones isolated from subjects with severe HA have not yet been reported. The TCRs that correspond to FVIII epitopes are largely unknown, with the exception of one that was sequenced and used to engineer antigen-specific T-regulatory cells.32 Earlier studies analyzed TCR β-chain variable-region (TCRBV) repertoires of inhibitor-positive subjects after stimulation of peripheral blood mononuclear cells (PBMCs) with FVIII33 or during ITI.34
Herein, experiments to define FVIII epitopes recognized by T cells from a severe HA subject with a multiexon deletion and a persistent high-titer inhibitor were carried out. This subject had MHCII alleles HLA-DRA*01-DRB1*01:01 (DRB1*01:01) and HLA-DRA*01-DRB1*10:01 (DRB1*10:01). We hypothesized that he would respond to the same DRB1*01:01-restricted epitope in FVIII2194-2213 identified previously in 2 mild HA subjects with missense mutation A2201P.28,29 Immunosequencing35 of TCRB genes in clones, polyclonal lines, and PBMCs isolated from these subjects was carried out to characterize the TCRB repertoires of their FVIII-specific CD4 T cells.
Materials and methods
Subjects and blood samples
Subjects were enrolled in “Genetic Studies in Hemophilia and von Willebrand Disease” (GS1) and provided informed consent according to the Principles of Helsinki. Institutional review board protocols were approved by the Seattle Children’s Hospital, University of Washington, and/or Uniformed Services University of the Health Sciences institutional review boards. Blood samples were obtained from an adult severe HA subject, GS1-56A, who had a persistent high-titer inhibitor with a peak titer of >2000 BU/mL measured ∼1 year prior to enrollment. His HLA-DRB1 genes were DRB1*01:01, DRB1*10:01. Exons 7 to 13 in his F8 gene were deleted, and he had failed ITI therapy. FVIII antigen was undetectable in his plasma (supplemental Data, available on the Blood Web site). Mild HA subjects GS1-17A28,29,36 and GS1-32A29,36 with missense substitution FVIII-A2201P and T-cell clones isolated from these subjects36 were described previously. T-cell lines were isolated from a subsequent blood sample collected from GS1-17A ∼5 years after his initial 250 BU/mL inhibitor was detected, at which time the titer had decreased to 2 to 13 BU/mL. Blood samples were also obtained from HLA-DRB1*01:01-mismatched and matched healthy controls. PBMCs were obtained from all samples by Ficoll-Paque PLUS (GE Healthcare, Piscataway, NJ) underlay and either frozen in 7% dimethyl sulfoxide (DMSO; Sigma-Aldrich, St. Louis, MO) in fetal bovine serum (FBS) (HyClone) or used immediately.
Epitope mapping
Overlapping 20-mer peptides (with a 12-amino-acid overlap) spanning the FVIII-A2, C1, and C2 domains were from Mimotopes (Clayton, VIC, Australia). Their sequences and the peptide composition of pools used for epitope mapping are in supplemental Table 1. Control tetanus toxoid (TT) peptides to detect DRB1*01:0137 and DRB1*10:01-restricted responses38 were either from Mimotopes or the Benaroya Research Institute Tetramer Core (Seattle, WA). Lyophilized peptides were suspended in 100% DMSO or 80% DMSO/20% water. Monomeric and tetrameric HLA-DR proteins were from the Benaroya Research Institute Tetramer Core. T-cell epitopes in FVIII were predicted using ProPred,39 NetMHCIIpan,40 and Immune Epitope Database41,42 MHCII-binding computer algorithms. Affinities of peptides for recombinant DRB1*01:01 and DRB1*10:01 proteins were determined by competition assays.28,30 TGEM was performed as described.28-30 Cells from GS1-56A were stimulated initially with pools of 5 FVIII peptides having sequences spanning the FVIII-A2, C1, and C2 domains. As positive controls, aliquots were stimulated with TT peptides previously shown to evoke DRB1*01:01 and DRB1*10:01-restricted T-cell responses.37,38 After 2 weeks of expansion, the cells were analyzed using DRB1*01:01 and DRB1*10:01 tetramers loaded with the same peptide pools used for stimulation. Tetramer-positive responses were then “decoded” by a second round of staining using tetramers carrying the individual peptides comprising the peptide pool. All flow cytometric data were analyzed using FlowJo software (FlowJo LLC, Ashland, OR).
T-cell clones, assays, and TCRB sequencing
T-cell clones were isolated by single-cell sorting of GS1-56A cells stained with DRB1*01:01 tetramers loaded with C2-peptide-pool 1 or FVIII2194-2213 using a fluorescence-activated cell sorter (FACS) Aria (BD Biosciences, San Jose, CA).29,43 Polyclonal T-cell lines were isolated from GS1-17A and GS1-56A by sorting 250 tetramer-positive cells per well following the same method used for single-cell sorting. T-cell clones and lines were expanded by stimulation with irradiated PBMCs from an HLA-mismatched individual plus phytohemagglutinin (Remel, Lenexa, KS) in the presence of natural human IL-2 (Hemagen Diagnostics Inc., Columbia, MD) in 15% human serum T-cell medium (supplemental Data).
T-cell clones and lines were incubated with phycoerythrin (PE)–labeled tetramers loaded with relevant or irrelevant peptides at 37°C for 1 hour in T-cell medium, then labeled with fluorescein isothiocyanate (FITC)–anti-human CD25 (Biolegend, San Diego, CA), PerCP-anti-human CD3 (BD Biosciences), and allophycocyanin (APC)–anti-human CD4 (eBioscience, San Diego, CA) immunoglobulin Gs (IgGs) for 20 minutes at 4°C. Cells were washed with Ca++/Mg++-free Dulbecco’s phosphate-buffered saline (ThermoFisher Scientific), 1% FBS, and 0.1% sodium azide and analyzed on a FACSCaliber (BD Biosciences). Proliferation of cells in response to relevant or irrelevant peptides was measured with a [3H]thymidine incorporation assay.29 Secretion of interferon-γ (IFN-γ), interleukin-4 (IL-4), IL-17A, and IL-21 in cell supernatants was measured by sandwich enzyme-linked immunosorbent assays (ELISAs).30,36
RNA was isolated from T-cell clones using an RNeasy mini kit (Qiagen, Valencia, CA). For 56A clones, complementary DNA (cDNA) was generated from 250 ng total RNA using a QuantiTect reverse transcription kit (Qiagen). TCRB-CDR3 regions were sequenced by Adaptive Biotechnologies (Seattle, WA).18,44,45 For 17A and 32A clones, cDNA was prepared by reverse transcription of 1 μg total RNA with random hexamers (supplemental Data). TCRBV typing was performed using multiplex polymerase chain reaction (PCR).46 PCR bands were sequenced by BigDye Terminator Cycle Sequencing (Applied Biosystems) using the TCRBC primer46 and the specific TCRBV primer.46
FVIII-specific T-cell line populations were sorted based on their avidity for the DRB1*01:01-FVIII2194-2213 tetramer. Cells were labeled by incubating 107 cells in 2.5 mL of T-cell medium with 50 μL PE-DRB1*01:01-FVIII2194-2213 tetramer for 1 hour, then with 31 μL FITC-anti-human CD4 IgG (eBioscience) for 20 minutes at 4°C. Cells were washed with Ca++/Mg++-free Dulbecco’s phosphate-buffered saline and 1% FBS and resuspended in 1 mL of the same buffer. Cell populations were sorted on a FACSAria. Genomic DNA was isolated from cell pellets, and TCRB-CDR3 deep sequencing was performed by Adaptive Biotechnologies.18,44,45
Results
Epitope mapping
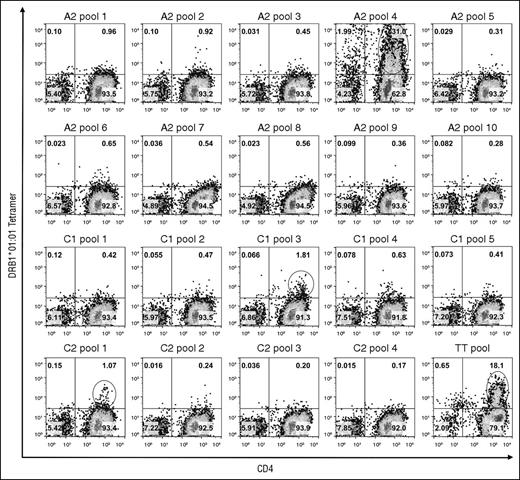
PBMCs obtained from 2 blood samples collected from GS1-56A over 2 months were used to search for T-cell epitopes in the FVIII-A2, C1, and C2 domains restricted to either of his HLA-DRA-DRB1 alleles. Tetramer staining of cells stimulated with 20 pools of overlapping FVIII peptides using DRB1*01:01 and DRB1*10:01 tetramers is shown in Figure 1 and supplemental Figure 1, respectively. Six peptide-loaded tetramers bound to ≥1% of CD4+ T cells. An additional 12 tetramers bound to >0.5% and <1.0% of the cells. These staining patterns were consistent with nonspecific or low-avidity tetramer binding. Staining of positive control TT peptide-stimulated cells with the corresponding DRB1*01:01 and DRB1*10:01 tetramers demonstrated large populations of tetramer-positive CD4+ cells.
TGEM of CD4 T cells from subject GS1-56A using DRB1*01:01 tetramers loaded with pooled FVIII peptides. CD4 T cells were stimulated with pools of peptides having overlapping sequences spanning the FVIII-A2, C1, and C2 domains and then incubated with DRB1*01:01 tetramers loaded with the corresponding peptide pools on day 14. As a positive control, cells from the same subject were stimulated with peptides corresponding to known DRB1*01:01-restricted TT epitopes37 : TT585-605, TT666-685, and TT674-693. Plots show binding of CD3+ lymphocytes to APC-anti-CD4 IgG and PE-tetramer. The name of the peptide pool used for staining is indicated above each plot. The cutoff used for selecting pools for staining using individual peptides (decoding) was 1.0% CD4+Tetramer+ cells. These populations are circled. Pseudocolor plots with percentages of cells in each quadrant were created with FlowJo v10.0.7.
TGEM of CD4 T cells from subject GS1-56A using DRB1*01:01 tetramers loaded with pooled FVIII peptides. CD4 T cells were stimulated with pools of peptides having overlapping sequences spanning the FVIII-A2, C1, and C2 domains and then incubated with DRB1*01:01 tetramers loaded with the corresponding peptide pools on day 14. As a positive control, cells from the same subject were stimulated with peptides corresponding to known DRB1*01:01-restricted TT epitopes37 : TT585-605, TT666-685, and TT674-693. Plots show binding of CD3+ lymphocytes to APC-anti-CD4 IgG and PE-tetramer. The name of the peptide pool used for staining is indicated above each plot. The cutoff used for selecting pools for staining using individual peptides (decoding) was 1.0% CD4+Tetramer+ cells. These populations are circled. Pseudocolor plots with percentages of cells in each quadrant were created with FlowJo v10.0.7.
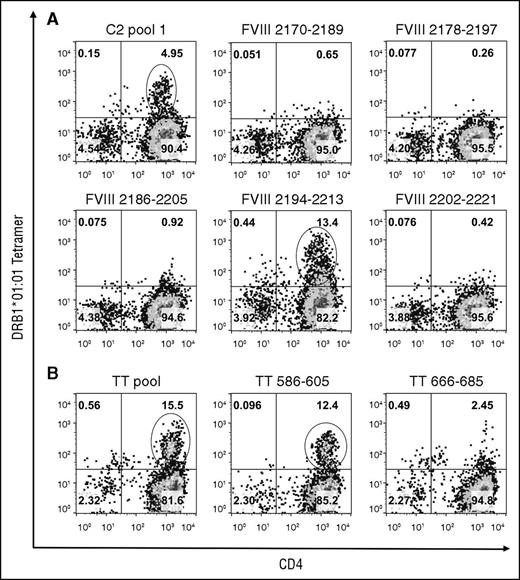
The CD4+ T cells were stained after 4 more days in culture using tetramers loaded with the individual peptides comprising the pools that produced ≥1% tetramer-positive staining. Decoding of the DRB1*01:01-FVIII-C2 pool 1 tetramer staining identified a cell population that bound to the DRB1*01:01-FVIII2194-2213 tetramer (Figure 2). The DRB1*01:01-FVIII2186-2205 tetramer bound fewer cells with lower avidity. Decoding of the other pools did not identify additional epitopes (supplemental Figure 2). An unusually high percentage of cells bound to DRB1*01:01 and DRB1*10:01 tetramers loaded with the A2 pool 4 peptides, which included FVIII508-527, and with FVIII508-527 alone, in a pattern indicative of nonspecific staining. FVIII508-527 binding to DRB1*01:01 and DRB1*10:01 was tested experimentally yielding 50% inhibitory concentration values >20 μM, which is considerably higher than the 1000 nM affinity threshold defined for MHCII binding,47-49 whereas FVIII2194-2213 bound to DRB1*01:01 with an 50% inhibitory concentration of 0.33 μM (supplemental Figure 3).
Decoding of GS1-56A pooled-peptide tetramer-positive staining results. CD4 T cells stimulated with FVIII-C2-pool 1 peptides (A) or with positive control DRB1*01:01-restricted TT peptides (B) were stained on day 18 using tetramers loaded with the corresponding peptide pool and with the individual peptides comprising the pool. Plots show binding of CD3+ lymphocytes to APC-anti-CD4 IgG and PE-tetramer. The name of the peptide pool or individual peptide is indicated above each plot. Pseudocolor plots with percentages of cells in each quadrant were created with FlowJo v10.0.7. Decoding identified strong HLA-DRB1*01:01-restricted T-cell responses to FVIII2194-2213 and to TT586-605. Decoding of A2-pool 4 and C1-pool 3 tetramer-positive staining results (Figure 1) indicated both were because of nonspecific binding (supplemental Data).
Decoding of GS1-56A pooled-peptide tetramer-positive staining results. CD4 T cells stimulated with FVIII-C2-pool 1 peptides (A) or with positive control DRB1*01:01-restricted TT peptides (B) were stained on day 18 using tetramers loaded with the corresponding peptide pool and with the individual peptides comprising the pool. Plots show binding of CD3+ lymphocytes to APC-anti-CD4 IgG and PE-tetramer. The name of the peptide pool or individual peptide is indicated above each plot. Pseudocolor plots with percentages of cells in each quadrant were created with FlowJo v10.0.7. Decoding identified strong HLA-DRB1*01:01-restricted T-cell responses to FVIII2194-2213 and to TT586-605. Decoding of A2-pool 4 and C1-pool 3 tetramer-positive staining results (Figure 1) indicated both were because of nonspecific binding (supplemental Data).
The ProPred and Immune Epitope Database programs predicted that 28 and 15 core FVIII sequences, respectively, could bind DRB1*01:01 (supplemental Tables 2-3). DRB1*10:01 predictions were available only using NetMHCIIpan, which predicted 30 core regions (supplemental Table 4). The FVIII sequence identified by TGEM was among the predicted DRB1*01:01-restricted epitopes. Peptide FVIII508-527 was not predicted to bind DRB1*01:01 or DRB1*10:01.
T-cell clones, assays, and TCRB sequencing
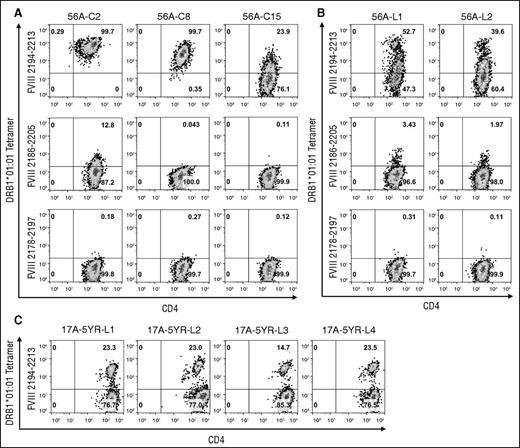
Identification of FVIII2194-2213 as containing a DRB1*01:01-restricted T-cell epitope recognized by this subject was further validated by generating T-cell clones and polyclonal lines expanded from sorted tetramer-positive cells. The clones showed a wide range of median fluorescence intensity (MFI) values (Table 150,51 ) indicating different tetramer-binding avidities. Clone 56A-C2 bound with high avidity to the DRB1*01:01-FVIII2194-2213 tetramer and with low avidity to the DRB1*01:01 tetramer loaded with the overlapping peptide FVIII2186-2205 (Figure 3A). Clones 56A-C8 and 56A-C15 bound with medium and low avidity, respectively, to the DRB1*01:01-FVIII2194-2213 tetramer. Clones were not stained with the irrelevant tetramer DRB1*01:01-FVIII2178-2197. Two lines were derived that showed similar staining patterns to the DRB1*01:01-FVIII2194-2213 tetramer with high (fluorescent intensity >400 units) and medium (∼150-300 fluorescent units) avidity populations in addition to a large low-avidity population (Figure 3B). An additional blood sample was obtained from GS1-17A 5 years after he developed an inhibitor. Four T-cell lines that contained a population of cells (15% to 23%) that bound to the DRB1*01:01-FVIII2194-2213 tetramer were isolated (Figure 3C).
DRB1*01:01-FVIII2194-2213 tetramer staining avidity and TCRB gene sequence analysis for T-cell clones isolated from severe HA subject GS1-56A
| Clone* . | Tetramer staining† . | TCRBV gene‡ . | TCRBD gene‡ . | TCRBJ gene‡ . | V-D-J junction‡,§ . | Sequence identifier|| . |
|---|---|---|---|---|---|---|
| 56A-C1 | 974 | 27*01 | Not identified | 1-1*01 | CASSFDPLNTEAFF | Seq01 |
| 56A-C2 | 643 | 27*01 | Not identified | 1-1*01 | CASSFDPLNTEAFF | Seq01 |
| 56A-C3 | 7 | 18*01 | 2*01 or 2*02¶ | 2-7*01 | CASSAGLAETYEQYF | Seq02 |
| 56A-C4 | 869 | 27*01 | Not identified | 1-1*01 | CASSFDPLNTEAFF | Seq01 |
| 56A-C6 | 896 | 27*01 | Not identified | 1-1*01 | CASSFDPLNTEAFF | Seq01 |
| 56A-C7 | 613 | 27*01 | Not identified | 1-1*01 | CASSFDPLNTEAFF | Seq01 |
| 56A-C8# | 202 | 7-8*01 or *03 | 2*01 or 2*02 | 2-3*01 | CASSSAPRLTSGRTDTQYF | Seq03 |
| 56A-C9 | 708 | 27*01 | Not identified | 1-1*01 | CASSFDPLNTEAFF | Seq01 |
| 56A-C10 | 3 | 3-1 or 3-2 | 1*01 | 1-5*01 | CASSQGHGNQPQHF | Seq04 |
| 56A-C11 | 909 | 27*01 | Not identified | 1-1*01 | CASSFDPLNTEAFF | Seq01 |
| 56A-C12 | 907 | 27*01 | Not identified | 1-1*01 | CASSFDPLNTEAFF | Seq01 |
| 56A-C13 | 26 | 20-1** | 1*01 | 2-7*01 | CSARGQGAYEQYF | Seq05 |
| 56A-C14 | 852 | 27*01 | Not identified | 1-1*01 | CASSFDPLNTEAFF | Seq01 |
| 56A-C15 | 5 | 9†† | 1*01 | 1-2*01 | CASSVAGTGPQVLYGYTF | Seq06 |
| 56A-C16 | 648 | nd | nd | nd | nd | |
| 56A-C17 | 804 | nd | nd | nd | nd | |
| 56A-C18 | 402 | 27*01 | Not identified | 1-1*01 | CASSFDPLNTEAFF | Seq01 |
| Clone* . | Tetramer staining† . | TCRBV gene‡ . | TCRBD gene‡ . | TCRBJ gene‡ . | V-D-J junction‡,§ . | Sequence identifier|| . |
|---|---|---|---|---|---|---|
| 56A-C1 | 974 | 27*01 | Not identified | 1-1*01 | CASSFDPLNTEAFF | Seq01 |
| 56A-C2 | 643 | 27*01 | Not identified | 1-1*01 | CASSFDPLNTEAFF | Seq01 |
| 56A-C3 | 7 | 18*01 | 2*01 or 2*02¶ | 2-7*01 | CASSAGLAETYEQYF | Seq02 |
| 56A-C4 | 869 | 27*01 | Not identified | 1-1*01 | CASSFDPLNTEAFF | Seq01 |
| 56A-C6 | 896 | 27*01 | Not identified | 1-1*01 | CASSFDPLNTEAFF | Seq01 |
| 56A-C7 | 613 | 27*01 | Not identified | 1-1*01 | CASSFDPLNTEAFF | Seq01 |
| 56A-C8# | 202 | 7-8*01 or *03 | 2*01 or 2*02 | 2-3*01 | CASSSAPRLTSGRTDTQYF | Seq03 |
| 56A-C9 | 708 | 27*01 | Not identified | 1-1*01 | CASSFDPLNTEAFF | Seq01 |
| 56A-C10 | 3 | 3-1 or 3-2 | 1*01 | 1-5*01 | CASSQGHGNQPQHF | Seq04 |
| 56A-C11 | 909 | 27*01 | Not identified | 1-1*01 | CASSFDPLNTEAFF | Seq01 |
| 56A-C12 | 907 | 27*01 | Not identified | 1-1*01 | CASSFDPLNTEAFF | Seq01 |
| 56A-C13 | 26 | 20-1** | 1*01 | 2-7*01 | CSARGQGAYEQYF | Seq05 |
| 56A-C14 | 852 | 27*01 | Not identified | 1-1*01 | CASSFDPLNTEAFF | Seq01 |
| 56A-C15 | 5 | 9†† | 1*01 | 1-2*01 | CASSVAGTGPQVLYGYTF | Seq06 |
| 56A-C16 | 648 | nd | nd | nd | nd | |
| 56A-C17 | 804 | nd | nd | nd | nd | |
| 56A-C18 | 402 | 27*01 | Not identified | 1-1*01 | CASSFDPLNTEAFF | Seq01 |
nd, not determined.
56A-C5 is not included in the list because these cells did not expand sufficiently to permit tetramer staining.
Tetramer staining was quantitated from the MFI of staining with DRB1*01:01-FVIII2194-2213 tetramer minus MFI of staining with an irrelevant tetramer (DRB1*01:01-FVIII2178-2197). Results are from a single determination that occurred while screening clones that were expanding after single cell sorting.
Nucleotide sequences were analyzed using Adaptive Biotechnologies ImmunoSEQ Analyzer version 2.0. Gene names were determined using international ImMunoGeneTics information system/V-QUEry and STandardization (IMGT/V-QUEST) version 3.3.4.50 TCR nomenclature is in accordance with the international reference IMGT/GENE-DB.51
V-D-J junction sequence corresponds to the third hypervariable region of the TCR beginning with the second conserved cysteine encoded by the 3′ portion of the V-region and ends with the conserved phenylalanine encoded by the 5′ portion of the J-region.
Sequences were given a numerical identifier for labeling purposes. Analyzed nucleotide sequences are in supplemental Table 6.
Multiple gene names result from >1 known annotated V, D, or J gene sequence matching equally well with the aligned sequence.
56A-C8 expressed a second TCRB gene that corresponded to TCRBV27*01, TCRBD2*01 or *02, TCRBJ2-1*01 and is out of frame.
Sequence is identical to 20-1*01, *02, *03, *04, and *05 alleles.
Sequence is identical to 9*01, *02, and *03 alleles.
Tetramer staining of T-cell clones and lines isolated from severe HA subject GS1-56A and mild HA subject GS1-17A. (A) Staining of T-cell clones from subject GS1-56A with the DRB1*01:01-FVIII2194-2213 tetramer, DRB1*01:01-FVIII2186-2205 tetramer, and DRB1*01:01-FVIII2178-2197 tetramer. The clones shown are representative of those with high-, medium-, and low-avidity binding to the DRB1*01:01-FVIII2194-2213 tetramer. (B) Staining of 2 T-cell lines from subject GS1-56A with the same tetramers in panel A. (C) Staining of 4 T-cell lines from subject GS1-17A with the DRB1*01:01-FVIII2194-2213 tetramer. Polyclonal lines from both subjects were isolated by sorting of 250 cells labeled with the PE-DRB1*01:01-FVIII2194-2213 tetramer. All plots show binding of CD3+ lymphocytes to anti-CD4 IgG (FITC label panels A and B, APC label panel C) and PE-tetramer. Pseudocolor plots with percentages of cells in each quadrant were created with FlowJo v10.0.7.
Tetramer staining of T-cell clones and lines isolated from severe HA subject GS1-56A and mild HA subject GS1-17A. (A) Staining of T-cell clones from subject GS1-56A with the DRB1*01:01-FVIII2194-2213 tetramer, DRB1*01:01-FVIII2186-2205 tetramer, and DRB1*01:01-FVIII2178-2197 tetramer. The clones shown are representative of those with high-, medium-, and low-avidity binding to the DRB1*01:01-FVIII2194-2213 tetramer. (B) Staining of 2 T-cell lines from subject GS1-56A with the same tetramers in panel A. (C) Staining of 4 T-cell lines from subject GS1-17A with the DRB1*01:01-FVIII2194-2213 tetramer. Polyclonal lines from both subjects were isolated by sorting of 250 cells labeled with the PE-DRB1*01:01-FVIII2194-2213 tetramer. All plots show binding of CD3+ lymphocytes to anti-CD4 IgG (FITC label panels A and B, APC label panel C) and PE-tetramer. Pseudocolor plots with percentages of cells in each quadrant were created with FlowJo v10.0.7.
Three clones and 1 line with cells having a range of DRB1*01:01-FVIII2194-2213 tetramer avidities were examined for proliferation and cytokine secretion in response to FVIII2194-2213. The high- and medium-avidity clones 56A-C2 and 56A-C8 proliferated with 50% effective concentration values of ∼0.2 μM and ∼50 μM, respectively (Figure 4A). The low-avidity clone 56A-C15 did not proliferate over background levels. T-cell line 56A-L2 had a 50% effective concentration value of ∼0.2 μM, but the proliferation level at each concentration tested was lower. IFN-γ and IL-4, but not IL-17A and IL-21, were detected in cell supernatants for 56A-C2, 56A-C8, and 56A-L2 (Figure 4B-C), and their IFN-γ:IL-4 ratio varied (Figure 4D). 56A-C2 secreted large amounts of IFN-γ and IL-4 with an IFN-γ:IL-4 ratio of ∼3. 56A-C8 secreted mostly IL-4 with IFN-γ:IL-4 ratios <<1. The T-cell line 56A-L2 secreted predominantly IFN-γ with IFN-γ:IL-4 ratios of ∼40.
Proliferation and cytokine secretion of subject GS1-56A T cells in response to FVIII2194-2213. Three T-cell clones and 1 line (10 000 cells) were stimulated with FVIII2194-2213 or irrelevant peptide (FVIII2178-2197) at concentrations from 0.01 to 100 μM presented by 100 000 irradiated PBMCs from a DRB1*01:01-typed donor. Forty-eight hours later, 50 μL of cell supernatant was removed from each well for cytokine analysis and replaced with 1 μCi [3H]thymidine. Cells were harvested after 18 hours of further incubation, and [3H]thymidine incorporation was measured. Cell supernatants in quadruplicate wells were pooled for measurement of IFN-γ, IL-4, IL-17A, and IL-21 by sandwich ELISAs using purified recombinant cytokines to generate standard curves. (A) Proliferation results are expressed as averages of quadruplicate determinations ± standard deviations. Proliferation in response to the FVIII2178-2197 peptide was negligible when compared with proliferation in the absence of peptide. Background proliferation levels: 56A-C2, 171.0 ± 69.9 counts per minute (cpm); 56A-C8, 84.2 ± 10.9 cpm; 56A-C15, 99.4 ± 7.2 cpm; and 56A-L2, 209.0 ± 61.4 cpm. (B) IFN-γ secretion levels. (C) IL-4 secretion levels. (D) IFN-γ:IL-4 ratios. IL-17A and IL-21 secretion were not detected.
Proliferation and cytokine secretion of subject GS1-56A T cells in response to FVIII2194-2213. Three T-cell clones and 1 line (10 000 cells) were stimulated with FVIII2194-2213 or irrelevant peptide (FVIII2178-2197) at concentrations from 0.01 to 100 μM presented by 100 000 irradiated PBMCs from a DRB1*01:01-typed donor. Forty-eight hours later, 50 μL of cell supernatant was removed from each well for cytokine analysis and replaced with 1 μCi [3H]thymidine. Cells were harvested after 18 hours of further incubation, and [3H]thymidine incorporation was measured. Cell supernatants in quadruplicate wells were pooled for measurement of IFN-γ, IL-4, IL-17A, and IL-21 by sandwich ELISAs using purified recombinant cytokines to generate standard curves. (A) Proliferation results are expressed as averages of quadruplicate determinations ± standard deviations. Proliferation in response to the FVIII2178-2197 peptide was negligible when compared with proliferation in the absence of peptide. Background proliferation levels: 56A-C2, 171.0 ± 69.9 counts per minute (cpm); 56A-C8, 84.2 ± 10.9 cpm; 56A-C15, 99.4 ± 7.2 cpm; and 56A-L2, 209.0 ± 61.4 cpm. (B) IFN-γ secretion levels. (C) IL-4 secretion levels. (D) IFN-γ:IL-4 ratios. IL-17A and IL-21 secretion were not detected.
The TCRB-CDR3 regions of the 56A T-cell clones were sequenced to determine the V-, D-, and J-genes and their junction amino acid sequences (Table 1). Ten clones had an identical TCRB-CDR3 sequence with the VJ-genes TCRBV27*01, TCRBJ1-1*01. A D-gene was not identified by IMGT/V-QUEST for this sequence. Staining by the DRB1*01:01-FVIII2194-2213 tetramer produced MFI values from 402 to 974. A second TCRB-CDR3 sequence was identified for clone 56A-C8, which had an MFI of 202 for DRB1*01:01-FVIII2194-2213 tetramer staining. Four clones with low-avidity binding (MFI 3-26) to DRB1*01:01-FVIII2194-2213 tetramer expressed 4 additional distinct TCRB-CDR3 genes.
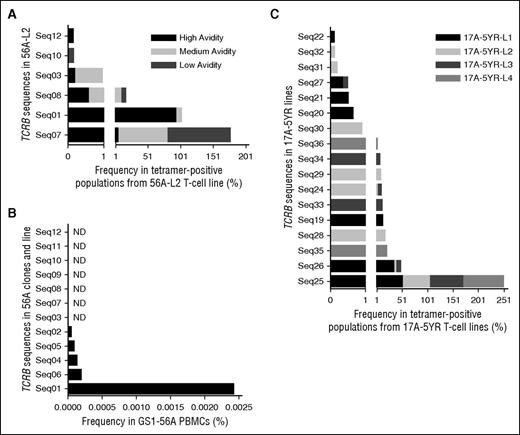
TCRB-CDR3 deep sequencing of the T-cell line 56A-L2 was performed. Cells that bound DRB1*01:01-FVIII2194-2213 tetramer with high, medium, and low avidities were sorted separately (supplemental Figure 4). The total numbers of sequences identified are shown in supplemental Table 5, with the most frequent sequences listed in Table 2. The polyclonal line consisted of 3 dominant sequences: Seq01, Seq07, and Seq08. Seq01 comprised 94% of the high-avidity population (Figure 5A) and was identical to that found in all 10 high-avidity clones. Seq07 and Seq08 were most abundant in the low- and medium-avidity populations, respectively, with neither represented among the clones. Seq03, found in clone 56A-C8, was most abundant in the medium-avidity population. None of the TCRB sequences for the low-avidity clones were found in the polyclonal line.
Deep TCRB sequencing of tetramer-sorted cell populations from T-cell line 56A-L2
| Cells* . | Frequency (%) . | Count . | TCRBV gene† . | TCRBD gene† . | TCRBJ gene† . | Junction sequence . | Sequence identifier‡ . |
|---|---|---|---|---|---|---|---|
| Unsorted | 82.65 | 226 589 | 24-1*01 | 1 or 2§ | 2-3*01 | CATSEVTSTDTQYF | Seq07 |
| Unsorted | 11.45 | 31 403 | 27*01 | Not identified | 1-1*01 | CASSFDPLNTEAFF | Seq01 |
| Unsorted | 3.78 | 10 368 | 20-1|| | 1*01 | 2-7*01 | CSAKGQGPYEQYF | Seq08 |
| Unsorted | 0.15 | 421 | 19¶ | 2*01 or 2*02 | 1-1*01 | CASSVAGGLNTEAFF | Seq09 |
| Unsorted | 0.13 | 361 | 24-1*01 | 1*01 | 2-3*01 | CATSEVNSTDTQYV | Seq10 |
| Unsorted | 0.13 | 349 | 27*01 | 1*01 | 2-7*01 | CASSLRTGGSYYEQYF | Seq11 |
| Unsorted | 0.12 | 326 | 7-8# | 2*01 or 2*02 | 2-3*01 | CASSSAPRLTSGRTDTQYF | Seq03 |
| High | 93.60 | 56 880 | 27*01 | Not identified | 1-1*01 | CASSFDPLNTEAFF | Seq01 |
| High | 4.89 | 2971 | 24-1*01 | 1 or 2 | 2-3*01 | CATSEVTSTDTQYF | Seq07 |
| High | 0.57 | 344 | 20-1|| | 1*01 | 2-7*01 | CSAKGQGPYEQYF | Seq08 |
| High | 0.19 | 113 | 7-8# | 2*01 or 2*02 | 2-3*01 | CASSSAPRLTSGRTDTQYF | Seq03 |
| High | 0.14 | 84 | 27*01 | 1*01 | 1-1*01 | CASSFDQLNTEAFV | Seq12 |
| Medium | 77.28 | 281 766 | 24-1*01 | 1 or 2 | 2-3*01 | CATSEVTSTDTQYF | Seq07 |
| Medium | 10.67 | 38 897 | 20-1|| | 1*01 | 2-7*01 | CSAKGQGPYEQYF | Seq08 |
| Medium | 8.26 | 30 122 | 27*01 | Not identified | 1-1*01 | CASSFDPLNTEAFF | Seq01 |
| Medium | 0.77 | 2799 | 7-8# | 2*01 or 2*02 | 2-3*01 | CASSSAPRLTSGRTDTQYF | Seq03 |
| Medium | 0.11 | 419 | 24-1*01 | 1*01 | 2-3*01 | CATSEVNSTDTQYV | Seq10 |
| Low** | 94.31 | 143 552 | 24-1*01 | 1 or 2 | 2-3*01 | CATSEVTSTDTQYF | Seq07 |
| Low | 4.78 | 7276 | 20-1|| | 1*01 | 2-7*01 | CSAKGQGPYEQYF | Seq08 |
| Low | 0.26 | 403 | 27*01 | Not identified | 1-1*01 | CASSFDPLNTEAFF | Seq01 |
| Low | 0.15 | 229 | 24-1*01 | 1*01 | 2-3*01 | CATSEVNSTDTQYV | Seq10 |
| Cells* . | Frequency (%) . | Count . | TCRBV gene† . | TCRBD gene† . | TCRBJ gene† . | Junction sequence . | Sequence identifier‡ . |
|---|---|---|---|---|---|---|---|
| Unsorted | 82.65 | 226 589 | 24-1*01 | 1 or 2§ | 2-3*01 | CATSEVTSTDTQYF | Seq07 |
| Unsorted | 11.45 | 31 403 | 27*01 | Not identified | 1-1*01 | CASSFDPLNTEAFF | Seq01 |
| Unsorted | 3.78 | 10 368 | 20-1|| | 1*01 | 2-7*01 | CSAKGQGPYEQYF | Seq08 |
| Unsorted | 0.15 | 421 | 19¶ | 2*01 or 2*02 | 1-1*01 | CASSVAGGLNTEAFF | Seq09 |
| Unsorted | 0.13 | 361 | 24-1*01 | 1*01 | 2-3*01 | CATSEVNSTDTQYV | Seq10 |
| Unsorted | 0.13 | 349 | 27*01 | 1*01 | 2-7*01 | CASSLRTGGSYYEQYF | Seq11 |
| Unsorted | 0.12 | 326 | 7-8# | 2*01 or 2*02 | 2-3*01 | CASSSAPRLTSGRTDTQYF | Seq03 |
| High | 93.60 | 56 880 | 27*01 | Not identified | 1-1*01 | CASSFDPLNTEAFF | Seq01 |
| High | 4.89 | 2971 | 24-1*01 | 1 or 2 | 2-3*01 | CATSEVTSTDTQYF | Seq07 |
| High | 0.57 | 344 | 20-1|| | 1*01 | 2-7*01 | CSAKGQGPYEQYF | Seq08 |
| High | 0.19 | 113 | 7-8# | 2*01 or 2*02 | 2-3*01 | CASSSAPRLTSGRTDTQYF | Seq03 |
| High | 0.14 | 84 | 27*01 | 1*01 | 1-1*01 | CASSFDQLNTEAFV | Seq12 |
| Medium | 77.28 | 281 766 | 24-1*01 | 1 or 2 | 2-3*01 | CATSEVTSTDTQYF | Seq07 |
| Medium | 10.67 | 38 897 | 20-1|| | 1*01 | 2-7*01 | CSAKGQGPYEQYF | Seq08 |
| Medium | 8.26 | 30 122 | 27*01 | Not identified | 1-1*01 | CASSFDPLNTEAFF | Seq01 |
| Medium | 0.77 | 2799 | 7-8# | 2*01 or 2*02 | 2-3*01 | CASSSAPRLTSGRTDTQYF | Seq03 |
| Medium | 0.11 | 419 | 24-1*01 | 1*01 | 2-3*01 | CATSEVNSTDTQYV | Seq10 |
| Low** | 94.31 | 143 552 | 24-1*01 | 1 or 2 | 2-3*01 | CATSEVTSTDTQYF | Seq07 |
| Low | 4.78 | 7276 | 20-1|| | 1*01 | 2-7*01 | CSAKGQGPYEQYF | Seq08 |
| Low | 0.26 | 403 | 27*01 | Not identified | 1-1*01 | CASSFDPLNTEAFF | Seq01 |
| Low | 0.15 | 229 | 24-1*01 | 1*01 | 2-3*01 | CATSEVNSTDTQYV | Seq10 |
Cells were sorted from this polyclonal line based on their binding avidity for the DRB1*01:01-FVIII2194-2213 tetramer into high-, medium-, and low-avidity populations (P2, P3, and P4 gates, respectively, in supplemental Figure 4); 200 000 cells were collected from each population except for the smaller high-avidity population, which consisted of 112 000 cells. Genomic DNA was isolated from cells, and deep TCRB-CDR3 sequencing was performed by Adaptive Biotechnologies. Sequences were analyzed with ImmunoSEQ Analyzer version 2.0. Productive sequences with a frequency ≥0.1% are shown.
Gene names for nucleotide sequences were determined using IMGT/V-QUEST version 3.3.4.50
Sequences were given a numerical identifier for labeling purposes. Analyzed nucleotide sequences are in supplemental Table 6.
Multiple gene names result from >1 known annotated V, D, or J gene sequence matching equally well with the aligned sequence.
Sequence was identical to 20-1*01, *02, *03, *04, and *05 alleles.
Sequence was identical to 19*01, *02, and *03 alleles.
Sequence was identical to 7-8*01 and *03 alleles.
Results shown are for the low-avidity CD4 P4 gate in supplemental Figure 4. TCRB sequences identified in the low-avidity CD4-low P5 gate were similar, as the 3 most frequent sequences were identical with similar frequencies.
Dominant TCRB-CDR3 sequences in T-cell lines and PBMCs. The frequency of each TCRB-CDR3 sequence per sorted population or sample was determined using ImmunoSEQ Analyzer version 2.0. Sequence numerical identifiers reference sequences in Tables 1, 2, and 4. (A) Frequency of specific TCRB sequences in sorted populations from the 56A-L2 T-cell line with high, medium, and low avidity for the DRB1*01:01-FVIII2194-2213 tetramer. The x-axis in panels A and C is presented as 2 segments with different scales in order to clearly see the results for the low frequency sequences. (B) Frequency of the 12 TCRB sequences identified in 56A clones and 56A-L2 line in GS1-56A PBMCs. Seven sequences were not detected (ND). (C) Frequency of specific TCRB sequences in DRB1*01:01-FVIII2194-2213 tetramer-positive sorted cell populations collected from 4 17A T-cell lines isolated at the 5-year postinhibitor time point.
Dominant TCRB-CDR3 sequences in T-cell lines and PBMCs. The frequency of each TCRB-CDR3 sequence per sorted population or sample was determined using ImmunoSEQ Analyzer version 2.0. Sequence numerical identifiers reference sequences in Tables 1, 2, and 4. (A) Frequency of specific TCRB sequences in sorted populations from the 56A-L2 T-cell line with high, medium, and low avidity for the DRB1*01:01-FVIII2194-2213 tetramer. The x-axis in panels A and C is presented as 2 segments with different scales in order to clearly see the results for the low frequency sequences. (B) Frequency of the 12 TCRB sequences identified in 56A clones and 56A-L2 line in GS1-56A PBMCs. Seven sequences were not detected (ND). (C) Frequency of specific TCRB sequences in DRB1*01:01-FVIII2194-2213 tetramer-positive sorted cell populations collected from 4 17A T-cell lines isolated at the 5-year postinhibitor time point.
TCRB-CDR3 deep sequencing of PBMCs from GS1-56A was performed, identifying 108 259 unique sequences (supplemental Table 5) ranging in frequency from 0.00004127% to 2.26%. Five of 12 sequences in the 56A T-cell clones and line were detected, with Seq01 being >10-fold more prevalent than the other sequences detected in PBMCs (Figure 5B). Seq07 and Seq08 were not detected, suggesting these clones were present at very low frequency in vivo.
TCRB genes in clones from mild HA subjects GS1-17A28,36 and GS1-32A,29,36 which had the same epitope specificity as the clones and line from the severe HA subject, were sequenced (Table 352 ). Five unique TCRB genes were identified among 13 clones isolated from GS1-17A at 2 different time points following his initial inhibitor. At the 19-week time point, all 5 clones had V-gene 20-1, although differences in the D- and J-genes produced 3 unique junction sequences. These sequences differed from the 2 sequences found at his 21-month time point. All 6 clones from GS1-32A, who had a very low-titer inhibitor, had the same TCRB sequence, which was distinct from those of the 17A clones. In summary, each HA subject showed a distinct TCRB gene usage.
TCRB gene sequence analysis for T-cell clones isolated from mild HA subjects GS1-17A and GS1-32A
| T-cell clone . | TCRBV gene* . | TCRBD gene* . | TCRBJ gene* . | Junction sequence* . | Sequence identifier† . |
|---|---|---|---|---|---|
| 17A-19WK-5‡ | 20-1§ | 1*01 | 1-2*01 | CSAHTRANYGYTF | Seq13 |
| 17A-19WK-10 | 20-1§ | 2*01 or *02 | 2-5*01 | CSAITGTSGETQYF | Seq14 |
| 17A-19WK-11 | 20-1§ | 2*01 or *02 | 2-7*01 | CSARGPSNGYEQYF | Seq15 |
| 17A-19WK-22 | 20-1§ | 2*01 or *02 | 2-7*01 | CSARGPSNGYEQYF | Seq15 |
| 17A-19WK-33 | 20-1§ | 2*01 or *02 | 2-7*01 | CSARGPSNGYEQYF | Seq15 |
| 17A-21MO-3 | 6-6*01 | 1*01 | 2-2*01 | CASSRQDTGELFF | Seq16 |
| 17A-21MO-4|| | 6-6*01 or *03 | Not identified | Not identified | Not identified | |
| 17A-21MO-5 | 6-6*01 or *03 | 1*01 | 2-2*01 | CASSRQDTGELFF | Seq16 |
| 17A-21MO-6 | 6-6*01 | 1*01 | 2-2*01 | CASSRQDTGELFF | Seq16 |
| 17A-21MO-7 | 6-6*01 | 1*01 | 2-2*01 | CASSRQDTGELFF | Seq16 |
| 17A-21MO-8¶ | 5-1*01 | 1*01 | 1-2*01 | CASSLDGRANYGYTF | Seq17 |
| 17A-21MO-11¶ | 5-1*01 | 1*01 | 1-2*01 | CASSLDGRANYGYTF | Seq17 |
| 17A-21MO-12¶ | 5-1*01 | 1*01 | 1-2*01 | CASSLDGRANYGYTF | Seq17 |
| 32A-5 | 6-1*01 | 1*01 | 2-7*01 | CASSDGQPIYEQYF | Seq18 |
| 32A-14 | 6-1*01 | 1*01 | 2-7*01 | CASSDGQPIYEQYF | Seq18 |
| 32A-15 | 6-1*01 | 1*01 | 2-7*01 | CASSDGQPIYEQYF | Seq18 |
| 32A-16 | 6-1*01 | 1*01 | 2-7*01 | CASSDGQPIYEQYF | Seq18 |
| 32A-18 | 6-1*01 | 1*01 | 2-7*01 | CASSDGQPIYEQYF | Seq18 |
| 32A-21 | 6-1*01 | 1*01 | 2-7*01 | CASSDGQPIYEQYF | Seq18 |
| T-cell clone . | TCRBV gene* . | TCRBD gene* . | TCRBJ gene* . | Junction sequence* . | Sequence identifier† . |
|---|---|---|---|---|---|
| 17A-19WK-5‡ | 20-1§ | 1*01 | 1-2*01 | CSAHTRANYGYTF | Seq13 |
| 17A-19WK-10 | 20-1§ | 2*01 or *02 | 2-5*01 | CSAITGTSGETQYF | Seq14 |
| 17A-19WK-11 | 20-1§ | 2*01 or *02 | 2-7*01 | CSARGPSNGYEQYF | Seq15 |
| 17A-19WK-22 | 20-1§ | 2*01 or *02 | 2-7*01 | CSARGPSNGYEQYF | Seq15 |
| 17A-19WK-33 | 20-1§ | 2*01 or *02 | 2-7*01 | CSARGPSNGYEQYF | Seq15 |
| 17A-21MO-3 | 6-6*01 | 1*01 | 2-2*01 | CASSRQDTGELFF | Seq16 |
| 17A-21MO-4|| | 6-6*01 or *03 | Not identified | Not identified | Not identified | |
| 17A-21MO-5 | 6-6*01 or *03 | 1*01 | 2-2*01 | CASSRQDTGELFF | Seq16 |
| 17A-21MO-6 | 6-6*01 | 1*01 | 2-2*01 | CASSRQDTGELFF | Seq16 |
| 17A-21MO-7 | 6-6*01 | 1*01 | 2-2*01 | CASSRQDTGELFF | Seq16 |
| 17A-21MO-8¶ | 5-1*01 | 1*01 | 1-2*01 | CASSLDGRANYGYTF | Seq17 |
| 17A-21MO-11¶ | 5-1*01 | 1*01 | 1-2*01 | CASSLDGRANYGYTF | Seq17 |
| 17A-21MO-12¶ | 5-1*01 | 1*01 | 1-2*01 | CASSLDGRANYGYTF | Seq17 |
| 32A-5 | 6-1*01 | 1*01 | 2-7*01 | CASSDGQPIYEQYF | Seq18 |
| 32A-14 | 6-1*01 | 1*01 | 2-7*01 | CASSDGQPIYEQYF | Seq18 |
| 32A-15 | 6-1*01 | 1*01 | 2-7*01 | CASSDGQPIYEQYF | Seq18 |
| 32A-16 | 6-1*01 | 1*01 | 2-7*01 | CASSDGQPIYEQYF | Seq18 |
| 32A-18 | 6-1*01 | 1*01 | 2-7*01 | CASSDGQPIYEQYF | Seq18 |
| 32A-21 | 6-1*01 | 1*01 | 2-7*01 | CASSDGQPIYEQYF | Seq18 |
Gene names and junction sequences were determined from nucleotide sequences using IMGT/V-QUEST version 3.3.4.50 IMGT/V-QUEST uses IMGT/Junction Analysis52 for analysis of the junction.
Sequences were given a numerical identifier for labeling purposes. Analyzed nucleotide sequences are in supplemental Table 6.
TCRB V-D-J genes and junction sequence for this clone were described previously.32
Sequence matched equally well to 20-1*01, *02, *04, *05, and *06 alleles.
17A-21MO-4 multiplex PCR product typed as TCRBV13 using the method and nomenclature in Akatsuka et al.46 This PCR product sequenced with the TCRBC primer whereas sequencing failed repeatedly with the TCRBV13 primer.
Clone has 2 TCRBV cDNA products. The second product was identified as TCRBV7-3*02, TCRBD2*01 or *02, TCRBJ2-3*01 and contains an out-of-frame junction.
The tetramer-positive populations from the 17A lines and the tetramer-negative population from line 17A-5YR-L1 were sorted (supplemental Figure 5) and TCRB-CDR3 deep sequenced (supplemental Table 5). Table 4 lists the sequences with a frequency ≥0.1%. Two sequences (Seq25, Seq26) were shared by the tetramer-positive populations from all 4 lines, one of which was the dominant sequence (51% to 77%) in all the lines (Figure 5C). The subdominant sequences varied between the lines. Some sequences in the tetramer-negative population also appeared in the tetramer-positive population, albeit at lower frequencies. The dominant TCRB sequences identified at the 5-year time point differed from those in samples isolated 19 weeks and 21 months postinhibitor development.
Deep TCRB sequencing of tetramer-sorted cell populations from GS1-17A T-cell lines
| Cells* and frequency (%) . | Count . | TCRBV gene† . | TCRBD gene† . | TCRBJ gene† . | Junction sequence . | Sequence identifier‡ . |
|---|---|---|---|---|---|---|
| 17A-5YR-L1 Tet-neg (323 000) | ||||||
| 69.83 | 468 811 | 6-6§ | 2*02 | 2-5*01 | CASSDGLAGGWETQYF | Seq19 |
| 15.98 | 107 291 | 7-9|| | 2*01 | 1-1*01 | CASSPWKVTGNTEAFF | Seq20 |
| 11.89 | 79 822 | 12-3 or 12-4¶ | 2*01 | 2-7*01 | CASSFSQPGRRDEQYF | Seq21 |
| 0.64 | 4282 | 24-1*01 | 2*01 | 2-1*01 | CATSDLGTSGYNEQFF | Seq22 |
| 0.14 | 957 | 6-6§ | 1*01 | 1-2*01 | CASSYSDRGYGYTF | Seq23 |
| 0.13 | 846 | 7-2# | 1*01 | 2-7*01 | CASSLGAGGEQYF | Seq24 |
| 17A-5YR-L1 Tet-pos (307 000) | ||||||
| 50.99 | 134 032 | 20-1** | 1*01 | 2-2*01 | CSARYFGPPKAGELFF | Seq25 |
| 34.53 | 90 759 | 20-1** | 1*01 | 1-6*02 | CSARVQGGNSPLHF | Seq26 |
| 11.81 | 31 040 | 6-6§ | 2*02 | 2-5*01 | CASSDGLAGGWETQYF | Seq19 |
| 0.64 | 1691 | 7-9|| | 2*01 | 1-1*01 | CASSPWKVTGNTEAFF | Seq20 |
| 0.50 | 1306 | 12-3 or 12-4¶ | 2*01 | 2-7*01 | CASSFSQPGRRDEQYF | Seq21 |
| 0.35 | 907 | 19†† | 1*01 | 1-4*01 | CASRDPHRVEKLFF | Seq27 |
| 0.10 | 269 | 24-1*01 | 2*01 | 2-1*01 | CATSDLGTSGYNEQFF | Seq22 |
| 17A-5YR-L2 Tet-pos-med‡‡(102 301) | ||||||
| 56.02 | 152 869 | 20-1** | 1*01 | 2-2*01 | CSARYFGPPKAGELFF | Seq25 |
| 16.78 | 45 777 | 18*01 | Not identified | 2-1*01 | CASSPLPYSYNEQFF | Seq28 |
| 7.45 | 20 322 | 7-2# | 1*01 | 1-1*01 | CASSAGFPTEAFF | Seq29 |
| 7.10 | 19 366 | 20-1** | 1*01 | 1-6*02 | CSARVQGGNSPLHF | Seq26 |
| 4.16 | 11 351 | 7-2# | 1*01 | 2-7*01 | CASSLGAGGEQYF | Seq24 |
| 0.90 | 2444 | 5-1*01 or *02 | 1*01 | 1-2*01 | CASSLAGREGGYTF | Seq30 |
| 0.18 | 486 | 20-1** | 1*01 | 2-5*01 | CSARWTPGQGGETQYF | Seq31 |
| 0.11 | 296 | 3-1 or 3-2a | 1*01 | 1-1*01 | CASSQAGGNTEAFF | Seq32 |
| 17A-5YR-L3 Tet-pos (178 500) | ||||||
| 65.62 | 193 633 | 20-1** | 1*01 | 2-2*01 | CSARYFGPPKAGELFF | Seq25 |
| 10.46 | 30 873 | 19†† | 1*01 | 1-2*01 | CASSTGTTSNYGYTF | Seq33 |
| 6.13 | 18 075 | 5-1*01 or *02 | 1*01 | 2-1*01 | CASSFQGVSYNEQFF | Seq34 |
| 4.86 | 14 327 | 20-1** | 1*01 | 1-6*02 | CSARVQGGNSPLHF | Seq26 |
| 4.79 | 14 127 | 7-2# | 1*01 | 2-7*01 | CASSLGAGGEQYF | Seq24 |
| 0.14 | 407 | 19†† | 1*01 | 1-4*01 | CASRDPHRVEKLFF | Seq27 |
| 17A-5YR-L4 Tet-pos (238 283) | ||||||
| 76.58 | 285 747 | 20-1** | 1*01 | 2-2*01 | CSARYFGPPKAGELFF | Seq25 |
| 19.78 | 73 815 | 6-1*01 | 2*02 | 1-1*01 | CASSARADTEAFF | Seq35 |
| 1.45 | 5397 | 4-2*01 or *02 | 2*02 | 2-1*01 | CASSQEKREFDEQFF | Seq36 |
| 0.46 | 1729 | 20-1** | 1*01 | 1-6*02 | CSARVQGGNSPLHF | Seq26 |
| Cells* and frequency (%) . | Count . | TCRBV gene† . | TCRBD gene† . | TCRBJ gene† . | Junction sequence . | Sequence identifier‡ . |
|---|---|---|---|---|---|---|
| 17A-5YR-L1 Tet-neg (323 000) | ||||||
| 69.83 | 468 811 | 6-6§ | 2*02 | 2-5*01 | CASSDGLAGGWETQYF | Seq19 |
| 15.98 | 107 291 | 7-9|| | 2*01 | 1-1*01 | CASSPWKVTGNTEAFF | Seq20 |
| 11.89 | 79 822 | 12-3 or 12-4¶ | 2*01 | 2-7*01 | CASSFSQPGRRDEQYF | Seq21 |
| 0.64 | 4282 | 24-1*01 | 2*01 | 2-1*01 | CATSDLGTSGYNEQFF | Seq22 |
| 0.14 | 957 | 6-6§ | 1*01 | 1-2*01 | CASSYSDRGYGYTF | Seq23 |
| 0.13 | 846 | 7-2# | 1*01 | 2-7*01 | CASSLGAGGEQYF | Seq24 |
| 17A-5YR-L1 Tet-pos (307 000) | ||||||
| 50.99 | 134 032 | 20-1** | 1*01 | 2-2*01 | CSARYFGPPKAGELFF | Seq25 |
| 34.53 | 90 759 | 20-1** | 1*01 | 1-6*02 | CSARVQGGNSPLHF | Seq26 |
| 11.81 | 31 040 | 6-6§ | 2*02 | 2-5*01 | CASSDGLAGGWETQYF | Seq19 |
| 0.64 | 1691 | 7-9|| | 2*01 | 1-1*01 | CASSPWKVTGNTEAFF | Seq20 |
| 0.50 | 1306 | 12-3 or 12-4¶ | 2*01 | 2-7*01 | CASSFSQPGRRDEQYF | Seq21 |
| 0.35 | 907 | 19†† | 1*01 | 1-4*01 | CASRDPHRVEKLFF | Seq27 |
| 0.10 | 269 | 24-1*01 | 2*01 | 2-1*01 | CATSDLGTSGYNEQFF | Seq22 |
| 17A-5YR-L2 Tet-pos-med‡‡(102 301) | ||||||
| 56.02 | 152 869 | 20-1** | 1*01 | 2-2*01 | CSARYFGPPKAGELFF | Seq25 |
| 16.78 | 45 777 | 18*01 | Not identified | 2-1*01 | CASSPLPYSYNEQFF | Seq28 |
| 7.45 | 20 322 | 7-2# | 1*01 | 1-1*01 | CASSAGFPTEAFF | Seq29 |
| 7.10 | 19 366 | 20-1** | 1*01 | 1-6*02 | CSARVQGGNSPLHF | Seq26 |
| 4.16 | 11 351 | 7-2# | 1*01 | 2-7*01 | CASSLGAGGEQYF | Seq24 |
| 0.90 | 2444 | 5-1*01 or *02 | 1*01 | 1-2*01 | CASSLAGREGGYTF | Seq30 |
| 0.18 | 486 | 20-1** | 1*01 | 2-5*01 | CSARWTPGQGGETQYF | Seq31 |
| 0.11 | 296 | 3-1 or 3-2a | 1*01 | 1-1*01 | CASSQAGGNTEAFF | Seq32 |
| 17A-5YR-L3 Tet-pos (178 500) | ||||||
| 65.62 | 193 633 | 20-1** | 1*01 | 2-2*01 | CSARYFGPPKAGELFF | Seq25 |
| 10.46 | 30 873 | 19†† | 1*01 | 1-2*01 | CASSTGTTSNYGYTF | Seq33 |
| 6.13 | 18 075 | 5-1*01 or *02 | 1*01 | 2-1*01 | CASSFQGVSYNEQFF | Seq34 |
| 4.86 | 14 327 | 20-1** | 1*01 | 1-6*02 | CSARVQGGNSPLHF | Seq26 |
| 4.79 | 14 127 | 7-2# | 1*01 | 2-7*01 | CASSLGAGGEQYF | Seq24 |
| 0.14 | 407 | 19†† | 1*01 | 1-4*01 | CASRDPHRVEKLFF | Seq27 |
| 17A-5YR-L4 Tet-pos (238 283) | ||||||
| 76.58 | 285 747 | 20-1** | 1*01 | 2-2*01 | CSARYFGPPKAGELFF | Seq25 |
| 19.78 | 73 815 | 6-1*01 | 2*02 | 1-1*01 | CASSARADTEAFF | Seq35 |
| 1.45 | 5397 | 4-2*01 or *02 | 2*02 | 2-1*01 | CASSQEKREFDEQFF | Seq36 |
| 0.46 | 1729 | 20-1** | 1*01 | 1-6*02 | CSARVQGGNSPLHF | Seq26 |
Cells were sorted with the DRB1*01:01-FVIII2194-2213 tetramer into populations that were stained (Tet-pos) or not stained (Tet-neg) with tetramer (supplemental Figure 5). The number of cells collected from each population is indicated in parentheses. Genomic DNA was isolated from cells, and deep TCRB-CDR3 sequencing was performed by Adaptive Biotechnologies. Sequences were analyzed with ImmunoSEQ Analyzer version 2.0. Productive sequences with a frequency ≥0.1% are shown.
Gene names for nucleotide sequences were determined using IMGT/V-QUEST version 3.3.5.50
Sequences were given a numerical identifier for labeling purposes. Analyzed nucleotide sequences are in supplemental Table 6.
Sequence matched equally well to 6-6*01, *02, *03, and *04 alleles.
Sequence matched equally well to 7-9*01, *02, *03, *04, and *05 alleles.
Sequence matched equally well to 12-3*01, 12-4*01, and 12-4*02 alleles.
Sequence matched equally well to 7-2*01, *02, *03, and *04 alleles.
Sequence matched equally well to 20-1*01, *02, *03, *04, and *05 alleles.
Sequence matched equally well to 19*01, *02, and *03 alleles.
Two tetramer-positive populations were observed at the time of sorting the 17A-5YR-L2 cell line. These populations were sorted and labeled medium and high, corresponding to their respective tetramer avidities. The high-avidity population (19 801 cells) was smaller than the medium-avidity population, and their sequence compositions were very similar. Therefore, only the medium-avidity population results are shown here.
Sequence matched equally well to 3-1*01 or *02, 3-2*01, *02, and *03 alleles.
Discussion
FVIII inhibitors make standard prophylaxis with FVIII impossible. New treatments to promote immune tolerance to FVIII in individuals with an inhibitor or at increased risk for inhibitor development are therefore badly needed.1 To facilitate their development, a better understanding of the anti-FVIII immune response, particularly in those who have developed a high-titer inhibitor refractive to ITI therapy, is required. Severe HA subject GS1-56A had failed ITI therapy and his high-titer inhibitor had persisted for >10 years. Mild HA subject GS1-17A was enrolled upon initial inhibitor detection,28 after which he contributed blood samples for 5 years, and his brother (GS1-32A) had a clinically insignificant inhibitor response, identified by concentrating and then assaying his IgG.29
Identification of HLA-restricted T-cell epitopes is a first essential step in characterizing cellular responses to FVIII, and isolation/characterization of FVIII-specific T-cell clones and polyclonal lines provides phenotypic and mechanistic information. An understanding of the specific MHCII-FVIII peptide-TCR complexes associated with inhibitor development and maintenance will facilitate the development of new therapies, which may include removal of immunodominant epitopes through FVIII sequence modification53 and targeted immunotherapies to promote antigen-specific immune tolerance to FVIII.32,54 Herein, we advance our understanding of these processes by characterizing T-cell clones and polyclonal lines isolated from 3 HA subjects who responded to the same HLA-restricted epitope in FVIII.
Interestingly and unexpectedly, only 1 HLA-DRA-DRB1-restricted T-cell epitope was recognized within FVIII-A2, C1, and C2 domains by the severe HA subject, who had a deletion of F8 gene exons 7 to 13 that eliminated almost the entire A2 domain coding region. Thus FVIII-A2 peptides would definitely be foreign and possibly immunogenic. Furthermore, FVIII antigen was not detected when his plasma was tested by ELISA using monoclonal antibody ESH-8, which recognizes a C2 domain epitope,55,56 suggesting that most or all of the FVIII protein would be foreign to his immune system. Additional epitopes may have been identified if FVIII-A1, B, and A3 domain peptides had also been tested; however, blood volumes were not sufficient for comprehensive TGEM. The experimental identification of only 1 T-cell epitope contrasts with the large number of potential FVIII epitopes predicted by computer algorithms, as has been shown in previous studies.30,43
Comprehensive epitope mapping using splenic cells from humanized HLA-DRA-DRB1*1501 transgenic FVIII-knockout mice and peptides spanning the full-length FVIII sequence identified 8 T-cell epitopes restricted to this allele.27 Unlike the severe HA subject studied here, these mice were not subjected to an ITI protocol. The mechanism of action of ITI is not well understood.57 T-cell tolerance to antigens may result from clonal deletion, clonal anergy, and/or induction of regulatory cells.58-60 Pautard et al investigated whether ITI deleted FVIII-specific T cells by attempting to expand FVIII-specific T-cell lines from blood of HA subjects who had been successfully tolerized via ITI.61 FVIII-specific T cells were expanded from 1 of 5 desensitized patients, indicating that clonal deletion is not absolutely required for functional immune tolerance to FVIII. Although the severe HA subject studied here failed ITI, some FVIII-specific clones may have been deleted or anergized, thus decreasing the number of epitopes contributing to his ongoing immune response. Six-day proliferation assays19 testing his CD4 responses to wild-type vs epitope-modified FVIII proteins53 (R.A.E. and K.P.P., manuscript in preparation), and enzyme-linked immunospot assays testing for IFN-γ responses to these proteins, were inconclusive (supplemental Figures 6-7), likely because of the low precursor frequency of FVIII-specific cells. However, these negative results indicated that he did not have robust T-cell responses to additional epitopes in FVIII.
The DRB1*01:01-restricted epitope was located within FVIII-C2 residues 2194 to 2213. It has been hypothesized that the FVIII-C2 domain may not be recognized as foreign to the immune system of almost all HA patients because of intracellular expression of the F8B gene, which encompasses the entire FVIII-C2 sequence. Intracellular synthesis of a partial FVIII and of the FVIIIB proteins has been reported in PBMCs and liver tissue from intron-22-inversion HA subjects.62,63 However, recent proteomics studies have not confirmed FVIII or FVIIIB expression in PBMCs or in thymus peptides associated with HLA-DR.64,65 The isolation reported herein of FVIII-C2-specific T-effector clones and lines from a severe HA subject whose F8 mutation did not affect the F8B region demonstrates directly that the FVIII-C2 domain is a target for T cells in HA subjects and thereby argues against widespread central tolerance to this domain.
T cells from severe HA subject GS1-56A recognized the same DRB1*01:01-restricted epitope to which mild HA subjects with an FVIII-A2201P missense mutation responded,28,29 indicating that immune responses associated with HLA can be assigned to specific FVIII sequences. FVIII589-608 has been identified as a DRB1*11:01-restricted epitope in 2 mild HA subjects with an FVIII-R593C missense mutation,30 providing further evidence of consistent HLA-restricted immune responses to specific FVIII epitopes. Additional epitope mapping studies of anti-FVIII T-cell responses for other MHCII are needed to determine whether therapies based on immunodominant epitopes will be feasible. Such studies may improve estimates of inhibitor risk, particularly for patients with missense mutations.
Differences were observed in the avidities of T-cell clones and polyclonal T cells for tetramers loaded with FVIII2194-2213 and FVIII2186-2205. Sequencing demonstrated distinct TCRB genes among the clones. Clones with the same TCRB-CDR3 sequence also had different tetramer avidities. Tetramer avidity for an αβ TCR is a combined effect of TCR-α and β chain sequences and TCR surface density.66 TCRs can vary in their contacts with specific MHCII-peptide complexes.67 For example, crystal structures of 2 autoimmune TCRs bound to an MHCII-peptide complex revealed an N-terminal peptide-TCR binding topology, which was proposed as a mechanism permitting autoreactive T cells to escape deletion.68,69 Differences in Th phenotypes among the clones described here were also observed. Clones from GS1-17A were Th17/Th1 or Th1/Th2 polarized at 19 weeks after inhibitor development and were Th2 polarized at 21 months postinhibitor.36 Clones from GS1-32A, who had a clinically insignificant inhibitor history, were all Th1 polarized.36 Clones from GS1-56A having high tetramer avidities were Th1/Th2 polarized, whereas the 1 56A clone with medium tetramer avidity was Th2 polarized. The 56A T-cell line, which contained cells with a range of tetramer avidities, was Th1 polarized. These results indicate that TCR binding avidity and Th phenotypes may contribute to different clinical outcomes.
The TCRB repertoire of cells recognizing the DRB1*01:01-restricted epitope was examined by sequencing of clones and high-throughput immunosequencing35 of polyclonal lines. The stepwise approach developed here of isolating antigen-specific T-cell lines, followed by sorting of specific populations and immunosequencing, allowed for an in-depth analysis of TCRB repertoires recognizing an immunodominant CD4+ T-cell epitope, similar to earlier studies of antigen-specific CD8+ T cells.70 TCRB repertoires of the FVIII-specific T cells were limited to a few dominant genes. TCRB gene usage changed over time in the mild HA subject whose T-cell responses were analyzed over 5 years. However, public T-cell clones were not identified among these 3 subjects. Focused TCR repertoires have been observed and studied extensively in CD8+ T cells responding to viral antigens.70,71 Interestingly, a dominant TCRB gene in the high-tetramer-avidity population isolated from GS1-56A was also detectable in his PBMCs, indicating that PBMCs could be tested in future studies to determine if this immunodominant sequence is shared with other inhibitor-positive individuals.
Only 1 immunodominant epitope was identified in GS1-56A using tetramers loaded with peptides spanning 700 residues of the FVIII sequence, and the clonal diversity of CD4 T cells recognizing this epitope was extremely low. This suggests that patients with a persistent high-titer inhibitor (ie, those with the most compelling need of new therapies to induce tolerance) have an oligoclonal T-cell response to a limited number of T-cell epitopes in FVIII. The present results suggest that novel therapies targeting immunodominant epitopes and/or T-cell clones may eventually improve success rates in tolerizing HA patients to FVIII.72,73 Future studies will include temporal analysis of blood samples from subjects undergoing ITI and mapping of epitopes restricted to additional MHCII alleles.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank Charles Cooper, Mark Bray, and Colette Norby-Slycord for enrolling subjects; Xiaoping Wu for cell sorting and scientific input; Shelley Fletcher for F8 genotyping and sequencing; Lonna Pells for TCRBV typing; Catherine Sanders for advising on ImmunoSEQ TCRB sequencing; and David Scott for critical reading of the manuscript. The authors are grateful to all subjects for their generous blood donations.
This work was supported by grants from Bayer (K.P.P.); the National Heart, Lung, and Blood Institute, National Institutes of Health (grants 1RC2-HL101851 and 1R01-HL130448 [K.P.P.], and R01-HL07109 [A.R.T.]); and Uniformed Services University of the Health Sciences (USUHS) startup funding (K.P.P.).
The opinions or assertions contained herein are the private ones of the author and are not to be construed as official or reflecting the views of the Department of Defense or the USUHS.
Authorship
Contribution: R.A.E., P.P., E.A.J., F.A., A.R.T., and K.P.P. conceived of and designed experiments; R.A.E., E.A.J., and D.G. performed experiments; A.R.T., D.C.M., and K.P.P. enrolled subjects; E.A.J. contributed reagents; R.A.E., P.P., E.A.J., D.G., and K.P.P. analyzed data; R.A.E. and K.P.P. wrote the manuscript; and all authors reviewed the manuscript.
Conflict-of-interest disclosure: K.P.P., R.A.E., and E.A.J. are inventors on FVIII patents. P.P. and F.A. are employees of Bayer Healthcare LLC. The remaining authors declare no competing financial interests.
Correspondence: Kathleen P. Pratt, Uniformed Services University of the Health Sciences, Department of Medicine A3075, 4301 Jones Bridge Rd, Bethesda, MD 20814; e-mail: kathleen.pratt@usuhs.edu.

![Figure 4. Proliferation and cytokine secretion of subject GS1-56A T cells in response to FVIII2194-2213. Three T-cell clones and 1 line (10 000 cells) were stimulated with FVIII2194-2213 or irrelevant peptide (FVIII2178-2197) at concentrations from 0.01 to 100 μM presented by 100 000 irradiated PBMCs from a DRB1*01:01-typed donor. Forty-eight hours later, 50 μL of cell supernatant was removed from each well for cytokine analysis and replaced with 1 μCi [3H]thymidine. Cells were harvested after 18 hours of further incubation, and [3H]thymidine incorporation was measured. Cell supernatants in quadruplicate wells were pooled for measurement of IFN-γ, IL-4, IL-17A, and IL-21 by sandwich ELISAs using purified recombinant cytokines to generate standard curves. (A) Proliferation results are expressed as averages of quadruplicate determinations ± standard deviations. Proliferation in response to the FVIII2178-2197 peptide was negligible when compared with proliferation in the absence of peptide. Background proliferation levels: 56A-C2, 171.0 ± 69.9 counts per minute (cpm); 56A-C8, 84.2 ± 10.9 cpm; 56A-C15, 99.4 ± 7.2 cpm; and 56A-L2, 209.0 ± 61.4 cpm. (B) IFN-γ secretion levels. (C) IL-4 secretion levels. (D) IFN-γ:IL-4 ratios. IL-17A and IL-21 secretion were not detected.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/128/16/10.1182_blood-2015-11-682468/4/m_2043f4.jpeg?Expires=1769825123&Signature=tg~yTddaKV3mquZ4ZNrnHGJsd1KDMeZKT1im7qKB8g7v3rrhtUFdoia~3eihPzOqjC0tZzEO7HRH9Ff69R7Dfs7kAZ8mWv9dYqWdy-MrMdyFSiSWW8BA~dFoCk-nHaMt~VS-6lmuB3JmHQwThG8Dug7qMHLemIaKlxhteisKIqF7g6yb3TuBhoZI8cAkc1d95nHG2RgVJQBYFsVhuC9fYUJdeydnZmwOBaR6AkDhL9xVnQA8WXGM-dt93yLlYDysN1ems7i5AtyxQ~rndDu9sWlljp-LHfQcGcm2BqOl1H27CHpkClzbdgIgDAFHcx7xlq7aqcONbw1z2f2h2aK4NA__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)