Abstract
Chromosomal abnormalities (CA) play a pivotal role in predicting MM patients risk. Accordingly, well-defined CA have been associated with higher risk of transformation in smoldering MM (SMM), have been recently incorporated to the revised ISS, and may even guide treatment decisions; furthermore, because high-risk CA can be present in minor subclones at diagnosis it is recommended to reassess them at the time of relapse. Altogether, this implies that throughout their clinical course MM patients go through several bone marrow (BM) aspirates for cytogenetic risk assessment, and less invasive methods to screen for CA [ie. peripheral blood (PB) liquid biopsies] would be mostly welcomed providing they are fully comparable to its assessment in BM clonal plasma cells (PCs).
Here, using the recently developed and sensitive next generation flow (NGF) method to monitor MRD in MM (median limit of detection, 3x10-6), we assessed the applicability of screening for CA in the PB, by determining the frequency of MGUS and MM patients with detectable CTCs and their respective absolute numbers. Using NGF, we detected CTCs in 22/37 (60%) MGUS cases, 9/12 (75%) SMM, 24/25 (96%) newly diagnosed MM, and 22/26 (85%) relapsed MM. Median numbers of CTCs/µL in the PB of MGUS, SMM, newly diagnosed and relapsed MM were 0.04, 0.1, 0.5 and 0.6, respectively. Thus, we noted highly significant differences (P<.001) between benign and malignant disease stages, with a 12.5-fold increment in CTCs/µL between MGUS and MM, and 5-fold between SMM and MM.
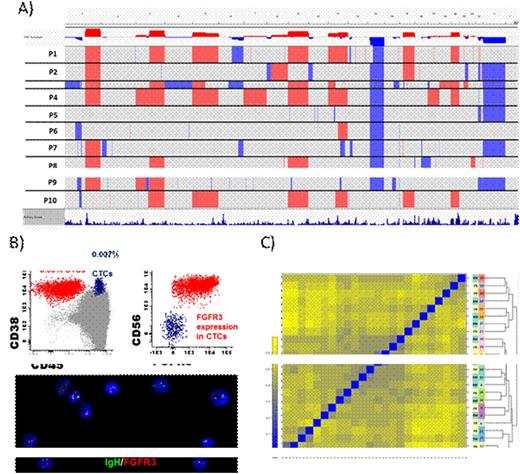
By using whole exome sequencing, we reported recently that the concordance in CA between CTCs and BM clonal PCs was typically <50% due to the limited amount of DNA extracted from low numbers of CTCs, which required whole genome amplification. Accordingly, we investigated in 18 patient paired BM and PB (~5mL) samples (2 MGUS, 2 SMM and 14 MM) whether after NGF sorting and using the Affymetrix CytoScan HD (which requires significantly less DNA to provide molecular karyotypes), we could determine the pattern of CA in BM clonal PCs by screening that of CTCs. In 10 patients in which CTCs were successfully characterized, we obtained 100% concordance between BM clonal PCs and CTCs at the chromosome level (including trisomies of chromosomes 3, 5 and 21 which have been described to modulate the prognosis of patients with high-risk MM), for chromosomal arms, and for interstitial gains or losses (Panel A). Importantly, there was also 100% concordance with FISH performed on CD138+ BM clonal PCs for del(1p), +1q, del(13q) and del(17p) CA. In the remaining cases, ultra-low numbers of sorted CTCs (≤5.000) precluded their successful characterization, which could be readily overcome by using larger volumes of PB (ie. >5mL). Because the CytoScan HD does not assess the t(4;14), we included FGFR3 into the NGF panel of mAb in order to detect its surface expression in patients harboring the t(4;14). Similarly to that described above, we observed 100% concordance between FGFR3 expression in CTCs and the presence of t(4;14) in CD138+ BM clonal PCs, as confirmed by FISH (Panel B).
Since gene expression profiling (GEP) has also been used to predict risk in MM, we investigated in 12 patient paired BM and PB samples whether after NGF sorting and using the Affymetrix Human Gene 2.0 ST Array, we could determine patients risk by screening the GEP of CTCs. Unsupervised analysis showed correct clustering between GEP of BM clonal PCs and CTCs in 9/12 cases, demonstrating that individual patient signatures typically outweighed GEP of MM clones located in different tissues (Panel C). Thus, CTCs could be used to predict risk according to well-established GEP signatures.
In conclusion, by using sensitive NGF we showed that CTCs are present in 60% of MGUS and 87% of MM, which unravels high feasibility for liquid biopsying these patients. By also using NGF to sort CTCs, as well as commercially available, cost-effective and standardized arrays to determine CA and gene expression profiles, we demonstrated for the first time that the genetic features of BM clonal PCs can be determined with great accuracy by studying CTCs. Altogether, this combined approach could offer to many MM patients a less invasive cytogenetic characterization at certain stages of their clinical course.
Paiva:Celgene: Honoraria, Research Funding; Janssen: Honoraria; Takeda: Honoraria, Research Funding; Sanofi: Consultancy, Research Funding; EngMab: Research Funding; Amgen: Honoraria; Binding Site: Research Funding.
Author notes
Asterisk with author names denotes non-ASH members.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal