To the editor:
Chronic myeloid leukemia (CML) represents a distinct subtype of myeloproliferative neoplasm originated from a deregulated hematopoietic stem cell, in which a constitutively activated breakpoint cluster region-Abelson (BCR-ABL) fusion kinase plays a crucial role.1 Although its clinical outcome has been markedly improved by the introduction of tyrosine kinase inhibitors (TKIs), this has not necessarily been translated into the permanent cure of the disease because of the presence of quiescent BCR-ABL–positive stem cells.1 Meanwhile, 2% to 15% of the patients successfully treated by TKIs have been reported to develop clonal hematopoiesis as evidenced by clonal chromosomal abnormalities, such as +8, −7/7q-, and -Y, after complete cytogenetic remission (CCR) is obtained.2-4 However, it remains unknown whether clonal hematopoiesis is a general phenomenon in the remission marrows of CML patients. To gain insights into leukemogenetic backgrounds of CML, we enrolled a total of 20 patients with CML in the chronic phase, who had achieved CCR after TKI treatment, and compared clonal burdens of somatic mutations in bone marrow (BM) samples before and after the treatment.
After approval by the Bioethics Committee at Dokkyo Medical University Hospital, written informed consent was obtained from all patients. Whole exome sequencing (WES) of genomic DNA was performed as described previously,5 using nails and/or buccal mucosa cells as germ-line controls. All the candidate mutations were validated by Sanger sequencing or independent deep sequencing using nonamplified DNA. Targeted-capture sequencing of 104 genes implicated in myeloid leukemogenesis was also performed according to the previous report.6
Our cohort included 12 males and 8 females (Table 1). The median age was 54.0 (range, 24-88) years. Fifteen patients were newly diagnosed, whereas the other 5 had a history of previous therapy. Conventional cytogenetic analysis confirmed either standard or complex type of Ph1 translocation in all cases. After collection of BM cells in tumor phases, all the patients were treated with TKIs. BM cells in CCR were also collected with a median therapeutic duration of 26 (range, 6-110) months. Levels of BCR-ABL transcripts were below sensitivity of qRT-PCR assays in 15 patients (Table 1). Germ-line controls (buccal mucosa cells from DU01 to DU08 and nails from all the patients) were collected in the remission phase. WES was performed at a median coverage of 104×.
Clinical information on patients and results of mutation analyses before and after TKI therapies
| Patient no. . | Sex . | Before TKI therapy . | After TKI therapy . | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Age (y) . | Previous therapy . | Karyotype . | BCR-ABL isoform . | FISH (B-A: %) . | qRT-PCR (B-A: ×10−6 copies per copy 18S rRNA)* . | No. of mutations . | TKI . | Duration of TKI (mo) . | Karyotype . | FISH (B-A: %) . | qRT-PCR (B-A: ×10−6 copies per copy 18S rRNA)* . | No. of mutations . | ||
| 1 | M | 32 | None | 46, XY, add(9)(q34), add(17)(q11.2), der(22)t(9;22)(q34;q11.2) [20] | b2a2 | 98.0 | n.d. | 10 | Ima | 26 | 46, XY [20] | 0 | 0.01 | 1 |
| 2 | F | 58 | IFN | 46, XX, t(9;22)(q34;q11.2) [5] / 46, XX [15] | b2a2 | n.d. | n.d. | 4 | Ima | 97 | 46, XX [20] | 0 | n.d. | 1 |
| 3 | F | 49 | None | 46, XX, t(9;22)(q34;q11.2) [20] | b3a2 | n.d. | 35.8 | 10 | Ima, Dasa | 69 | 48, XX,+5,+5 [1]/46, XX [19] | 0 | <0.01 | 0 |
| 4 | F | 49 | None | 46, XX, t(9;22)(q34;q11.2) [17]/46,XX [3] | b3a2 | n.d. | n.d. | 3 | Nilo | 18 | 46, XX [20] | 0 | n.d. | 0 |
| 5 | F | 58 | HU, IFN | 46, XX, t(9;22)(q34;q11.2) [6]/46,XX [14] | b3a2 | 25.8 | n.d. | 9 | Ima | 75 | 46, XX [20] | 0 | <0.01 | 1 |
| 6 | M | 88 | None | 46, XY, t(9;22)(q34;q11.2) [20] | b3a2 | 92.0 | n.d. | 13 | Nilo | 12 | 45, X, -Y [6]/46, XY [14] | n.d. | n.d.† | 3 |
| 7 | M | 57 | None | 46, XY, t(9;22)(q34;q11.2) [20] | b2a2 | 95.0 | 41.0 | 14 | Ima | 21 | 46, XY [20] | n.d. | <0.01 | 0 |
| 8 | F | 33 | None | 46, XX, t(9;22;15)(q34;q11.2;q26) [18] / 46, XX [2] | b2a2 | 98.0 | 17.3 | 7 | Nilo | 6 | 46, XX [20] | 0 | <0.01 | 0 |
| 9 | M | 30 | None | 46, XY, t(9;22)(q34;q11,2) [20] | b3a2 | 97.0 | 64.2 | 3 | Ima | 24 | 46, XY [20] | 0 | <0.01 | 0 |
| 10 | M | 39 | None | 46, XY, t(9;22)(q34;q11,2) [20] | b3a2 | 99.0 | 36.7 | 3 | Ima | 32 | 46, XY [20] | 0 | <0.01 | 0 |
| 11 | F | 62 | None | 46, XX, t(9;22)(q34;q11,2) [20] | b2a2 | 99.0 | 14.0 | 17 | Ima | 23 | 46, XX [20] | 0 | 0.01 | 0 |
| 12 | M | 40 | None | 46, XY, t(9;22)(q34;q11,2)[19] / 46, idem, add(3)(q26) [1] | b2a2 | 99.0 | 15.6 | 4 | Dasa | 12 | 46, XY [3] | 0 | 0.02 | 0 |
| 13 | M | 35 | HU, IFN | 46, XY, t(9;22)(q34;q11,2) [20] | b3a2 | 91.9 | n.d. | 2 | Ima | 69 | 46, XY [20] | 0 | <0.01 | 0 |
| 14 | M | 55 | MCNU, IFN | 46, XY, t(9;22)(q34;q11,2) [13]/46,XY [7] | b3a2 | 82.0 | n.d. | 4 | Ima, Nilo | 43 | 46, XY [20] | 0 | <0.01 | 7 |
| 15 | F | 60 | None | 46, XX, der(9)inv(9)(p12q13)t(9;22)(q34;q11.2), der(22)t(9;22) [20] | b3a2 | n.d. | 81.3 | 9 | Ima | 17 | 46, XX, inv(9)(p12q13) [20] | 0 | <0.01 | 0 |
| 16 | M | 54 | None | 46, XY, t(9;22)(q34;q11,2) [19]/46, XY [1] | b2a2 | 96.0 | 12.9 | 11 | Ima, Nilo | 26 | 46, XY [20] | n.d. | <0.01 | 0 |
| 17 | M | 24 | None | 46, XY, t(1;8)(q42;p23), t(9;22)(q34;q11.2) [20] | b2a2 | n.d. | 59.9 | 1 | Ima | 48 | 46. XY [20] | 0 | <0.01 | 1 |
| 18 | F | 67 | None | 46, XX, t(9;22)(q34;q11,2) [20] | b3a2 | n.d. | 16.0 | 13 | Nilo | 21 | 46, XX [20] | 0 | <0.01 | 0 |
| 19 | M | 67 | HU | 46, XY, t(9;22)(q34;q11,2) [20] | b3a2 | 96.0 | 28.1 | 8 | Ima | 110 | 46, XY [20] | 0 | 0.01 | 0 |
| 20 | M | 54 | None | 46, XY, t(9;22)(q34;q11,2) [20] | b3a2 | n.d. | 81.2 | 11 | Ima, Nilo | 30 | 46, XY [20] | 0 | <0.01 | 0 |
| Patient no. . | Sex . | Before TKI therapy . | After TKI therapy . | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Age (y) . | Previous therapy . | Karyotype . | BCR-ABL isoform . | FISH (B-A: %) . | qRT-PCR (B-A: ×10−6 copies per copy 18S rRNA)* . | No. of mutations . | TKI . | Duration of TKI (mo) . | Karyotype . | FISH (B-A: %) . | qRT-PCR (B-A: ×10−6 copies per copy 18S rRNA)* . | No. of mutations . | ||
| 1 | M | 32 | None | 46, XY, add(9)(q34), add(17)(q11.2), der(22)t(9;22)(q34;q11.2) [20] | b2a2 | 98.0 | n.d. | 10 | Ima | 26 | 46, XY [20] | 0 | 0.01 | 1 |
| 2 | F | 58 | IFN | 46, XX, t(9;22)(q34;q11.2) [5] / 46, XX [15] | b2a2 | n.d. | n.d. | 4 | Ima | 97 | 46, XX [20] | 0 | n.d. | 1 |
| 3 | F | 49 | None | 46, XX, t(9;22)(q34;q11.2) [20] | b3a2 | n.d. | 35.8 | 10 | Ima, Dasa | 69 | 48, XX,+5,+5 [1]/46, XX [19] | 0 | <0.01 | 0 |
| 4 | F | 49 | None | 46, XX, t(9;22)(q34;q11.2) [17]/46,XX [3] | b3a2 | n.d. | n.d. | 3 | Nilo | 18 | 46, XX [20] | 0 | n.d. | 0 |
| 5 | F | 58 | HU, IFN | 46, XX, t(9;22)(q34;q11.2) [6]/46,XX [14] | b3a2 | 25.8 | n.d. | 9 | Ima | 75 | 46, XX [20] | 0 | <0.01 | 1 |
| 6 | M | 88 | None | 46, XY, t(9;22)(q34;q11.2) [20] | b3a2 | 92.0 | n.d. | 13 | Nilo | 12 | 45, X, -Y [6]/46, XY [14] | n.d. | n.d.† | 3 |
| 7 | M | 57 | None | 46, XY, t(9;22)(q34;q11.2) [20] | b2a2 | 95.0 | 41.0 | 14 | Ima | 21 | 46, XY [20] | n.d. | <0.01 | 0 |
| 8 | F | 33 | None | 46, XX, t(9;22;15)(q34;q11.2;q26) [18] / 46, XX [2] | b2a2 | 98.0 | 17.3 | 7 | Nilo | 6 | 46, XX [20] | 0 | <0.01 | 0 |
| 9 | M | 30 | None | 46, XY, t(9;22)(q34;q11,2) [20] | b3a2 | 97.0 | 64.2 | 3 | Ima | 24 | 46, XY [20] | 0 | <0.01 | 0 |
| 10 | M | 39 | None | 46, XY, t(9;22)(q34;q11,2) [20] | b3a2 | 99.0 | 36.7 | 3 | Ima | 32 | 46, XY [20] | 0 | <0.01 | 0 |
| 11 | F | 62 | None | 46, XX, t(9;22)(q34;q11,2) [20] | b2a2 | 99.0 | 14.0 | 17 | Ima | 23 | 46, XX [20] | 0 | 0.01 | 0 |
| 12 | M | 40 | None | 46, XY, t(9;22)(q34;q11,2)[19] / 46, idem, add(3)(q26) [1] | b2a2 | 99.0 | 15.6 | 4 | Dasa | 12 | 46, XY [3] | 0 | 0.02 | 0 |
| 13 | M | 35 | HU, IFN | 46, XY, t(9;22)(q34;q11,2) [20] | b3a2 | 91.9 | n.d. | 2 | Ima | 69 | 46, XY [20] | 0 | <0.01 | 0 |
| 14 | M | 55 | MCNU, IFN | 46, XY, t(9;22)(q34;q11,2) [13]/46,XY [7] | b3a2 | 82.0 | n.d. | 4 | Ima, Nilo | 43 | 46, XY [20] | 0 | <0.01 | 7 |
| 15 | F | 60 | None | 46, XX, der(9)inv(9)(p12q13)t(9;22)(q34;q11.2), der(22)t(9;22) [20] | b3a2 | n.d. | 81.3 | 9 | Ima | 17 | 46, XX, inv(9)(p12q13) [20] | 0 | <0.01 | 0 |
| 16 | M | 54 | None | 46, XY, t(9;22)(q34;q11,2) [19]/46, XY [1] | b2a2 | 96.0 | 12.9 | 11 | Ima, Nilo | 26 | 46, XY [20] | n.d. | <0.01 | 0 |
| 17 | M | 24 | None | 46, XY, t(1;8)(q42;p23), t(9;22)(q34;q11.2) [20] | b2a2 | n.d. | 59.9 | 1 | Ima | 48 | 46. XY [20] | 0 | <0.01 | 1 |
| 18 | F | 67 | None | 46, XX, t(9;22)(q34;q11,2) [20] | b3a2 | n.d. | 16.0 | 13 | Nilo | 21 | 46, XX [20] | 0 | <0.01 | 0 |
| 19 | M | 67 | HU | 46, XY, t(9;22)(q34;q11,2) [20] | b3a2 | 96.0 | 28.1 | 8 | Ima | 110 | 46, XY [20] | 0 | 0.01 | 0 |
| 20 | M | 54 | None | 46, XY, t(9;22)(q34;q11,2) [20] | b3a2 | n.d. | 81.2 | 11 | Ima, Nilo | 30 | 46, XY [20] | 0 | <0.01 | 0 |
B-A, BCR-ABL; Dasa, dasatinib; FISH, fluorescence in situ hybridization; HU, hydroxyurea; IFN, interferon; in/del, insertion/deletion; Ima, imatinib; MCNU, methyl chloroethyl nitroso urea; n.d., not determined; Nilo, nilotinib; qRT-PCR, quantitative reverse transcription polymerase chain reaction; rRNA, ribosomal RNA.
qRT-PCR: levels of BCR-ABL messenger RNA (mRNA) in the BM quantified using real-time RT-PCR were indicated as ×10−6 copies per copy 18S rRNA.
Minimal residual BCR-ABL mRNA in the peripheral blood for patient no. 6 was below detection limit (<5 copies per 0.5 μg RNA) using transcription-mediated amplification followed by hybridization protection assay (Amp-CML).
A median of 8 (range, 1-17) somatic mutations were detected in samples before TKI treatment (supplemental Table 1; available on the Blood Web site). Older patients tended to carry more mutations than younger patients (correlation coefficient, 0.64; P = .002). In 17 patients, BCR-ABL was positive in most of the BM cells, indicating that observed mutations were owned by BCR-ABL–positive cells. By contrast, in the remaining patients (DU02, DU05, and DU14), low positivity of BCR-ABL precluded accurate determination of whether BCR-ABL–positive cells carried the mutations. As opposed to previous reports,7,8 only 3 patients were found to have mutations implicated in myeloid malignancies, including mutations of DOT1L in DU02, ATM in DU11, and PTPN11 in DU20,9-13 and additional mutations in ASXL1, STAG2, WAPAL, PDS5B, and FANCE14-17 with low variant allele frequencies (VAFs) were newly identified by targeted-capture sequencing in a single case (DU14).
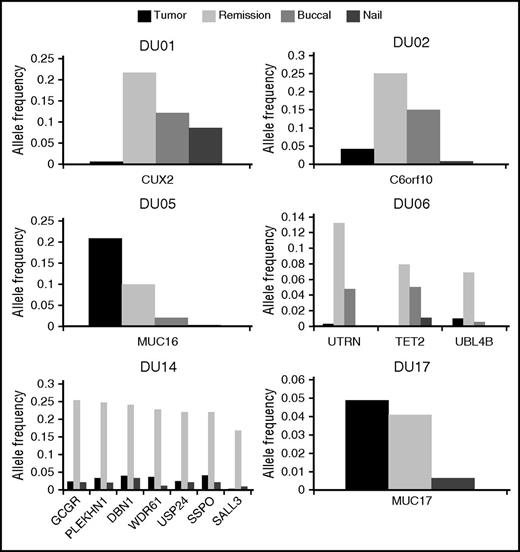
We also detected a total of 14 mutations in the remission marrow specimens from 6 patients (supplemental Table 2), 5 of whom tested by qRT-PCR showed an undetectable level of BCR-ABL transcripts. Among those mutations, only a MUC16 mutation in DU05 had been also detected in the corresponding tumor sample. The remaining 13 mutations were newly detected in the remission marrow from 5 patients (DU01, DU02, DU06, DU14, and DU17), including mutations in CUX2 (DU01), TET2 (DU06), and SALL3 (DU14). We could not detect any specific features with regard to age, type of BCR-ABL transcript, or type of TKIs among these patients, except that DU14 was the only patient previously treated with an alkylating agent. Although not detected by WES, all these mutations were demonstrated to be present also in tumor-phase samples using amplicon-based deep sequencing, albeit generally at much lower VAFs (<0.05) compared with those in remission samples (Figure 1).
Allele frequencies of mutations in the tumor and remission marrows, buccal mucosa cells, and nails. Among 20 patients, 6 patients (DU01, DU02, DU05, DU06, DU14, and DU17) exhibited gene mutations in the remission marrows. We analyzed VAFs of these mutations in the tumor and remission marrows, buccal mucosa cells, and nails by using deep sequencing. Horizontal and vertical axes indicate names of mutated genes and VAFs, respectively. Dark and light gray boxes indicate the tumor and remission marrows, and small dotted and patched boxes denote buccal mucosa cells and nails.
Allele frequencies of mutations in the tumor and remission marrows, buccal mucosa cells, and nails. Among 20 patients, 6 patients (DU01, DU02, DU05, DU06, DU14, and DU17) exhibited gene mutations in the remission marrows. We analyzed VAFs of these mutations in the tumor and remission marrows, buccal mucosa cells, and nails by using deep sequencing. Horizontal and vertical axes indicate names of mutated genes and VAFs, respectively. Dark and light gray boxes indicate the tumor and remission marrows, and small dotted and patched boxes denote buccal mucosa cells and nails.
Notably, according to the observed VAFs, the MUC16 mutation in DU05 was shared in 41.2% and 19.8% of tumor and remission samples, respectively. As BCR-ABL–positive cells comprised only 25.8% of the tumor sample in FISH analysis, we were not able to determine whether BCR-ABL had occurred in MUC16-mutation positive or negative cells. However, cells carrying the MUC16 mutation alone persisted even after BCR-ABL–positive cells were eliminated by TKI treatment. By contrast, VAFs in tumor samples were much lower (<0.05) for other mutations. These mutations continued to be detected in remission samples in the fraction comparable (DU17) to or substantially larger (all other patients) than that in tumor samples. Thus, it is likely that these mutations may have been present in BCR-ABL–negative clones.
To further investigate the origin of the mutations identified in remission samples from 6 patients, we also interrogated germ-line DNA obtained from nails and/or buccal mucosa for the relevant mutations. Amplicon-based deep sequencing reproducibly detected identical mutations in these nonhematopoietic tissues at varying VAFs in all 6 patients, although generally their VAFs were lower than those in the corresponding remission marrows (Figure 1). Although contamination of blood cells to buccal mucosa samples at low levels cannot be ruled out, these findings suggest that mutations detected in remission samples should represent somatic mosaicism rather than age-related clonal hematopoiesis recently described in the general population.18
This is the first study through comprehensive genome sequencing of BM cells in patients with CML before and after TKI therapy, albeit limitations resulting from small number and heterogeneity of the patients exist. In the tumor phase, BM cells demonstrated a variety of mutations, although epigenetic regulators were less frequently involved, compared with the previous report.7 All the mutations except for the MUC16 mutation disappeared after CCR. Our study could not find any clear predisposition to the generation of BCR-ABL gene fusion. Among 20 patients, 6 exhibited persistent or newly appeared gene mutations in the remission marrow. This observation may be consistent with previous reports in which somatic mosaicism in the hematopoietic cells was shown to be detected in a substantial number of individuals among the general population.19,20 These mutations in the remission marrow were also detected in the tumor marrow when analyzed retrospectively by deep sequencing, although their VAFs were much lower than those in the remission samples. Thus, it may be reasonably speculated that TKI treatment eliminated BCR-ABL–positive clones, which allowed severely suppressed normal cells, including mutation-carrying cells, in the tumor phase to rapidly expand and repopulate the entire marrow. This may have provided the mutated clone with a chance to occupy a substantial fraction of the remission marrow because of a bottleneck effect, especially after achieving undetectable levels of BCR-ABL transcripts. It would be worth noting that some of these mutations, such as the TET2 mutation found in DU06, could potentially predispose to the development of another hematological malignancy,21 which may necessitate careful monitoring of the emergence of such abnormal clones even once CCR has been successfully obtained after TKI treatment.
In summary, mutations of the genes commonly found in myelodysplastic syndrome and acute myelogenous leukemia are less prevalent in CML. Some cells carrying mosaic mutations can be expanded after the disappearance of BCR-ABL in the CCR phase of CML.
The online version of this article contains a data supplement.
Authorship
Acknowledgments: The authors thank Kyoko Tabei and Kazuhiko Iwasaki in the Division of Clinical Science, Research Support Center, Dokkyo Medical University for their special technical assistance. This work was supported in part by research funding from Kyowa Hakko Kirin Co. Ltd., Chugai Pharmaceutical Co. Ltd., Novartis Pharma K.K., Bristol-Myers Squibb, Pfizer, Astellas, Dainippon Sumitomo Pharma, Eisai, Takeda, and MSD K.K. (K.M.).
Contribution: K.M. and S.O. designed the research; Y.Nakamura and K.Y. conducted genomic analyses; K.C., H.T., Y.S., and S.M. performed bioinformatics; all the authors analyzed data; K.M. and M.I. wrote the manuscript; and H.M. and S.O. reviewed the manuscript.
Conflict-of-interest disclosure: K.M. has received honoraria from Kyowa Hakko Kirin, Co. Ltd. S.O. has participated on the advisory committee for Kan Research Institute Inc. The remaining authors declare no competing financial interests.
Correspondence: Kinuko Mitani, Department of Hematology and Oncology, Dokkyo Medical University School of Medicine, 880 Kitakobayashi, Mibu-machi, Shimotsuga-gun, Tochigi 321-0293, Japan; e-mail: kinukom-tky@umin.ac.jp.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal