Abstract
Aim: Evaluation of tandem minimal residual disease (MRD) assessment using multi-gene next generation sequencing (NGS) and multi-parameter flow cytometry (MFC) in acute myeloid leukemia (AML) pts undergoing allogeneic hematopoietic stem cell transplant (allo-HCT).
Methods: MRD was measured on the same bone marrow aspirate using 10-color MFC and a targeted myeloid specific 28-gene NGS panel pre and post allo-HCT in available samples from 122 consecutive pts with AML transplanted between 2014 and 2015. Any level of MRD measured by MFC in the blast compartment was regarded as positive, while somatic mutations detected above a pre-defined variant allele frequency (VAF) threshold on bulk marrow by multi-gene NGS were regarded as positive. Mutations identified on diagnostic or relapse samples were tracked throughout the disease course. FLT3-ITD and TKD mutations were detected in a stand-alone PCR based assay and VAF was not quantified.
Results: NGS (HR: 2.37 (95% CI 1.06-5.28) and HR: 3.23 (95% CI 1.21-8.62)) and MFC (HR: 2.44 (95% CI 1-5.97) and HR: 4.62 (95% CI 1.32-16.09)) predicted overall survival (OS) and time-to-relapse respectively with median observation time of 12 months among survivors. MRD detection using both assays was associated with relapse, with MRD detected by MFC being the most predictive (table 1). NGS was applicable to 85% of tested pts with probable pathogenic mutations seen at diagnosis, while all pts tested at diagnosis had abnormal blasts detected by MFC. Transplant factors including donor source, conditioning intensity, stem cell source and GVHD prophylaxis were not associated with transplant outcomes while complex and monosomal karyotype were associated with OS and time-to-relapse (table 1).
Pre allo-HCT concordance rate of MRD detection using the two assays was 70% (table 2). 12 (20%) pts had detectable MRD by MFC and not NGS. Five of these patients had NGS assessment at diagnosis and on manual review of NGS results 3 of these 5 had diagnostic mutations detected on pre allo-HCT samples at VAF below threshold to call mutations. Six (10%) were MFC negative but had detectable mutations by NGS, which were typically clonal hematopoiesis (CH) type mutations with VAF ranging between 3-20%, only 1/6 of these has relapsed post allo-HSCT.
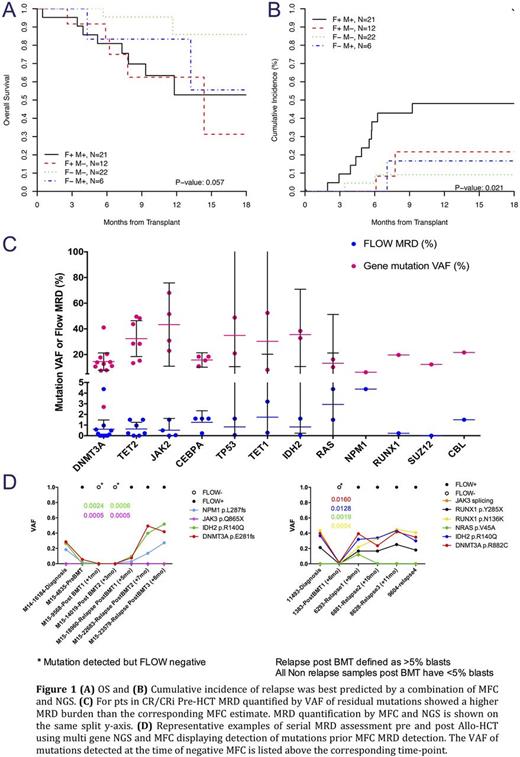
MRD pre allo-HCT using both MFC and NGS was associated with relapse; however, the risk was highest in pts who had pre transplant MRD detected concurrently using both techniques and cumulative incidence of relapse was lowest in those who were MRD negative using both techniques (Figure 1A & B).
No significant change in mutant DNMT3A, TET2 and JAK2 variant allele frequency (VAF) was seen between assessment at diagnosis and pre transplant, while a significant reduction in NPM1 and IDH VAF was noted. For pts in CR or CRi pre allo-HCT MRD burden quantified using MFC (on blasts) was markedly lower than the corresponding VAF of residual mutations (on bulk marrow) measured in the same sample (Figure 1C) with VAF of residual mutation ranging between 10-20% suggesting that a large bulk of cells at the time of CR were derived from the abnormal clone.
We tracked pathogenic mutations identified on diagnostic samples in pts who had marrow MFC and NGS MRD assessment after transplant. In pts who relapsed, multi-gene NGS detected mutations earlier than MFC although at very low allele burden (<1%) (Figure 3D). Low VAF (<2%) DNMT3A mutations were detected after transplant in 10/11 evaluable pts who had these mutations on pre transplant assessment while MFC was negative in 9/10 of those with low VAF DNMT3A mutations detected after transplant. Of the 10 pts with detectable DNMT3A mutations after transplant only 2 have relapsed and both had mutations in TP53. This suggests that the clinical significance of persistent low allelic burden DNMT3A mutations needs further clarification, as they may not be strongly predictive of relapse even if detected after transplant.
Conclusion: Despite different assay sensitivity MFC and multi gene NGS had a concordance of 70% for detecting pre allo-HCT MRD in AML.Assessing pre Allo-HCT MRD with MFC and multi-gene NGS improves the ability to predict relapse and OS. MFC had more universal applicability, while NGS could identify residual CH-type mutations with greater sensitivity than MFC and identify MRD at earlier time points than MFC post-HCT. Using a more extensive gene panel is likely to improve the sensitivity of MRD detection and may improve correlation with MFC
Levine:Novartis: Consultancy; Qiagen: Membership on an entity's Board of Directors or advisory committees. Roshal:BD Biosciences: Consultancy.
Author notes
Asterisk with author names denotes non-ASH members.
This icon denotes a clinically relevant abstract

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal