Key Points
Setdb1, an H3K9 histone methyltransferase, is essential for the maintenance of HSPCs.
Setdb1 restricts the activation of nonhematopoietic genes, such as gluconeogenic pathway genes, to maintain HSPCs.
Abstract
Setdb1, also known as Eset, is a methyltransferase that catalyzes trimethylation of H3K9 (H3K9me3) and plays an essential role in the silencing of endogenous retroviral elements (ERVs) in the developing embryo and embryonic stem cells (ESCs). Its role in somatic stem cells, however, remains unclear because of the early death of Setdb1-deficient embryos. We demonstrate here that Setdb1 is the first H3K9 methyltransferase shown to be essential for the maintenance of hematopoietic stem and progenitor cells (HSPCs) in mice. The deletion of Setdb1 caused the rapid depletion of hematopoietic stem and progenitor cells (HSPCs), as well as leukemic stem cells. In contrast to ESCs, ERVs were largely repressed in Setdb1-deficient HSPCs. A list of nonhematopoietic genes was instead ectopically activated in HSPCs after reductions in H3K9me3 levels, including key gluconeogenic enzyme genes fructose-1,6-bisphosphatase 1 (Fbp1) and Fbp2. The ectopic activation of gluconeogenic enzymes antagonized glycolysis and impaired ATP production, resulting in a compromised repopulating capacity of HSPCs. Our results demonstrate that Setdb1 maintains HSPCs by restricting the ectopic activation of nonhematopoietic genes detrimental to their function and uncover that the gluconeogenic pathway is one of the critical targets of Setdb1 in HSPCs.
Introduction
In mammals, DNA methylation and trimethylation at histone H3 lysine 9 (H3K9me3) are strongly associated with each other and cooperatively silence genes and retroelements.1 Although DNA methylation is considered to repress the expression of viral genes in differentiated cells, repression in pluripotent cells is mediated by DNA methylation and histone modifications.2 There are 5 members of the SUV39 family that catalyze H3K9 methylation: Suv39h1, Suv39h2, G9a, GLP, and Setdb1. Setdb1 (also known as Eset) is an H3K9 trimethyltransferase that is mainly localized in euchromatic regions. Homozygous mutations of Setdb1 have been shown to result in peri-implantation lethality between 3.5 and 5.5 dpc.3 Setdb1 represses subfamilies of endogenous retroviruses (ERVs) in a DNA methylation-independent manner in murine embryonic stem cells (ESCs) and the early embryo.2,4,5 Kap1 (also known as Trim28 and Tif1β), a scaffold protein targeted to specific sequences through KRAB zinc finger proteins, recruits Setdb1. Thus, Kap1 and Setdb1 cooperatively repress ERVs via the enrichment of H3K9me3 in ESCs.2,6 However, their role in adult somatic stem cells remained largely obscure.
It has been reported that the amount of euchromatin decreases while the layer of heterochromatin at the nuclear envelop increases during the differentiation of hematopoietic stem cells (HSCs). Prevention of heterochromatin formation by pharmacological inhibition of G9a, which catalyzes H3K9me2, results in delayed HSC differentiation. Thus, the proper transition from euchromatin to heterochromatin mediated by H3K9 methylation is required for efficient HSC differentiation.7 Of interest, however, is that G9a is dispensable for the maintenance of HSCs.8 We previously characterized the hematopoietic cell-specific deletion of Kap1 in mice and reported that Kap1 is essential for the maintenance of HSCs. In collaboration with heterochromatin protein 1 (HP1) proteins, Kap1 functions as a critical repressive machinery that targets genes not normally activated in the hematopoietic compartment, thereby maintaining the transcriptional signature specific to HSCs.9 These findings suggested the H3K9me3-mediated regulation of HSC functions and a role for Setdb1 in this process.
In the present study, we examined mice in which Setdb1 was deleted specifically in hematopoietic cells and found that Setdb1 is the first H3K9 methyltransferase shown to be essential for the maintenance of hematopoietic stem and progenitor cells (HSPCs). Comprehensive transcriptome and epigenomic analyses revealed that Setdb1 is mostly dispensable for the silencing of ERVs in HSPCs, but preferentially targets nonhematopoietic genes, the ectopic expression of which compromises the function of HSPCs. Among nonhematopoietic target genes, we demonstrate that the gluconeogenic pathway is one of the critical targets of Setdb1 in HSPCs.
Materials and methods
Metabolite extraction and metabolome analysis
Cells were washed twice by 5% mannitol solution (10 mL first and then 2 mL), treated with 800 µL of methanol, and left at rest for 30 seconds to inactivate enzymes. The cell extract was then treated with 550 µL Milli-Q water containing internal standards (H3304-1002; Human Metabolome Technologies [HMT]) and left at rest for another 30 seconds. The extract was obtained and centrifuged at 2300 × g and 4°C for 5 minutes, and then 800 µL upper aqueous layer was centrifugally filtered through a Millipore 5-kDa cutoff filter (UltrafreeMC-PLHCC; HMT) to remove macromolecules (9100 × g, 4°C, 120 min). The filtrate was centrifugally concentrated and resuspended in 50 µL Milli-Q water for metabolome analysis at HMT. Metabolome analysis was conducted by C-SCOPE package of HMT, using capillary electrophoresis time-of-flight mass spectrometry (CE-TOFMS) for cation analysis and CE-tandem mass spectrometry (CE-MS/MS) for anion analysis. Briefly, CE-TOFMS analysis was carried out using an Agilent CE capillary electrophoresis system equipped with an Agilent 6210 TOF mass spectrometer (Agilent Technologies). The spectrometer was scanned from m/z 50 to 1000.10,11 Peaks were extracted using MasterHands automatic integration software12 and MassHunter Quantitative Analysis B.04.00 (Agilent Technologies) to obtain peak information, including m/z, peak area, and migration time (MT). Signal peaks were annotated according to the HMT metabolite database on the basis of their m/z values with the MTs. Concentrations of metabolites were calculated by normalizing the peak area of each metabolite with respect to the area of the internal standard and by using standard curves with 3-point calibrations. Detected metabolites were plotted on metabolic pathway maps using VANTED software.13
RNA-sequencing
Total RNA was subjected to reverse transcription and amplification for 16 cycles, respectively, with SMARTer Ultra Low Input RNA Kit for Sequencing v3 (Clontech). After sonication with an ultrasonicator (Covaris), the libraries for RNA-seq were generated from 50 ng fragmented DNA with 8 cycles of amplification, using a NEBNext Ultra DNA Library Prep Kit (New England BioLabs). After the libraries were quantified, using a high-sensitivity Chip on Bioanalyzer (Agilent), the samples were subjected to sequencing with a Hiseq1500 (Illumina), and 61 cycles of the sequencing reactions were performed. TopHat2 (version 2.0.13; with default parameters) and Bowtie2 (version 2.1.0) were used to align to the reference mouse genome (mm9 from University of California, Santa Cruz Genome Browser; http://genome.ucsc.edu/). Then gene expression values were calculated as reads per kilobase of exon unit per million mapped reads, using cufflinks (version 2.2.1). Total read number per ERV family was determined as described.14 Proviruses with a Smith–Watterman score of at least 5000 (Repeatmasker) were used to properly annotate ERVs.
ChIP-sequencing
Chromatin immunoprecipitation (ChIP) assays were performed as previously described.15 Briefly, 1 × 105 pooled bone marrow (BM) Lin− Sca-1+ c-Kit+ (LSK) cells were subjected to immunoprecipitation, using anti-H3K27me3 (07449; Millipore) and anti-H3K9me3 (clone 2F3).16 Sheep anti-rabbit immunoglobulin G (IgG) and sheep anti-mouse IgG Dynabeads were used to capture anti-H3K27me3 and anti-H3K9me3 (IgG1), respectively. The RPM (reads per million mapped reads) values of the sequenced read were calculated every 2000-base pair bin with a shifting size of 200 base pairs, using bedtools. To evaluate the histone modification mark of each gene, the RPM values of the region from 4 kb upstream to 4 kb downstream of the transcription start site (TSS) of the immunoprecipitated samples were divided by RPM of corresponding input. To visualize with Integrative Genomics Viewer (http://www.broadinstitute.org/igv), the RPM values of the immunoprecipitated samples were normalized by subtracting the RPM values of the input samples in each bin and converted to a bigwig file, using the wigToBigWig tool.
Reduced representation bisulfite sequencing
Five hundred nanograms DNA was digested by MspI (Takara), and fragments cut into 150-250 bp were selectively extracted. Methylated adaptor-oligos (Illumina) were ligated to both ends of the fragments, followed by bisulfite conversion and polymerase chain reaction (PCR) amplification to prepare libraries for sequencing. The libraries were sequenced under single-end 50 bp protocol, using Hiseq1500 sequencer (Illumina). Differentially methylated nucleotides/regions were identified using methylKit.17
Accession numbers
Data were deposited in DNA Data Bank of Japan (RNA-sequence, DRA004191; ChIP-sequence, DRA002605; reduced representation bisulfite sequencing [RRBS], DRA002147, DRA002148, DRA002149, DRA002150, DRA002151, DRA002152).
Results
Deletion of Setdb1 leads to the rapid depletion of HSPCs and leukemic stem cells
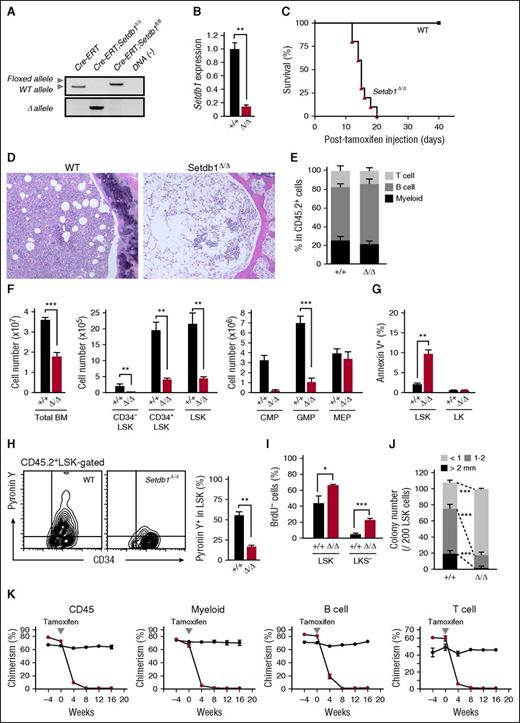
To delineate the function of Setdb1 in HSPCs, we conditionally deleted Setdb1 by crossing Setdb1fl/fl mice with Rosa:Cre-ERT (Cre-ERT) mice. We transplanted BM cells from Cre-ERT control and Cre-ERT;Setdb1fl/fl mice without competitor cells into lethally irradiated recipient mice. After confirming engraftment, we deleted Setdb1 by intraperitoneal injection of tamoxifen at 4 weeks after transplantation. We hereafter refer to the recipient mice reconstituted with Cre-ERT and Setdb1Δ/Δ cells as wild-type (WT) and Setdb1Δ/Δ mice, respectively. The deletion of Setdb1 was efficient, as judged by the genomic PCR and the reverse transcription (RT)-PCR of donor-derived BM progentor cells (Figure 1A-B). All Setdb1Δ/Δ mice died within 21 days of the initial injection of tamoxifen (Figure 1C) because of severe hematopoietic failure, as evident in BM sections (Figure 1D). Therefore, we subsequently examined hematopoiesis in Setdb1Δ/Δ mice 2 weeks after the initiation of the tamoxifen injection. Although the proportion of each hematopoietic lineage in the peripheral blood (PB) was not significantly changed in Setdb1Δ/Δ mice (Figure 1E), the numbers of BM cells and primitive hematopoietic cells, including CD34−Lin−Sca-1+c-Kit+ (CD34−LSK) HSCs, CD34+LSK multipotent progenitor cells, common myeloid progenitors, and granulocyte/macrophage progenitors (GMPs), but not megakaryocyte/erythrocyte progenitors were markedly lower in Setdb1Δ/Δ mice than in WT mice (Figure 1F). Apoptotic cell death was enhanced in Setdb1Δ/Δ LSK cells (Figure 1G), and mitochondrial reactive oxygen species levels were also increased in Setdb1Δ/Δ CD34−LSK HSCs and CD34+LSK multipotent progenitors (supplemental Figure 1, available on the Blood Web site). However, RNA content detected by Pyronin Y staining was significantly lower (Figure 1H), and the incorporation of BrdU was significantly less (Figure 1I), in Setdb1Δ/Δ LSK cells than in WT cells, suggesting impaired cell proliferation. Correspondingly, Setdb1Δ/Δ LSK cells gave rise to significantly smaller colonies than WT cells (Figure 1J). The heterozygous deletion of Setdb1 also mildly compromised the proliferation of HSPCs. Setdb1+/Δ LSK cells showed mildly impaired proliferation in culture with a significant reduction of LSK cells (supplemental Figure 2A-D). In addition, Setdb1+/Δ LSK cells generated fewer colonies, although the reduction in colony numbers was mild compared with Setdb1Δ/Δ LSK cells (supplemental Figure 2E).
Deletion of Setdb1 leads to rapid depletion of HSPCs in BM. (A) Deletion of Setdb1 in Cre-ERT;Setdb1fl/fl BM Lin-c-Kit+ hematopoietic progenitor cells were confirmed by genomic PCR 2 weeks after the first injection of tamoxifen. “WT,” “floxed,” and “∆” alleles indicate the wild-type Setdb1 allele, floxed Setdb1 allele, and floxed Setdb1 allele after the removal of exons 15 and 16 by Cre recombinase, respectively. (B) RT-PCR of Setdb1 expression in LSK HSPCs 2 weeks after the first injection of tamoxifen. mRNA levels were normalized to Hprt1 expression, and relative expression levels are shown as the mean ± SD of triplicate analyses. (C) Survival curve of recipient mice repopulated with BM cells from Cre-ERT (WT) and Cre-ERT;Setdb1fl/fl (Setdb1Δ/Δ) mice after the initial injection of tamoxifen (n = 10). BM cells from 8-week-old Cre-ERT and Cre-ERT;Setdb1fl/fl mice were transplanted into lethally irradiated recipient mice without competitor cells. At 4 weeks after transplantation, recipient mice were injected with tamoxifen for 5 consecutive days. (D) Representative hematoxylin and eosin staining of BM sections from WT and moribund Setdb1Δ/Δ mice in (C). (E) The proportions of Mac-1+ myeloid, B220+ B, and CD4+ or CD8+ T cells among CD45.2+ donor-derived hematopoietic cells in PB from WT and Setdb1Δ/Δ mice (n = 4) 2 weeks after the first injection of tamoxifen. Data are shown as the mean ± SD. (F) Absolute numbers of total BM cells, CD34−LSK HSCs, CD34+LSK multipotent progenitor cells, common myeloid progenitors, GMPs, and megakaryocyte/erythrocyte progenitors in the unilateral femur and tibia of WT and Setdb1Δ/Δ mice (n = 4) 2 weeks after the first injection of tamoxifen. Data are shown as the mean ± SD. (G) Percentage of Annexin V+ cells in BM LSK and LK cells in WT (n = 4) and Setdb1Δ/Δ mice (n = 6) 9 days after the first injection of tamoxifen. Data are shown as the mean ± SD. (H) Representative flow cytometry profiles of the incorporation of Pyronin Y by BM CD45.2+ LSK cells 2 weeks after the first injection of tamoxifen (left). The percentage of Pyronin Y+ cells in LSK cells in WT and Setdb1Δ/Δ mice (n = 4) are shown as the mean ± SD (right). (I) Cell cycle status of LSK and Lin−c-Kit+ Sca-1− (LKS−) committed progenitor cells evaluated by BrdU short-labeling assays. BrdU was administered 12 and 24 hours before the analysis to mark cells that entered S phase. Data are shown as the mean ± SD (n = 6). (J) Colony-forming assays with LSK cells from WT and Setdb1Δ/Δ mice 2 weeks after the initial injection of tamoxifen. The absolute numbers of colonies with the indicated size per 200 LSK cells are shown as the mean ± SD of triplicate cultures. (K) Competitive reconstitution assays. BM cells from 8-week-old CreERT and CreERT;Setdb1fl/fl mice (2 × 106 cells) were transplanted into lethally irradiated recipient mice with competitor BM cells (1 × 106 cells). At 8 weeks after transplantation, recipient mice were injected with tamoxifen for 5 consecutive days. The chimerism of CD45.2+ donor-derived cells in all CD45+ cells and in myeloid, B, and T cells in the PB of recipient mice is shown as the mean ± SD (n = 5). *P < .05; **P < .01; ***P < .001; ns, not significant.
Deletion of Setdb1 leads to rapid depletion of HSPCs in BM. (A) Deletion of Setdb1 in Cre-ERT;Setdb1fl/fl BM Lin-c-Kit+ hematopoietic progenitor cells were confirmed by genomic PCR 2 weeks after the first injection of tamoxifen. “WT,” “floxed,” and “∆” alleles indicate the wild-type Setdb1 allele, floxed Setdb1 allele, and floxed Setdb1 allele after the removal of exons 15 and 16 by Cre recombinase, respectively. (B) RT-PCR of Setdb1 expression in LSK HSPCs 2 weeks after the first injection of tamoxifen. mRNA levels were normalized to Hprt1 expression, and relative expression levels are shown as the mean ± SD of triplicate analyses. (C) Survival curve of recipient mice repopulated with BM cells from Cre-ERT (WT) and Cre-ERT;Setdb1fl/fl (Setdb1Δ/Δ) mice after the initial injection of tamoxifen (n = 10). BM cells from 8-week-old Cre-ERT and Cre-ERT;Setdb1fl/fl mice were transplanted into lethally irradiated recipient mice without competitor cells. At 4 weeks after transplantation, recipient mice were injected with tamoxifen for 5 consecutive days. (D) Representative hematoxylin and eosin staining of BM sections from WT and moribund Setdb1Δ/Δ mice in (C). (E) The proportions of Mac-1+ myeloid, B220+ B, and CD4+ or CD8+ T cells among CD45.2+ donor-derived hematopoietic cells in PB from WT and Setdb1Δ/Δ mice (n = 4) 2 weeks after the first injection of tamoxifen. Data are shown as the mean ± SD. (F) Absolute numbers of total BM cells, CD34−LSK HSCs, CD34+LSK multipotent progenitor cells, common myeloid progenitors, GMPs, and megakaryocyte/erythrocyte progenitors in the unilateral femur and tibia of WT and Setdb1Δ/Δ mice (n = 4) 2 weeks after the first injection of tamoxifen. Data are shown as the mean ± SD. (G) Percentage of Annexin V+ cells in BM LSK and LK cells in WT (n = 4) and Setdb1Δ/Δ mice (n = 6) 9 days after the first injection of tamoxifen. Data are shown as the mean ± SD. (H) Representative flow cytometry profiles of the incorporation of Pyronin Y by BM CD45.2+ LSK cells 2 weeks after the first injection of tamoxifen (left). The percentage of Pyronin Y+ cells in LSK cells in WT and Setdb1Δ/Δ mice (n = 4) are shown as the mean ± SD (right). (I) Cell cycle status of LSK and Lin−c-Kit+ Sca-1− (LKS−) committed progenitor cells evaluated by BrdU short-labeling assays. BrdU was administered 12 and 24 hours before the analysis to mark cells that entered S phase. Data are shown as the mean ± SD (n = 6). (J) Colony-forming assays with LSK cells from WT and Setdb1Δ/Δ mice 2 weeks after the initial injection of tamoxifen. The absolute numbers of colonies with the indicated size per 200 LSK cells are shown as the mean ± SD of triplicate cultures. (K) Competitive reconstitution assays. BM cells from 8-week-old CreERT and CreERT;Setdb1fl/fl mice (2 × 106 cells) were transplanted into lethally irradiated recipient mice with competitor BM cells (1 × 106 cells). At 8 weeks after transplantation, recipient mice were injected with tamoxifen for 5 consecutive days. The chimerism of CD45.2+ donor-derived cells in all CD45+ cells and in myeloid, B, and T cells in the PB of recipient mice is shown as the mean ± SD (n = 5). *P < .05; **P < .01; ***P < .001; ns, not significant.
We next transplanted BM cells from Cre-ERT control and Cre-ERT;Setdb1fl/fl mice with competitor cells into lethally irradiated WT recipient mice. After the deletion of Setdb1, the chimerism of Setdb1Δ/Δ hematopoietic cells significantly decreased in PB (Figure 1K), and Setdb1Δ/Δ LSK HSPCs in BM were completely depleted by 6 months (data not shown).
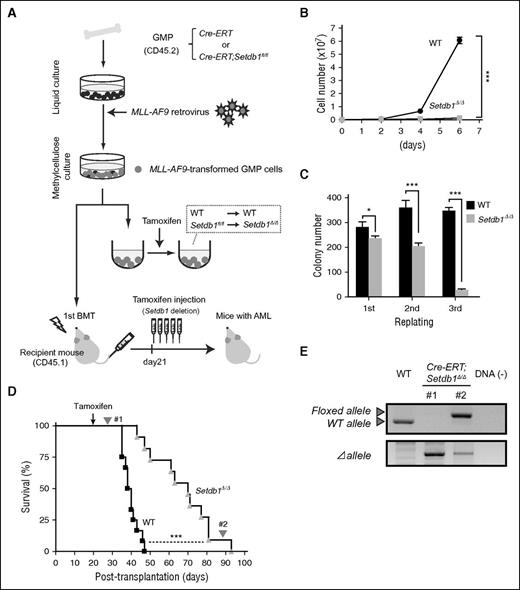
To test the role of Setdb1 in cancer stem cells, we used a myeloid leukemia model induced by the leukemic fusion gene MLL-AF9.18 We purified GMPs from Cre-ERT control and Cre-ERT;Setdb1fl/fl mice and transduced them with an MLL-AF9 retrovirus (Figure 2A). We found that the proliferation of transformed GMPs was severely compromised after the deletion of Setdb1 in liquid culture (Figure 2B) and in colony replating assays (Figure 2C). We then transplanted transformed GMPs into lethally irradiated recipient mice, along with wild-type BM cells for radioprotection. After confirming the development of overt leukemia, we injected tamoxifen from day 21 to delete Setdb1. Although the deletion of Setdb1 significantly prolonged the survival of recipient mice, all mice eventually died (Figure 2D). Genomic PCR demonstrated the efficient deletion of Setdb1 in leukemic cells immediately after the injection of tamoxifen. However, leukemic cells from moribund mice retained the floxed allele (Figure 2E), suggesting that only the leukemic cells that retained at least 1 floxed allele (escapers) gradually expanded and induced lethal leukemia. Moreover, SETDB1 knockdown significantly suppressed the growth of human leukemic cells (HL-60 and K562 cells) (supplemental Figure 3). These results indicate that Setdb1 also plays an essential role in leukemic stem cells.
Role of Setdb1 in leukemic stem cells. (A) A schematic diagram of the experimental process. GMPs from CreERT and CreERT;Setdb1fl/fl mice were transduced with MLL-AF9 and cultured in methylcellulose medium. To delete Setdb1 in vitro, MLL-AF9-transformed GMPs were transferred to liquid medium containing 200 nM 4-hydroxy tamoxifen (4-OHT). MLL-AF9-transformed GMPs were also transplanted into lethally irradiated recipient mice, together with WT BM cells for radioprotection. To delete Setdb1 in vivo, tamoxifen was intraperitoneally injected once a day for 5 consecutive days at 21 days after transplantation. (B) Growth of MLL-AF9-transformed GMPs after the deletion of Setdb1. MLL-AF9-transformed GMPs (1 × 104 cells each) were cultured in IMDM with 20% fetal calf serum, SCF, FP6, GM-CSF, and IL-3 (10 ng/mL each). Data are shown as the mean ± SD of triplicate cultures. (C) Replating efficiency of MLL-AF9-transformed GMPs after the deletion of Setdb1. MLL-AF9-transformed GMPs (1500 cells) were serially replated in methylcellulose medium containing 10 ng/mL SCF, 10 ng/mL FP6, 10 ng/mL GM-CSF, 10 ng/mL IL-3, and 100 nM 4-OHT. Data are shown as the mean ± SD of triplicate cultures. (D) Overall survival of mice injected with 4 × 105 WT or Setdb1Δ/ΔMLL-AF9-transformed cells compared by a Kaplan-Meier analysis (WT, n = 10; Setdb1Δ/Δ n = 9). (E) The efficiency of the deletion of Setdb1 in MLL-AF9-transformed GMPs was monitored. Genomic PCR data of leukemic cells immediately after the injection of tamoxifen and from moribund mice (#1 and #2, respectively, indicated in panel D). *P < .05; **P < .01; ***P < .001.
Role of Setdb1 in leukemic stem cells. (A) A schematic diagram of the experimental process. GMPs from CreERT and CreERT;Setdb1fl/fl mice were transduced with MLL-AF9 and cultured in methylcellulose medium. To delete Setdb1 in vitro, MLL-AF9-transformed GMPs were transferred to liquid medium containing 200 nM 4-hydroxy tamoxifen (4-OHT). MLL-AF9-transformed GMPs were also transplanted into lethally irradiated recipient mice, together with WT BM cells for radioprotection. To delete Setdb1 in vivo, tamoxifen was intraperitoneally injected once a day for 5 consecutive days at 21 days after transplantation. (B) Growth of MLL-AF9-transformed GMPs after the deletion of Setdb1. MLL-AF9-transformed GMPs (1 × 104 cells each) were cultured in IMDM with 20% fetal calf serum, SCF, FP6, GM-CSF, and IL-3 (10 ng/mL each). Data are shown as the mean ± SD of triplicate cultures. (C) Replating efficiency of MLL-AF9-transformed GMPs after the deletion of Setdb1. MLL-AF9-transformed GMPs (1500 cells) were serially replated in methylcellulose medium containing 10 ng/mL SCF, 10 ng/mL FP6, 10 ng/mL GM-CSF, 10 ng/mL IL-3, and 100 nM 4-OHT. Data are shown as the mean ± SD of triplicate cultures. (D) Overall survival of mice injected with 4 × 105 WT or Setdb1Δ/ΔMLL-AF9-transformed cells compared by a Kaplan-Meier analysis (WT, n = 10; Setdb1Δ/Δ n = 9). (E) The efficiency of the deletion of Setdb1 in MLL-AF9-transformed GMPs was monitored. Genomic PCR data of leukemic cells immediately after the injection of tamoxifen and from moribund mice (#1 and #2, respectively, indicated in panel D). *P < .05; **P < .01; ***P < .001.
Setdb1 is mostly dispensable for the silencing of ERVs in HSPCs
To investigate the effects of the Setdb1 deletion in HSPCs, we performed chromatin immunoprecipitation followed by the sequencing (ChIP-seq) of H3K9me3 and H3K9/K14 acetylation (ac) in WT and Setdb1Δ/Δ GMPs and RNA-sequencing (RNA-seq) of Setdb1Δ/Δ LSK cells and GMPs recovered from recipient mice 2 weeks after the deletion of Setdb1. Although western blot analysis did not reveal any significant difference in global H3K9me3 levels (supplemental Figure 4A), ChIP-seq detected changes in the levels of H3K9me3 between WT and Setdb1Δ/Δ GMPs (supplemental Figure 4B). Significant reductions were observed at ERV genome regions such as endogenous retrovirus-1 (ERV1) (class 1), intracisternal A-particle (IAP) (class 2), and endogenous retrovirus-like elements (class 3), and the promoters of coding genes (4.0 kb ± TSS). However, the reductions observed in H3K9me3 levels at ERV genome regions were milder than previously reported changes in ESCs2,4 (supplemental Figure 4B). Furthermore, RRBS revealed that DNA methylation was maintained at high levels in Setdb1Δ/Δ LSK cells and GMPs (supplemental Figure 4C). Correspondingly, the deletion of Setdb1 in HSPCs did not induce the high-level activation of ERVs (supplemental Figure 5). Although class 1 (ERV1) and class 2 (IAP and early transposon element), but not class 3 (mouse transposon A, murine endogenous retrovirus-L) and long interspersed elements, showed slight upregulation in a similar manner to Setdb1Δ/Δ ESCs, the levels of activation were very mild, particularly in LSK cells, compared with in Setdb1Δ/Δ ESCs, and fold upregulation was less than 2.0 on average, even in GMPs (supplemental Figure 5A). In Setdb1Δ/Δ ESCs and the developing brain, the activation of the long terminal repeat of ERVs, such as IAPs, has been shown to lead to the ectopic upregulation of neighboring genes, generating chimeric transcripts starting from ERVs.4,5 A RNA-seq analysis revealed no chimeric transcripts for genes highly derepressed in Setdb1Δ/Δ LSK cells and GMPs listed in Figure 3, such as Fbp1, Fbp2, and epithelial cell adhesion molecule (Epcam), even though these gene loci have a number of neighboring and intronic ERV sequences (supplemental Figure 5B). Chimeric transcripts were not detected, even with the selected ERV loci showing significant derepression in Setdb1Δ/Δ LSK cells and GMPs (data not shown). These results indicate that the effect of Setdb1 loss on the silencing of ERVs is limited in adult HSPCs.
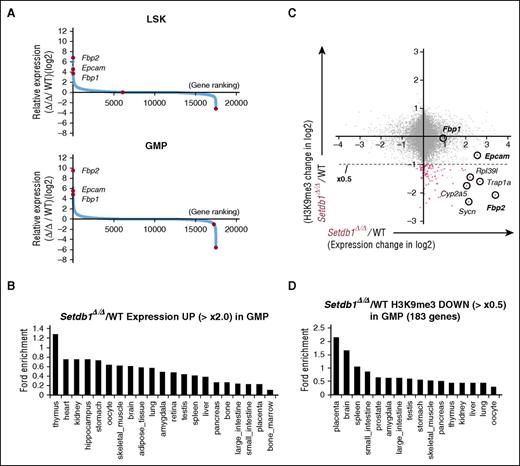
Nonhematopoietic genes are ectopically activated in Setdb1-deficient HSPCs. (A) Sequential plots of genes according to fold expression changes in Setdb1Δ/Δ LSK cells and GMPs from WT cells 2 weeks after the first injection of tamoxifen detected by RNA-seq. Representative genes with significant expression changes are indicated. (B) Enrichment of organ-specific genes in upregulated genes (≥×2.0) in Setdb1Δ/Δ GMPs. The enrichment of genes preferentially expressed in each organ and tissue in the above gene set was calculated by ExAtlas software (http://lgsun.grc.nia.nih.gov/exatlas/). (C) A scatter plot showing the correlation between H3K9me3 levels and the expression levels of RefSeq genes in Setdb1Δ/Δ GMPs relative to WT GMPs. The 183 genes showing reductions in H3K9me3 levels greater than 2-fold (below dotted line) are indicated in red dots. (D) Enrichment of organ-specific genes in 183 genes losing H3K9me3 greater than 2-fold in Setdb1Δ/Δ GMPs among genes that showed H3K9me3 enrichment greater than 2-fold over the input signal in (C). The enrichment of genes preferentially expressed in each organ and tissue in the above gene set was calculated by ExAtlas software, as in (B).
Nonhematopoietic genes are ectopically activated in Setdb1-deficient HSPCs. (A) Sequential plots of genes according to fold expression changes in Setdb1Δ/Δ LSK cells and GMPs from WT cells 2 weeks after the first injection of tamoxifen detected by RNA-seq. Representative genes with significant expression changes are indicated. (B) Enrichment of organ-specific genes in upregulated genes (≥×2.0) in Setdb1Δ/Δ GMPs. The enrichment of genes preferentially expressed in each organ and tissue in the above gene set was calculated by ExAtlas software (http://lgsun.grc.nia.nih.gov/exatlas/). (C) A scatter plot showing the correlation between H3K9me3 levels and the expression levels of RefSeq genes in Setdb1Δ/Δ GMPs relative to WT GMPs. The 183 genes showing reductions in H3K9me3 levels greater than 2-fold (below dotted line) are indicated in red dots. (D) Enrichment of organ-specific genes in 183 genes losing H3K9me3 greater than 2-fold in Setdb1Δ/Δ GMPs among genes that showed H3K9me3 enrichment greater than 2-fold over the input signal in (C). The enrichment of genes preferentially expressed in each organ and tissue in the above gene set was calculated by ExAtlas software, as in (B).
Nonhematopoietic genes are ectopically activated in Setdb1-deficient HSPCs
We next analyzed changes in the expression of NCBI reference sequence (RefSeq) genes.19 The genes derepressed (≥2-fold) in Setdb1Δ/Δ LSK cells and GMPs included many nonhematopoietic genes (Figure 3A; supplemental Figure 5B), such as Fbp2 and Fbp1, gluconeogenic enzyme genes specific to muscle and liver, respectively, and Epcam, an epithelial cell-specific cell surface glycoprotein that mediates Ca2+-independent homophilic cell–cell adhesion and regulates cell migration.20 A mega-analysis of the gene expression database using the online software tool ExAtlas21 revealed that derepressed genes (≥2-fold) were largely nonhematopoietic genes (Figure 3B). We compared ChIP-seq data for H3K9me3 levels with RNA-seq data in GMPs. Among the genes that showed H3K9me3 enrichment greater than 2-fold over the input signal at promoters (4.0 kb ± TSS), 183 genes (indicated in red dots in Figure 3C) reduced H3K9me3 levels by more than 2-fold in Setdb1Δ/Δ GMPs than in WT. These genes were assumed to be potential direct targets of Setdb1, showed a trend toward upregulation in expression levels (Figure 3C), and were also enriched for nonhematopoietic genes in the mega-analysis (Figure 3D).
Among these genes, Fbp2, but not Fbp1, showed a clear reduction in H3K9me3 levels at the promoter (Figure 4A) and DNA methylation levels around the TSS and in the gene body in Setdb1Δ/Δ GMPs (Figure 4B). Reductions in H3K9me3 levels were also confirmed in Setdb1Δ/Δ LSK cells and GMPs by manual ChIP assays (Figure 4C). In contrast, H3K9/14ac levels were increased in the absence of Setdb1 (Figures 4A,C). Although the Epcam promoter was marked with H3K9me3 at a low level, H3K9me3 was completely lost in Setdb1Δ/Δ GMPs (Figure 4A). These results suggest that Fbp2 and Epcam are direct targets of Setdb1.
Derepression of gluconeogenic genes and Epcam in Setdb1-deficient HSPCs. (A) Visualization of ChIP-sequence data of the levels of H3K9me3, H3K9/14ac, and H3K27me3 at the Fbp2, Fbp1, and Epcam gene loci in WT and Setdb1Δ/Δ GMPs, using the Integrative Genomics Viewer. Schematic diagrams of these gene loci indicate their genomic structures. Exons and untranslated regions are demarcated by large and small black boxes, respectively. (B) DNA methylation status at the Fbp2 and Fbp1 gene loci detected by RRBS in WT and Setdb1Δ/Δ ESCs and LSK cells and GMPs from WT and Setdb1Δ/Δ mice. The proportions of methylated cytosine are depicted. (C) Manual ChIP assays for H3K9me3 and H3K9/14Ac at the Fbp2 loci using anti-H3K9me3 and H3K9/14Ac antibodies. The relative amounts of immunoprecipitated DNA are depicted as a percentage of input DNA. Data are shown as the mean ± SD (n = 3). (D) Flow cytometric profiles of BM Setdb1Δ/Δ LSK cells and GMPs 2 weeks after the first injection of tamoxifen for Epcam expression.
Derepression of gluconeogenic genes and Epcam in Setdb1-deficient HSPCs. (A) Visualization of ChIP-sequence data of the levels of H3K9me3, H3K9/14ac, and H3K27me3 at the Fbp2, Fbp1, and Epcam gene loci in WT and Setdb1Δ/Δ GMPs, using the Integrative Genomics Viewer. Schematic diagrams of these gene loci indicate their genomic structures. Exons and untranslated regions are demarcated by large and small black boxes, respectively. (B) DNA methylation status at the Fbp2 and Fbp1 gene loci detected by RRBS in WT and Setdb1Δ/Δ ESCs and LSK cells and GMPs from WT and Setdb1Δ/Δ mice. The proportions of methylated cytosine are depicted. (C) Manual ChIP assays for H3K9me3 and H3K9/14Ac at the Fbp2 loci using anti-H3K9me3 and H3K9/14Ac antibodies. The relative amounts of immunoprecipitated DNA are depicted as a percentage of input DNA. Data are shown as the mean ± SD (n = 3). (D) Flow cytometric profiles of BM Setdb1Δ/Δ LSK cells and GMPs 2 weeks after the first injection of tamoxifen for Epcam expression.
To directly analyze the expression profile of HSCs, we performed single-cell expression profiling, using the Fluidigm system, because of the limited number of available Setdb1Δ/Δ HSCs. The expression of representative HSC genes was maintained well in Setdb1Δ/Δ CD34−LSK HSCs (supplemental Figure 6). As in LSK cells and GMPs, however, Epcam was derepressed at high levels in HSCs, whereas Fbp2 was not (supplemental Figure 6). A flow cytometric analysis readily detected the upregulation of Epcam in Setdb1Δ/Δ LSK cells and GMPs (Figure 4D).
Metabolic homeostasis is impaired in Setdb1-deficient HSPCs
Fbp1 and Fbp2 are enzymes that convert fructose-1,6-bisphosphate (F-1,6-BP) to fructose 6-phosphate (F6P) in gluconeogenesis and counteract the reaction catalyzed by phosphofructokinase (Pfk) in glycolysis (supplemental Figure 8).22 Although the Fbp1 and Fbp2 loci are in very close proximity (Figure 4A), they are differentially expressed in the gluconeogenic tissues (the liver and kidney and the muscle, respectively) (Figure 5A). In the absence of Setdb1, Fbp2 was highly derepressed in Setdb1Δ/Δ CD34+LSK cells and GMPs at levels similar to and greater than those in muscle, respectively (Figure 5A). Fbp2 protein was readily detected in Setdb1Δ/Δ GMPs by a western blot analysis (Figure 5C), and in Setdb1Δ/Δ LSK cells and GMPs but not CD34−LSK HSCs by an immunofluorescence staining (Figure 5D). Fbp2 appeared to be activated specifically in BM cells among major organs upon the deletion of Setdb1 (Figure 5B). Fbp1 was also derepressed in Setdb1Δ/Δ LSK cells and GMPs to a quarter of the level in the liver (Figure 5A). However, Fbp1 protein was not detected in Setdb1Δ/Δ GMPs (Figure 5C). These results indicate that both Fbp1 and Fbp2 were ectopically activated in Setdb1Δ/Δ HSPCs, but only Fbp2 was overexpressed at the protein level. Of interest, Fbp1 and Fbp2 were both maintained in a transcriptionally repressed state in Setdb1Δ/Δ CD34−LSK HSCs (Figure 5A; supplemental Figure 6).
Ectopic activation of Fbp2 in Setdb1-deficient HSPCs. (A) RT-PCR of Fbp1 and Fbp2 expression in the indicated cells from WT mice. mRNA levels were normalized to Hprt1 expression, and relative expression levels are shown as the mean ± SD of triplicate analyses. ND, not detected. **P < .01. (B) RT-PCR of Setdb1 and Fbp2 expression in the indicated organs from WT and Setdb1Δ/Δ mice 7 days after the first injection of tamoxifen. mRNA levels were normalized to Hprt1 expression, and relative expression levels are shown as the mean ± SD of triplicate analyses. **P < .01. (C) A western blot analysis of Fbp1 and Fbp2 protein expression in GMPs from WT and Setdb1Δ/Δ mice 2 weeks after the first injection of tamoxifen. Liver and muscle were used as references, and β-actin was probed as a loading control. The arrowhead indicates a cross-reactive Fbp1 signal in lane 1. (D) Immunostaining of Fbp2 in CD34−LSK HSCs, LSK HSPCs, and GMPs from WT and Setdb1Δ/Δ mice 2 weeks after the first injection of tamoxifen. Cells were counterstained with DAPI. Representative images are depicted.
Ectopic activation of Fbp2 in Setdb1-deficient HSPCs. (A) RT-PCR of Fbp1 and Fbp2 expression in the indicated cells from WT mice. mRNA levels were normalized to Hprt1 expression, and relative expression levels are shown as the mean ± SD of triplicate analyses. ND, not detected. **P < .01. (B) RT-PCR of Setdb1 and Fbp2 expression in the indicated organs from WT and Setdb1Δ/Δ mice 7 days after the first injection of tamoxifen. mRNA levels were normalized to Hprt1 expression, and relative expression levels are shown as the mean ± SD of triplicate analyses. **P < .01. (C) A western blot analysis of Fbp1 and Fbp2 protein expression in GMPs from WT and Setdb1Δ/Δ mice 2 weeks after the first injection of tamoxifen. Liver and muscle were used as references, and β-actin was probed as a loading control. The arrowhead indicates a cross-reactive Fbp1 signal in lane 1. (D) Immunostaining of Fbp2 in CD34−LSK HSCs, LSK HSPCs, and GMPs from WT and Setdb1Δ/Δ mice 2 weeks after the first injection of tamoxifen. Cells were counterstained with DAPI. Representative images are depicted.
Fbp1 and Fbp2 are the rate-limiting enzymes of gluconeogenesis. Fbp1 has recently been identified as a critical regulator of glucose metabolism in renal cell carcinoma.23 To elucidate the effect of the ectopic expression of Fbp2 in hematopoietic progenitor cells, we performed a metabolome analysis. The metabolites at the early steps of glycolysis, including glucose 6-phosphate, F6P, and F-1,6-BP, were all decreased in freshly isolated Setdb1Δ/Δ GMPs (Figure 6A; supplemental Figure 8). In contrast, pyruvic acid, a downstream metabolite, showed a mild reduction. Notably, F-1,6-BP levels relative to F6P levels (F-1,6-BP/F6P) were lower in Setdb1Δ/Δ GMPs and Setdb1Δ/ΔMLL-AF9-transformed GMPs, as well as in MLL-AF9-transformed GMPs overexpressing Fbp2 (Figure 6B), suggesting Fbp2 antagonizes the glycolytic flux in hematopoietic progenitor cells. In addition, the levels of ribose 5-phosphate (ribose 5-P), which is used in the synthesis of nucleotides and nucleic acids, and the ratio of reduced glutathione to oxidized glutathione (reduced glutathione/oxidized glutathione) were decreased in Setdb1Δ/Δ GMPs (Figure 6A; supplemental Figure 8). Given the reduction in glucose 6-phosphate levels, the entry metabolite of the pentose phosphate pathway, Fbp2 also affects the pentose phosphate pathway flux by regulating glycolysis similar to Fbp1 in renal cell carcinoma (supplemental Figure 8).23 We then investigated the consequence of compromised metabolism in Setdb1Δ/Δ HSPCs. ATP levels were significantly reduced not only in GMPs (Figure 6A,C) but also in LSK cells and MLL-AF9-transformed GMPs in the absence of Setdb1 (Figure 6C).
Metabolic homeostasis is impaired in Setdb1-deficient HSPCs. (A) Results of metabolome analyses of GMPs freshly isolated from Setdb1Δ/Δ mice 2 weeks after the first injection of tamoxifen. Relative values (WT values were set as 1) are shown as the mean ± SD (glucose 6-phosphate, F6P, F1,6BP, n = 4-6; pyruvic acid, n = 2; ribose-5-P, reduced glutathione/oxidized glutathione, n = 3; ATP, n = 9). (B) F1,6BP levels relative to F6P in GMPs (n = 4), MLL-AF9-transformed GMPs 48 hours after the tamoxifen treatment (n = 3), and MLL-AF9-transformed GMPs overexpressing Fbp2 (n = 1). (C) ATP levels in LSK cells and GMPs freshly isolated from Setdb1Δ/Δ mice 2 weeks after the first injection of tamoxifen and MLL-AF9-transformed GMPs 48 hours after the tamoxifen treatment. Data are shown as the mean ± SD (n = 3). (D) ATP levels in LSK cells transduced with an Fbp2 retrovirus in culture (left) and their growth in culture supplemented with SCF (10 ng/mL) and thrombopoietin (10 ng/mL) without or with IL-3 (10 ng/mL). mRNA levels of exogenous Setdb1 in LSK cells detected by RT-PCR were normalized to Hprt1 expression. (Right) Relative expression levels. Data are shown as the mean ± SD (n = 3). (E) Repopulating capacity of Fbp2-overexpressing HSCs in vivo. Fifty CD45.2 CD34−LSK HSCs were transduced either with a control or Fbp2 retrovirus, and then transplanted into lethally irradiated CD45.1 mice (n = 6) with 2 × 105 CD45.1 competitor cells. The chimerism of CD45.2+GFP+ cells derived from transduced HSCs as well as CD45.2+GFP− cells derived from nontransduced HSCs was monitored. At 16 weeks after primary transplantation, all BM cells from recipients were pooled and 1 × 107 cells were infused into the secondary recipients. The proportions of GFP+ cells in donor-derived cells are plotted in blue (control) and red (Fbp2) lines. *P < .05; **P < .01; ***P < .001.
Metabolic homeostasis is impaired in Setdb1-deficient HSPCs. (A) Results of metabolome analyses of GMPs freshly isolated from Setdb1Δ/Δ mice 2 weeks after the first injection of tamoxifen. Relative values (WT values were set as 1) are shown as the mean ± SD (glucose 6-phosphate, F6P, F1,6BP, n = 4-6; pyruvic acid, n = 2; ribose-5-P, reduced glutathione/oxidized glutathione, n = 3; ATP, n = 9). (B) F1,6BP levels relative to F6P in GMPs (n = 4), MLL-AF9-transformed GMPs 48 hours after the tamoxifen treatment (n = 3), and MLL-AF9-transformed GMPs overexpressing Fbp2 (n = 1). (C) ATP levels in LSK cells and GMPs freshly isolated from Setdb1Δ/Δ mice 2 weeks after the first injection of tamoxifen and MLL-AF9-transformed GMPs 48 hours after the tamoxifen treatment. Data are shown as the mean ± SD (n = 3). (D) ATP levels in LSK cells transduced with an Fbp2 retrovirus in culture (left) and their growth in culture supplemented with SCF (10 ng/mL) and thrombopoietin (10 ng/mL) without or with IL-3 (10 ng/mL). mRNA levels of exogenous Setdb1 in LSK cells detected by RT-PCR were normalized to Hprt1 expression. (Right) Relative expression levels. Data are shown as the mean ± SD (n = 3). (E) Repopulating capacity of Fbp2-overexpressing HSCs in vivo. Fifty CD45.2 CD34−LSK HSCs were transduced either with a control or Fbp2 retrovirus, and then transplanted into lethally irradiated CD45.1 mice (n = 6) with 2 × 105 CD45.1 competitor cells. The chimerism of CD45.2+GFP+ cells derived from transduced HSCs as well as CD45.2+GFP− cells derived from nontransduced HSCs was monitored. At 16 weeks after primary transplantation, all BM cells from recipients were pooled and 1 × 107 cells were infused into the secondary recipients. The proportions of GFP+ cells in donor-derived cells are plotted in blue (control) and red (Fbp2) lines. *P < .05; **P < .01; ***P < .001.
The ectopic overexpression of Fbp2 in HSPCs significantly reduced their ATP levels and significantly inhibited the cell growth of primitive HSPCs in liquid culture supplemented with stem cell factor (SCF) and thrombopoietin (S+T), as well as in that supplemented with multiple cytokines containing interleukin 3 (IL-3) (S+T+IL-3) (Figure 6D). In competitive repopulating assays, the repopulating capacity of CD45.2+ GFP+ HSPCs overexpressing Fbp2 was also found to be impaired, and these cells were eventually depleted in the secondary recipient mice (Figure 6E).
We attempted rescue experiments of Setdb1Δ/ΔMLL-AF9-transformed GMPs by exogenous Setdb1. We infected cells with retroviruses harboring either full-length Setdb1 or short-form Setdb1 truncated for the portion encoding the SET catalytic domain (ΔSET) and then deleted endogenous Setdb1 by adding tamoxifen in culture (supplemental Figure 7A,B). Full-length Setdb1 largely restored the proliferative capacity of Setdb1Δ/Δ cells and maintained the transcriptional repression of Fbp2 and ATP levels. In contrast, the effects of ΔSET were minimal, suggesting the requirement of the enzymatic activity of Setdb1 in gene silencing and maintenance of HSPCs. We further tested knockdown of Fpb2 and overexpression of Pfk1 in Setdb1Δ/ΔMLL-AF9-transformed GMPs. Both partially but significantly restored ATP levels. However, impaired cell growth was not rescued at all (supplemental Figure 7C,D).
Discussion
The histone methyltransferases responsible for H3K9me3, such as Suv39h and Setdb1, play key roles in the gene silencing of retroelements, including long interspersed elements and ERVs in cells of embryonic origin. In contrast, DNA methylation is mainly responsible for silencing in differentiated cells such as neural progenitor cells and fibroblasts, whereas histone methyltransferases are largely dispensable.2,24 Setdb1 was recently shown to contribute to the silencing of ERVs in B lymphocytes; however, the transcriptional activation of ERVs was dependent on the regulatory architecture of their long terminal repeats and the availability of corresponding B-cell-specific transcription factors such as Pax5.25 In the present study, we found that H3K9me3 and DNA methylation were not largely altered at ERV loci and derepression of ERVs was minimal in the absence of Setdb1. These results confirm previous findings obtained in neural progenitor cells and fibroblasts2,24 and highlight the important role of DNA methylation in the silencing of ERVs in HSPCs.
In the present study, we clearly demonstrated that Setdb1 is the first H3K9 histone methyltransferase to be identified as an essential regulator of HSPCs. G9a/GLP, which catalyzes H3K9me2, has been shown to promote progressive H3K9me2 patterning during HSPC lineage specification, and its inhibition delays HSPC lineage commitment.7,26 G9a is dispensable for HSPCs, but plays a critical role in the proliferation of acute myeloid leukemia by regulating HoxA9-dependent transcription.8 Thus, the role of Setdb1 in HSPCs is in marked contrast to that of G9a and corresponds to that of Kap1, a binding partner of Setdb1.9 In the absence of Setdb1, we found only a limited number of genes losing H3K9me3 levels in hematopoietic progenitors, which appeared to be mostly nonhematopoietic genes. However, not all of them became activated, which may have been a result of the absence of transcriptional factors compatible to their transcriptional regulatory architecture. It is also possible that the effects of Setdb1 loss on epigenome and transcription we observed in this study could be secondary to events resulting in loss of hematopoietic cells, rather than direct effects of Setdb1 loss. Nevertheless, among candidate Setdb1 target genes, a key gluconeogenic enzyme gene Fbp2 was strongly activated in both HSPCs and MLL-AF9-transformed GMPs, suggesting that derepression of Fbp2 is a direct effect of Setdb1 loss.
Gluconeogenesis occurs in a limited number of organs such as the liver, kidney, and muscle. Other organs, including HSPCs as well as cancer cells, favor glycolysis over the oxidative phosphorylation of glucose, and thus maintain Fbp1 and Fbp2, in a transcriptionally repressed state.27 FBP1 and FBP2 are strongly repressed, particularly in cancer cells, via mechanisms including the DNA hypermethylation of their promoters.23,28,29 FBP1 and FBP2 both antagonize glycolysis in cancer cells, thereby inhibiting the Warburg effect. In contrast, the loss of FBP1 or FBP2 in cancer cells enhances glycolysis, resulting in the maintenance of ATP production and proliferative capacity under hypoxic conditions.23,28,29 However, the regulation of gluconeogenic enzyme genes in HSPCs has not yet been characterized. In the present study, we showed that Setdb1-dependent H3K9me3, in concert with DNA methylation, targets Fbp1 and Fbp2. Because the Fbp2 promoter was marked with higher levels of H3K9me3 than the Fbp1 promoter, Fbp2 may be more dependent on Setdb1-dependent H3K9me3 for silencing, and the activation of Fbp1 could be an indirect event. Nevertheless, both Fbp1 and Fbp2 were maintained in a repressed state in CD34−LSK HSCs, even in the absence of Setdb1, suggesting that Fbp1 and Fbp2 are more dependent on DNA methylation for silencing in HSCs.
The consequence of the ectopic expression of Fbp2 protein in HSPCs was very similar to that in cancer cells (supplemental Figure 8). It antagonized glycolysis and attenuated the production of ATP, resulting in an impaired repopulating capacity in HSPCs. Fbp2 also affected the pentose phosphate pathway flux by antagonizing glycolysis similar to Fbp1.23 Therefore, Fbp2 is detrimental to HSPCs once activated, and has to be strongly repressed to maintain precise energy metabolism in HSPCs. Knockdown of Fpb2 and overexpression of Pfk1 in Setdb1Δ/ΔMLL-AF9-transformed GMPs partially but significantly restored ATP levels. Nonetheless, impaired cell growth was not rescued at all. These results support the notion that ectopic expression of Fbp2 compromises energy production via glycolysis in HSPCs, while at the same time strongly suggesting that gluconeogenesis pathway is not the only causative pathway compromised in the absence of Setdb1.
Although Fbp2 was maintained in a repressed state in Setdb1Δ/Δ HSCs, HSCs were depleted in a manner similar to HSPCs, suggesting that Setdb1 regulates multiple important targets in HSPCs. Although we could not directly evaluate apoptosis of HSCs because of the limited number of Setdb1Δ/Δ HSCs, we found that mitochondrial reactive oxygen species levels were significantly higher in Setdb1Δ/Δ HSCs. Together with the ectopic expression of Epcam in Setdb1Δ/Δ HSCs, multiple defects may be responsible for depletion of Setdb1Δ/Δ HSCs.
Although the epigenetic regulation of HSPCs has been extensively characterized, more information is needed. Together with our previous findings on Kap1-deficient HSPCs, the results of the present study clearly demonstrate that nonhematopoietic genes are permanently repressed in HSPCs via H3K9me-mediated silencing machineries in collaboration with DNA methylation. This silencing manner is in marked contrast to the transient as well as reversible silencing mediated by the polycomb-group complexes that maintain the multipotency of HSPCs.30 The precise mechanisms by which these differential-silencing processes occur requires further study.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank George R. Wendt for technical assistance and Ola Mohammed Kamel Rizq for critical review of the manuscript.
This work was supported in part by grants-in-aid for Scientific Research (#15639313) and Scientific Research on Innovative Areas “Stem Cell Aging and Disease” (#26115002) and “Genome Science” (#221S0002) from the Ministry of Education, Culture, Sports, Science and Technology (MEXT), Japan, Grant-in-Aid for Core Research for Evolutional Science and Technology from the Japan Science and Technology Corporation, and grants from the Uehara Foundation and Yasuda Foundation. S.K. is a research fellow supported by Japan Society for the Promotion of Science, MEXT, Japan.
Authorship
Contribution: S.K. and A.I. designed this study; S.K. performed experiments, analyzed data, and actively wrote the manuscript; M.O., K.T., S.Y., E.N., A.S., K.A., Y.K., S.M., Y.N.-T., H.M., F.A., and Y.S. performed experiments and analyzed data; T.C., H.N., and T.S. provided analysis tools and analyzed data; H.K. provided antibodies; S.Y. provided mice; and A.I. conceived of and directed the project, secured funding, and actively wrote the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Atsushi Iwama, Department of Cellular and Molecular Medicine, Graduate School of Medicine, Chiba University, 1-8-1 Inohana, Chuo-ku, Chiba 260-8670, Japan; e-mail: aiwama@faculty.chiba-u.jp.