Key Points
The RNA regulators PUMILIO sustain HSPC and acute myeloid leukemia cell growth by upregulating FOXP1 expression through direct binding to 2 FOXP1-3′UTR PUMILIO-binding elements.
FOXP1 mediates PUMILIO growth-promoting activities by repressing expression of p21CIP1 and p27KIP1cell cycle inhibitors.
Abstract
RNA-binding proteins (RBPs) have emerged as important regulators of invertebrate adult stem cells, but their activities remain poorly appreciated in mammals. Using a short hairpin RNA strategy, we demonstrate here that the 2 mammalian RBPs, PUMILIO (PUM)1 and PUM2, members of the PUF family of posttranscriptional regulators, are essential for hematopoietic stem/progenitor cell (HSPC) proliferation and survival in vitro and in vivo upon reconstitution assays. Moreover, we found that PUM1/2 sustain myeloid leukemic cell growth. Through a proteomic approach, we identified the FOXP1 transcription factor as a new target of PUM1/2. Contrary to its canonical repressive activity, PUM1/2 rather promote FOXP1 expression by a direct binding to 2 canonical PUM responsive elements present in the FOXP1-3′ untranslated region (UTR). Expression of FOXP1 strongly correlates with PUM1 and PUM2 levels in primary HSPCs and myeloid leukemia cells. We demonstrate that FOXP1 by itself supports HSPC and leukemic cell growth, thus mimicking PUM activities. Mechanistically, FOXP1 represses the expression of the p21−CIP1 and p27−KIP1 cell cycle inhibitors. Enforced FOXP1 expression reverses shPUM antiproliferative and proapoptotic activities. Altogether, our results reveal a novel regulatory pathway, underscoring a previously unknown and interconnected key role of PUM1/2 and FOXP1 in regulating normal HSPC and leukemic cell growth.
Introduction
During the past several years, significant strides have been made in our ability to isolate functional multipotent hematopoietic stem/progenitor cells (HSPCs), and also in our understanding of the regulatory processes responsible for their maintenance and growth. Investigations have largely focused on extracellular signals, and also on roles of transcription, chromatin, and noncoding RNA regulators.1-4 In contrast to the understanding of transcriptional networks regulating HSPC fate, the role of posttranscriptional events in this process remains poorly studied. The HOXB4 homeoprotein is a major expansion factor of mouse and human HSPCs, as we and others have shown.5,6 Besides, Notch signaling pathway activated by Δ-like (Dll)-1 or -4 ligands favors HSPC self-renewal,7-9 and combining HOXB4 and Notch activation has additive effects.10 We previously reported that the HOXB4 and Dll/Notch pathways activate expression of the 2 posttranscriptional regulators PUMILIO (PUM)-1 and PUM-2 in human and murine HSPCs.11,12 PUM1/2 belong to the PUF family of RNA-binding proteins (RBPs) that dictates stem cell fate in invertebrates by inhibiting expression/translation of target messenger RNAs (mRNAs).13 PUM1/2 also contribute to the self-renewal of human embryonic stem cells (ESCs)14,15 and human spermatogonial stem cells16 and favor expansion of cultured adipocyte-derived stem cells.17 Interestingly, PUM expression in mice is higher in HSPCs, as compared with more committed cells.18 The fact that PUM1 and PUM2 were at the crossroad of HOXB4 and Notch signaling in HSPCs prompted us to investigate their activities in this context. Here, we reveal that the canonical repressors PUM1/2 control expansion of HSPCs and leukemia cells by enhancing expression of the transcription factor FOXP1 posttranscriptionally. Our data identify PUM1/2 and FOXP1 as new regulators of human HSPC physiology as well as myeloid hemopathies. They provide new targets to amplify adult HSPCs in vitro for stem cell–based regenerative therapies.
Materials and methods
Cell collection, purification, and culture
Human cord blood units were collected according to institutional guidelines. Handling, characterization, and storage of patient samples were performed by the FILOtheque (BB-0033-00073), and Saint-Louis Hospital’s Tumor Biobank. Patients and healthy donors provided a written informed consent in accordance with the Declaration of Helsinki. CD34+ cells were purified and cultured in Iscove modified Dulbecco medium supplemented with BIT (bovine serum albumin [BSA], insulin, transferrin; StemCell Technologies, Vancouver, BC, Canada), human thrombopoietin (20 nM), human interleukin-3 (10 ng/mL), human stem cell factor (50 ng/mL), and human feline McDonough sarcoma-like tyrosine kinase 3 ligand (50 ng/mL) as previously described.19 CD34+ subpopulations were sorted on a fluorescence-activated cell sorter (FACS) AriaIII (Becton Dickinson) using antibodies listed in supplemental Table 1, available on the Blood Web site. Lin−Sca+Kit+ cells were purified from murine bone marrow (BM) and cultured in Iscove modified Dulbecco medium supplemented with 10% fetal calf serum, murine stem cell factor (50 ng/mL), human FMS-like tyrosine kinase 3 ligand (100 ng/mL), huIL-6 (50 ng/mL), and huIL-11 (10 ng/mL). Purification of murine lineage–negative (Lin−) subpopulations was performed as described,12 using antibodies listed in supplemental Table 1. Cell lines were cultured in αminimal essential medium supplemented with 10% fetal calf serum, l-glutamine (2 mM), 50 U/mL penicillin, and 50 µg/mL streptomycin. UT7-GM cell medium was supplemented with granulocyte-macrophage colony-stimulating factor (2.5 ng/mL). Murine HPC-7 cells were cultured as described.20 Clonogenic potential was evaluated according to manufacturer’s recommendations (MethoCult H4230, MethoCult M3234; StemCell Technologies, Vancouver, BC, Canada).
Constructions and lentiviral transduction are detailed in supplemental data.
In vivo hematopoietic reconstitution assays
Human HSPCs.
NOD-SCID/IL2Rγc−/− immunodeficient mice (NSG; originally from the Jackson Laboratory, Bar Harbor, ME) (8-12 weeks old) were bred at Commissariat à l’Energie Atomique animal facility (Fontenay-aux-Roses, Paris, France). Three-gray-irradiated NSG mice were transplanted using intrafemur route with a mixture of 7 × 104 shPUM-Tomato-vector-transduced CD34+ cells and 7 × 104 shCtrl-GFP-vector transduced CD34+ cells. Hematopoietic reconstitution was assessed 12 weeks after transplantation by labeling BM cells with human CD45 antibody. Percentages of Tomato+/shPUM- or GFP+/shCtrl-CD45+ cells were determined by flow cytometry. Mice were considered positive when at least 0.5% of human cells were detected in mouse BM.
Murine HSPCs.
C57BL/6-Ly5.2, C57Bl/6-Ly5.1 mice (8-14 weeks old) were purchased from Charles River (l’Arbresle, France), maintained at Cochin Institute facility (Paris, France) under pathogen-free conditions, and used for experiments according to guidelines from the Ethical Committee of the French Agriculture Department. C57Bl/6 (Ly5.2) mice were used as recipients, whereas LSKCD150+ cells were prepared from C57Bl/6 (Ly5.1) donors. Lethally irradiated recipients (9.5 Gy) were injected intravenously with 15 000 untransduced and transduced Ly5.1 cells (1:1), together with 1.5 × 105 Ly5.2 BM cells. Hematopoietic reconstitution was assessed 4 months after transplantation through quantification of GFP expression of CD45.1-phycoerythrin (PE)-labeled cells by flow cytometry.
Cell cycle, cell viability
Cell cycle was examined by propidium iodide labeling (Life Technologies) following manufacturer’s instructions using flow cytometry (AccuriC6; Becton Dickinson) and FlowJo software. Cell viability was assessed using PE-conjugated Annexin V labeling detection kit (BD Pharmingen).
Immunoprecipitation and western blot analysis
Cells were lysed at 4°C with lysis buffer (50 mM Tris-HCl pH 7.4, 150 mM NaCl, 1 mM EDTA, 0.5% NP-40, 0.25% sodium deoxycholate, 10% glycerol) supplemented with protease inhibitor cocktail (Roche). Immunoprecipitation (IP) was performed with 500 µg proteins and 5 µg of the indicated antibody. Immunoprecipitates or 20 to 50 µg of whole cell lysates were loaded on sodium dodecyl sulfate–polyacrylamide gel electrophoresis gels and transferred onto nitrocellulose membranes (Amersham Biosciences). Detection was performed using enhanced chemiluminescence (Amersham Biosciences). Images were captured using a CCD camera (Fuji-LAS4000; Fujifilm, Tokyo, Japan). The antibodies are listed in supplemental Table 2.
Stable isotope labeling with amino acids in cell-based quantitative proteomic approach is detailed in supplemental data.
Luciferase assays
MOLM-14 cells (106) were transfected with 300 pmol of siPUM1 (SR306467B; OriGene), siPUM2 (SR308223C; OriGene), or scrambled control small interfering RNA (siRNA) (SR30004; OriGene) using Lipofectamine 2000. After 24 hours, cells were transfected with 300 ng of psiCHECK-2 constructs by Nucleofection with Amaxa Cell Line Nucleofector Kit V (programT-003; Lonza). Twenty-four hours later, luciferase activity was measured using the Dual Luciferase Reporter Assay System (Promega).
Oligoribonucleotide pull-down assays
Streptavidin magnetic beads (Pierce) were presaturated overnight at 4°C with 50 µg/mL yeast transfer RNA (tRNA; Life Technologies) and 0.1 mg/mL RNase-free BSA (Ambion) in lysis buffer (25 mM Tris-HCl pH 7.4, 150 mM NaCl, 1 mM EDTA, 1% NP-40, 5% glycerol) and then washed. Cells were lysed in lysis buffer supplemented with protease and phosphatase inhibitors, 30 µg/mL tRNA and 400 U/mL RNaseOUT (Life Technologies). Lysates were subjected to 1 round of preclearing for 30 minutes at 4°C with streptavidin beads, before incubation with 50 pmol of the indicated biotin-labeled FOXP1-RNA probe wild-type (WT)1 (UAUUACGUUGUACAUAUAUCCCGAU) or MUT1 (UAUUACGUUCCCGUUAUAUCCCGAU) and 250 pmol of the indicated FOXP1-RNA competitor: WT1 (UACGUUGUACAUAUAUCC), MUT1 (UACGUUCCCGUUAUAUCC), WT2 (AAGGCUUAUGUACAUACGUGAAGAG), or MUT2 (AAGGCUUAUCCCGUUACGUGAAGAG) in RNA-binding buffer (10 mM HEPES [N-2-hydroxyethylpiperazine-N′-2-ethanesulfonic acid] pH 7, 1 mM EDTA, 150 mM KCl, 0.5% Triton X-100, 10% glycerol, 1 mM dithiothreitol, 15 µg/mL yeast tRNA, and 200 U/mL RNaseOUT) for 2.5 hours at 4°C. Presaturated streptavidin beads were added for 30 minutes at 4°C and then washed sequentially in RNA-binding buffer, lysis buffer, and lysis buffer with 600 mM NaCl, and eluted in Laemmli lysis buffer.
RNA immunoprecipitation
Dynabeads ProteinG (Life Technologies) were presaturated overnight at 4°C with 20 µg/mL yeast tRNA and 0.5 mg/mL RNase-free BSA in 50 mM Tris-HCl pH 7.4, 150 mM KCl, 1% NP-40 buffer and then washed and incubated at 4°C for 4 hours with antibodies against PUM1 or PUM2 or control goat immunoglobulin G (IgG). Cells were lysed in lysis buffer (25 mM Tris-HCl pH 7.4, 150 mM KCl, 2 mM EDTA, 0.5% NP-40) supplemented with protease and phosphatase inhibitors, and 100 U/mL RNaseOUT. PUM-associated RNAs were immunoprecipitated overnight at 4°C with the indicated preformed bead-antibody complexes (5 µg of antibody per 500 µg of proteins), then washed in wash buffer (50 mM Tris-HCl pH 7.4, 150 mM KCl, 0.5% NP-40) and in TE (10 mM Tris-HCl pH 7, 1 mM EDTA), and resuspended in resuspension buffer (50 mM Tris-HCl pH 7.4, 5 mM EDTA, 10 mM dithiothreitol, 1% sodium dodecyl sulfate, 400 U/mL RNaseOUT). Aliquots (1:10) were removed for immunoblotting analysis. RNAs were purified using TriPURE (Roche), treated with DNase I (DNase I amplification grade, 18068-015; ThermoFisher-Scientific), and analyzed by quantitative reverse transcription polymerase chain reaction (RT-qPCR).
RNA extraction and quantitative real-time PCR
Total RNA was extracted with PureLink RNA Mini Kit (Life Technologies) according to manufacturer’s instructions. Contaminating DNA was eliminated with DNase I treatment. RNA (25-300 ng) was reverse-transcribed with Maxima First Strand cDNA Synthesis Kit (Thermo Fischer Scientific). qPCR was performed using primers indexed in supplemental Table 3, with the SYBR Green Master Mix (Thermo Fischer Scientific) in a LightCycler 480 (Roche). Expression levels were normalized to GAPDH and/or HPRT and/or Cyclophilin A expression and analyzed using ΔΔCt method.
Statistical analysis
All analyses were performed using GraphPad Prism software. Data are presented as the mean ± standard error of the mean (SEM). When distribution was normal (assessed with a Shapiro-Wilk test), the 2-tailed Student t test was used for group comparisons. In the other cases, the Mann-Whitney test was used. The Pearson coefficient was calculated to determine the correlation between the normally distributed PUM mRNA and FOXP1 mRNA expressions in acute myeloid leukemia (AML). Statistics were carried out on a minimum of 3 independent experiments.
Results
PUM1 and PUM2 control human and murine HSPC expansion
To assess the role of PUM1/2 in human HSPCs, we first analyzed PUM1 and PUM2 expression in various stem/progenitor subpopulations. PUM1 and PUM2 expression was higher in primitive CD34+CD38lowCD90+ hematopoietic stem cells (HSC), as compared with multipotent progenitors (MPPs), and to more committed granulocyte macrophagic progenitors (GMP), common myeloid progenitors (CMPs), and megakaryocyte-erythroid progenitors (MEP) (Figure 1A). We knocked down (KD) PUM1 or PUM2 in human hematopoietic cells using different small hairpin (sh)RNAs (Figure 1B; supplemental Figure 1A). PUM1 or PUM2 KD strongly inhibited CD34+ HSPC expansion in vitro (Figure 1C; supplemental Figure 1B). It first enhanced proportion of HSPCs in G0/G1 phase, as evidenced at day 3 posttransduction (Figure 1D) and only later on induced apoptosis (Figure 1E;supplemental Figure 1C) without modification of cell differentiation (supplemental Figure 1D and data not shown). Moreover, PUM1 or PUM2 KD decreased the number and size of colony-forming cells (CFCs) without modifying the proportions of the different CFC subtypes (Figure 1F; supplemental Figure 1E). The in vivo engraftment potential of human PUM1 or PUM2 KD HSPCs was further investigated using competitive transplantation in NSG mice. Five of 9 mice transplanted with an equal mixture of shPUM1/Tomato+ (Tom+) and control shCtrl/GFP+ cells and 6 of 8 mice having received an equal mixture of shPUM2/Tom+ and shCtrl/GFP+ cells displayed human shCtrl/GFP+ cells 12 weeks later in BM, whereas none of them exhibited human shPUM1/Tom+ or shPUM2/Tom+ cells (Figure 1G). Analysis of homing capacity of shRNA-transduced cells rules out any homing defect of PUM1 or PUM2 KD cells (supplemental Figure 1F).
PUM1 and PUM2 contribute to human HSPC expansion. (A) RT-qPCR analysis of hPUM1 and hPUM2 transcripts in human stem/progenitor cell subpopulations: HSC (CD34+CD38lowCD90+), MPP (CD34+CD38lowCD90−), CMP (CD34+CD38+CD45RA−IL-3Rαlow), MEP (CD34+CD38+IL-3Rα−CD45RA+), and GMP (CD34+CD38+IL-3Rα+CD45RA+). Results are normalized to GAPDH expression and expressed relative to PUM expression in HSCs (n = 3). (B-F) Cord blood CD34+ cells were transduced or not (nt) with lentiviral vectors encoding shRNA targeting PUM1 (shPUM1-a, sequence #a in supplemental Table 8), PUM2 (shPUM2-a, sequence #a in supplemental Table 8), or luciferase as control (shCtrl), and were maintained in culture posttransduction. (B) Immunoblot analysis of the indicated proteins at day 4 posttransduction. Actin was used as loading control. (C) Cell expansion analysis along time of culture posttransduction (n = 4). (D) Cell cycle analysis upon propidium iodide labeling at day 3 posttransduction (n = 4). (E) Annexin V labeling at day 5 posttransduction (n = 4). (F) At day 7 posttransduction, shRNA/GFP+ output cells were seeded in methylcellulose medium and numbering of CFC was assessed after 2 weeks (n = 3); representative micrographs of colonies are presented on the right side (×20). BFU-E (burst forming unit-erythroid), GM-CFC (granulomacrophagic colony forming cell). (G) Cord blood CD34+ cells from the same batch were transduced with lentiviral vectors encoding shRNA targeting PUM1 (shPUM1-a), PUM2 (shPUM2-a), or luciferase as control (shCtrl). Three days posttransduction, sublethally irradiated NSG-immunodeficient mice received a 1:1 mixture of sorted shPUM/Tomato+ CD34+ cells and shCtrl/GFP+ CD34+ cells. Presence of GFP+ and Tom+ cells among human BM CD45+ cells of engrafted mice was scored 12 weeks later by FACS analysis. Each symbol represents data from a single chimeric mouse. Data are expressed as mean ± SEM. *P < .05; **P < .01; ***P < .001 (Student t test).
PUM1 and PUM2 contribute to human HSPC expansion. (A) RT-qPCR analysis of hPUM1 and hPUM2 transcripts in human stem/progenitor cell subpopulations: HSC (CD34+CD38lowCD90+), MPP (CD34+CD38lowCD90−), CMP (CD34+CD38+CD45RA−IL-3Rαlow), MEP (CD34+CD38+IL-3Rα−CD45RA+), and GMP (CD34+CD38+IL-3Rα+CD45RA+). Results are normalized to GAPDH expression and expressed relative to PUM expression in HSCs (n = 3). (B-F) Cord blood CD34+ cells were transduced or not (nt) with lentiviral vectors encoding shRNA targeting PUM1 (shPUM1-a, sequence #a in supplemental Table 8), PUM2 (shPUM2-a, sequence #a in supplemental Table 8), or luciferase as control (shCtrl), and were maintained in culture posttransduction. (B) Immunoblot analysis of the indicated proteins at day 4 posttransduction. Actin was used as loading control. (C) Cell expansion analysis along time of culture posttransduction (n = 4). (D) Cell cycle analysis upon propidium iodide labeling at day 3 posttransduction (n = 4). (E) Annexin V labeling at day 5 posttransduction (n = 4). (F) At day 7 posttransduction, shRNA/GFP+ output cells were seeded in methylcellulose medium and numbering of CFC was assessed after 2 weeks (n = 3); representative micrographs of colonies are presented on the right side (×20). BFU-E (burst forming unit-erythroid), GM-CFC (granulomacrophagic colony forming cell). (G) Cord blood CD34+ cells from the same batch were transduced with lentiviral vectors encoding shRNA targeting PUM1 (shPUM1-a), PUM2 (shPUM2-a), or luciferase as control (shCtrl). Three days posttransduction, sublethally irradiated NSG-immunodeficient mice received a 1:1 mixture of sorted shPUM/Tomato+ CD34+ cells and shCtrl/GFP+ CD34+ cells. Presence of GFP+ and Tom+ cells among human BM CD45+ cells of engrafted mice was scored 12 weeks later by FACS analysis. Each symbol represents data from a single chimeric mouse. Data are expressed as mean ± SEM. *P < .05; **P < .01; ***P < .001 (Student t test).
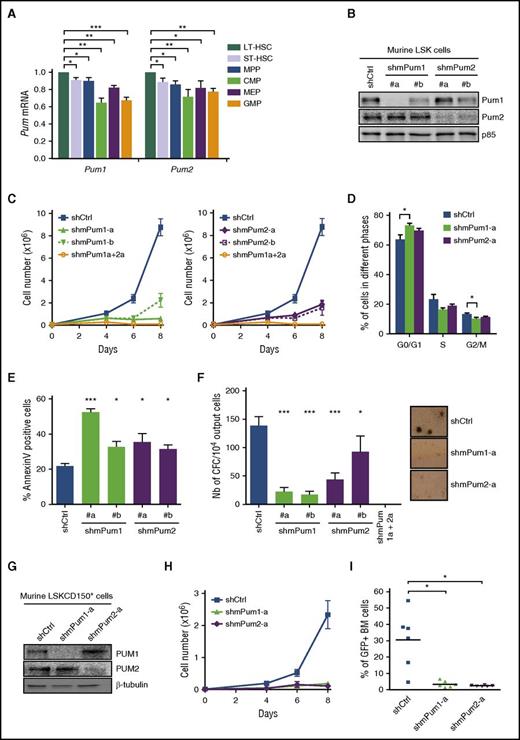
To better explore the functionality of the HSPC compartment, we extended our experiments to the murine model. Analysis of Pum1 and Pum2 expression in various mouse stem/progenitor subpopulations led to the same trend observed with human cells, with a higher expression in the more primitive compartments (Figure 2A). As for human HSPCs, murine HSPC-enriched Lin−Sca+cKit+ (LSK) cells transduced with shRNAs engineered to reduce murine (m) Pum1 or Pum2 (Figure 2B) stopped growing after 7 days in liquid culture, in contrast to cells expressing shCtrl; simultaneous PUM1 and PUM2 KD led to an even stronger reduced cell expansion (Figure 2C). Five days posttransduction, shmPum/GFP+/LSK cells displayed a significantly increased proportion of cells in G0/G1 phase (Figure 2D); cell apoptosis was evident 2 days later, as assessed by increased Annexin V labeling (Figure 2E), with no change in cell differentiation (not shown). Consequently, Pum1 or Pum2 KD triggered a drastic reduction of clonogenic potential (Figure 2F), with no colony growth upon simultaneous PUM1 and PUM2 KD. To investigate PUM activity in more immature HSPCs, we KD PUM1 or PUM2 in LSKCD150+ cells (Figure 2G), which similarly led to a strong reduction in cell expansion (Figure 2H). Furthermore, in long-term competitive hematopoietic reconstitution assays, recipient mice only harbored rare shmPum1/GFP+ or shmPum2/GFP+ cells in BM, as opposed to shCtrl/GFP+ cells (Figure 2I), indicating that murine Pum1 or Pum2 KD HSPCs lost their ability to reconstitute hematopoiesis after myeloablation. Altogether, these results indicate that PUM1 and PUM2 are essential to both murine and human HSPC growth.
PUM1 and PUM2 contribute to murine HSPC expansion. (A) RT-qPCR analysis of mPum1 and mPum2 transcripts in mouse stem/progenitor subpopulations: LT-HSC (long-term HSC, Lin−Sca+cKit+CD34−CD150+), ST-HSC (short-term HSC, Lin−Sca+cKit+CD34+CD150+), MPP (Lin−Sca+cKit+CD34+CD150−), CMP (Lin−Sca−cKit+CD34+CD16−), MEP (Lin−Sca−cKit+CD34−CD16−), and GMP (Lin−Sca−cKit+CD34+CD16+). Results are normalized to Gapdh expression and expressed relative to Pum expression in LT-HSCs. (B-F) LSK cells were transduced with lentiviral vectors encoding the indicated shRNA targeting mPum1 (shmPum1) or mPum2 (shmPum2) or both (shmPum1a+2a) or luciferase as control (shCtrl) (sequences detailed in supplemental Table 8). shRNA/GFP+ LSK cells were sorted 2 days after transduction and maintained in liquid culture. (B) Immunoblot analysis of the indicated proteins at day 7 posttransduction. p85-PI3kinase was used as loading control. (C) Cell expansion analysis along time of culture posttransduction (n = 3). (D) Cell cycle analysis upon propidium iodide labeling at day 5 posttransduction (n = 4). (E) Apoptosis analysis at day 7 posttransduction through AnnexinV-PE/7AAD labeling (n = 4). (F) At day 7 posttransduction, shRNA/GFP+ output LSK-derived cells were seeded in methylcellulose medium and numbering of CFC was assessed after 7 days (n = 5); representative micrographs of colonies are presented on the right side (×20). (G-I) Murine LSKCD150+ cells were transduced with lentiviral vectors encoding shRNA targeting mPum1 (shmPum1-a) or mPum2 (shmPum2-a) or luciferase as control (shCtrl). shRNA/GFP+ cells were sorted 2 days after transduction and maintained in liquid culture. (G) Immunoblot analysis of the indicated proteins at day 7 posttransduction. β-Tubulin was used as loading control. (H) Cell expansion analysis along time of culture posttransduction (n = 3). (I) Three days posttransduction, lethally irradiated C57BL/6-Ly5.2 mice received 15 000 sorted shRNA/GFP+ LSKCD150+ cells from C57Bl/6 (Ly5.1) donors, in competition with the same amount of GFP− LSKCD150+ cells from Ly5.1 mice, together with 1.5 × 105 Ly5.2 BM cells. Presence of GFP+ cells in CD45.1+ BM cells of engrafted mice was assessed 4 months later by flow cytometry (at least 105 events). Each symbol represents data from a single chimeric mouse. (Figure representative of 1 experiment out of 2). Data are expressed as mean ± SEM. *P < .05; **P < .01; ***P < .001 (Student t test).
PUM1 and PUM2 contribute to murine HSPC expansion. (A) RT-qPCR analysis of mPum1 and mPum2 transcripts in mouse stem/progenitor subpopulations: LT-HSC (long-term HSC, Lin−Sca+cKit+CD34−CD150+), ST-HSC (short-term HSC, Lin−Sca+cKit+CD34+CD150+), MPP (Lin−Sca+cKit+CD34+CD150−), CMP (Lin−Sca−cKit+CD34+CD16−), MEP (Lin−Sca−cKit+CD34−CD16−), and GMP (Lin−Sca−cKit+CD34+CD16+). Results are normalized to Gapdh expression and expressed relative to Pum expression in LT-HSCs. (B-F) LSK cells were transduced with lentiviral vectors encoding the indicated shRNA targeting mPum1 (shmPum1) or mPum2 (shmPum2) or both (shmPum1a+2a) or luciferase as control (shCtrl) (sequences detailed in supplemental Table 8). shRNA/GFP+ LSK cells were sorted 2 days after transduction and maintained in liquid culture. (B) Immunoblot analysis of the indicated proteins at day 7 posttransduction. p85-PI3kinase was used as loading control. (C) Cell expansion analysis along time of culture posttransduction (n = 3). (D) Cell cycle analysis upon propidium iodide labeling at day 5 posttransduction (n = 4). (E) Apoptosis analysis at day 7 posttransduction through AnnexinV-PE/7AAD labeling (n = 4). (F) At day 7 posttransduction, shRNA/GFP+ output LSK-derived cells were seeded in methylcellulose medium and numbering of CFC was assessed after 7 days (n = 5); representative micrographs of colonies are presented on the right side (×20). (G-I) Murine LSKCD150+ cells were transduced with lentiviral vectors encoding shRNA targeting mPum1 (shmPum1-a) or mPum2 (shmPum2-a) or luciferase as control (shCtrl). shRNA/GFP+ cells were sorted 2 days after transduction and maintained in liquid culture. (G) Immunoblot analysis of the indicated proteins at day 7 posttransduction. β-Tubulin was used as loading control. (H) Cell expansion analysis along time of culture posttransduction (n = 3). (I) Three days posttransduction, lethally irradiated C57BL/6-Ly5.2 mice received 15 000 sorted shRNA/GFP+ LSKCD150+ cells from C57Bl/6 (Ly5.1) donors, in competition with the same amount of GFP− LSKCD150+ cells from Ly5.1 mice, together with 1.5 × 105 Ly5.2 BM cells. Presence of GFP+ cells in CD45.1+ BM cells of engrafted mice was assessed 4 months later by flow cytometry (at least 105 events). Each symbol represents data from a single chimeric mouse. (Figure representative of 1 experiment out of 2). Data are expressed as mean ± SEM. *P < .05; **P < .01; ***P < .001 (Student t test).
PUM1 and PUM2 promote leukemic cell growth
Having established the major role of PUM1/2 in normal HSPCs, we investigated PUM1 and PUM2 activities in myeloid leukemia. We analyzed expression of PUM1 and PUM2 transcripts in primary AML cell samples categorized according to the French-American-British classification and compared with healthy CD34+ cells. AML0-2 cells exhibited higher levels of PUM1 and PUM2 mRNA, as compared with control CD34+ HSPCs (Figure 3A). PUM1 or PUM2 KD was induced in 3 different AML cell lines (MOLM-14, MV4-11, UT7-GM). In all, PUM1/2 loss hampered cell growth (Figure 3B-C; supplemental Figure 2A-E) by keeping cells in G0/G1 phase (Figure 3D; supplemental Figure 2F), and induced apoptosis later on (Figure 3E; supplemental Figure 2C,G). The consequences of PUM1 or PUM2 KD were also studied in 6 primary AML samples (supplemental Table 4). Again, loss of PUM1 or PUM2 inhibited proliferation of every AML cell (Figure 3F). Collectively, our results demonstrate that both PUM1 and PUM2 are crucial regulators of normal HSPC and myeloid leukemia cell growth.
PUM1 and PUM2 contribute to human leukemic cell growth. (A) RT-qPCR analysis of hPUM1 and hPUM2 mRNA expression in AML patient samples grouped according to the French-American-British classification, and in CD34+ cells from healthy donors (peripheral blood or cord blood or BM). Each symbol represents data from a single patient. CD34+: 11 samples; AML0: 5 samples; AML1: 16 samples; AML2: 26 samples; AML3: 7 samples; AML4: 6 samples; AML5: 4 samples. Results are normalized to GAPDH, HPRT, and Cyclophilin A transcript levels. (B-E) MOLM-14 leukemia cells were transduced or not (nt) with the indicated shRNA-encoding vectors. (B) Immunoblot analysis of the indicated proteins at day 3 posttransduction. Actin was used as loading control. (C) Cell expansion analysis along time of culture posttransduction (n = 3). (D) Cell cycle analysis upon propidium iodide labeling at day 3 posttransduction (n = 3). (E) Annexin V labeling at day 4 posttransduction (n = 3). (F) Primary AML patient samples were transduced with shCtrl-, shPUM1-, or shPUM2-encoding vectors, and cultured onto MS5 stromal cells in CD34+ growth medium. At day 10, the cultures were disrupted by vigorous pipetting and scraping, and nucleated cells were counted by FACS. Results are expressed relative to shCtrl-transduced cell numbering (n = 6). Each symbol represents data from a single patient. Data are expressed as mean ± SEM. Ns, not significant. *P < .05; **P < .01; ***P < .001 (Mann-Whitney test in panel A; Student t test in panels C-F).
PUM1 and PUM2 contribute to human leukemic cell growth. (A) RT-qPCR analysis of hPUM1 and hPUM2 mRNA expression in AML patient samples grouped according to the French-American-British classification, and in CD34+ cells from healthy donors (peripheral blood or cord blood or BM). Each symbol represents data from a single patient. CD34+: 11 samples; AML0: 5 samples; AML1: 16 samples; AML2: 26 samples; AML3: 7 samples; AML4: 6 samples; AML5: 4 samples. Results are normalized to GAPDH, HPRT, and Cyclophilin A transcript levels. (B-E) MOLM-14 leukemia cells were transduced or not (nt) with the indicated shRNA-encoding vectors. (B) Immunoblot analysis of the indicated proteins at day 3 posttransduction. Actin was used as loading control. (C) Cell expansion analysis along time of culture posttransduction (n = 3). (D) Cell cycle analysis upon propidium iodide labeling at day 3 posttransduction (n = 3). (E) Annexin V labeling at day 4 posttransduction (n = 3). (F) Primary AML patient samples were transduced with shCtrl-, shPUM1-, or shPUM2-encoding vectors, and cultured onto MS5 stromal cells in CD34+ growth medium. At day 10, the cultures were disrupted by vigorous pipetting and scraping, and nucleated cells were counted by FACS. Results are expressed relative to shCtrl-transduced cell numbering (n = 6). Each symbol represents data from a single patient. Data are expressed as mean ± SEM. Ns, not significant. *P < .05; **P < .01; ***P < .001 (Mann-Whitney test in panel A; Student t test in panels C-F).
PUM1/2 promote FOXP1 expression
To decipher how PUMs contribute to HSPC expansion, we set up a stable isotope labeling with amino acids in cell culture-based global quantitative proteomic analysis to identify PUM1/2 targets. We used the murine HPC-7 multipotent and stem cell factor–dependent progenitor cell line that recapitulates typical HSPC transcription factor network.21 Loss of PUM1 or PUM2 inhibited HPC-7 cell survival, as we observed in human cell lines (supplemental Figure 3A-B). We compared the proteome of shCtrl-, shmPum1-, or shmPum2-transduced HPC-7 cells and looked for common changes in Pum1 and Pum2 KD cells (supplemental Tables 5 and 6). We found that both Pum1 and Pum2 KD strongly reduced expression of the Forkhead transcription factor Foxp1, whose transcript displays 2 Pumilio-binding elements (PBE) in its 3′ untranslated region (UTR). Lower expression of FOXP1 was confirmed in primary human CD34+ HSPCs upon PUM1 or PUM2 KD, both FOXP1 mRNA and protein being decreased (Figure 4A-B). Conversely, FOXP1 levels were higher in the human HSC compartment that exhibited high PUM1 and PUM2 levels, as compared with MPP, GMP, CMP, and MEP (Figure 4C). In the 3 AML cell lines (MOLM-14, MV4-11, UT7-GM), we observed that shPUM1 or shPUM2 induced loss of FOXP1 protein and mRNA expression (Figure 4D-E; supplemental Figure 4). Highly significant correlative expression between PUM and FOXP1 transcripts was further observed in primary AML samples (Figure 4F-G), and strong cooccurrence was evidenced from public TCGA dataset (supplemental Table 7). Altogether, these results indicate that expression of FOXP1 transcription factor is regulated by, and correlates with, PUM1 and PUM2 levels in HSPCs and myeloid leukemia cells.
PUM1 and PUM2 regulate FOXP1 expression in CD34+HSPCs and leukemic cells. (A) Immunoblot analysis of the indicated proteins in CD34+ HSPCs transduced or not (nt) with the indicated shRNA-encoding vectors at day 4 post-transduction. Actin was used as loading control. (B) RT-qPCR analysis of hFOXP1 mRNA in shRNA-transduced CD34+ cells at day 4 post-transduction. Results are normalized to GAPDH mRNA expression, and expressed relative to shCtrl-expressing cells (n = 3). (C) RT-qPCR analysis of hPUM1, hPUM2 and hFOXP1 transcripts in the indicated human stem/progenitor subpopulations (see Figure 1A). Results are normalized to GAPDH expression, and expressed relative to expression in HSC (n = 3). (D) Immunoblot analysis of the indicated proteins in shRNA-transduced MOLM-14 cells at day 3 post-transduction. Actin was used as loading control. (E) RT-qPCR analysis of hFOXP1 mRNA in shRNA-transduced MOLM-14 cells at day 3 post-transduction. Results are normalized to GAPDH expression and expressed relative to shCtrl-expressing cells (n = 3). (F) RT-qPCR analysis of FOXP1 expression in AML patient samples grouped according to FAB classification, and in CD34+ cells from healthy donors. Each symbol represents data from a single patient. CD34+: 11 samples; AML0: 4 samples; AML1: 9 samples; AML2: 17 samples; AML3: 6 samples; AML4: 6 samples; AML5: 4 samples. Results are normalized to GAPDH, HPRT and Cyclophilin A mRNA levels. (G) Correlation between PUM1 or PUM2 and FOXP1 mRNA expressions in AML samples. r = Pearson correlation’s coefficient. Data are expressed as mean ± SEM. ns: not significant, * P < .05; **P < .01; ***P < .001 [Mann-Whitney test in (F); Student t test in (B, C, E)].
PUM1 and PUM2 regulate FOXP1 expression in CD34+HSPCs and leukemic cells. (A) Immunoblot analysis of the indicated proteins in CD34+ HSPCs transduced or not (nt) with the indicated shRNA-encoding vectors at day 4 post-transduction. Actin was used as loading control. (B) RT-qPCR analysis of hFOXP1 mRNA in shRNA-transduced CD34+ cells at day 4 post-transduction. Results are normalized to GAPDH mRNA expression, and expressed relative to shCtrl-expressing cells (n = 3). (C) RT-qPCR analysis of hPUM1, hPUM2 and hFOXP1 transcripts in the indicated human stem/progenitor subpopulations (see Figure 1A). Results are normalized to GAPDH expression, and expressed relative to expression in HSC (n = 3). (D) Immunoblot analysis of the indicated proteins in shRNA-transduced MOLM-14 cells at day 3 post-transduction. Actin was used as loading control. (E) RT-qPCR analysis of hFOXP1 mRNA in shRNA-transduced MOLM-14 cells at day 3 post-transduction. Results are normalized to GAPDH expression and expressed relative to shCtrl-expressing cells (n = 3). (F) RT-qPCR analysis of FOXP1 expression in AML patient samples grouped according to FAB classification, and in CD34+ cells from healthy donors. Each symbol represents data from a single patient. CD34+: 11 samples; AML0: 4 samples; AML1: 9 samples; AML2: 17 samples; AML3: 6 samples; AML4: 6 samples; AML5: 4 samples. Results are normalized to GAPDH, HPRT and Cyclophilin A mRNA levels. (G) Correlation between PUM1 or PUM2 and FOXP1 mRNA expressions in AML samples. r = Pearson correlation’s coefficient. Data are expressed as mean ± SEM. ns: not significant, * P < .05; **P < .01; ***P < .001 [Mann-Whitney test in (F); Student t test in (B, C, E)].
FOXP1 mediates PUM1/2 cell growth activity
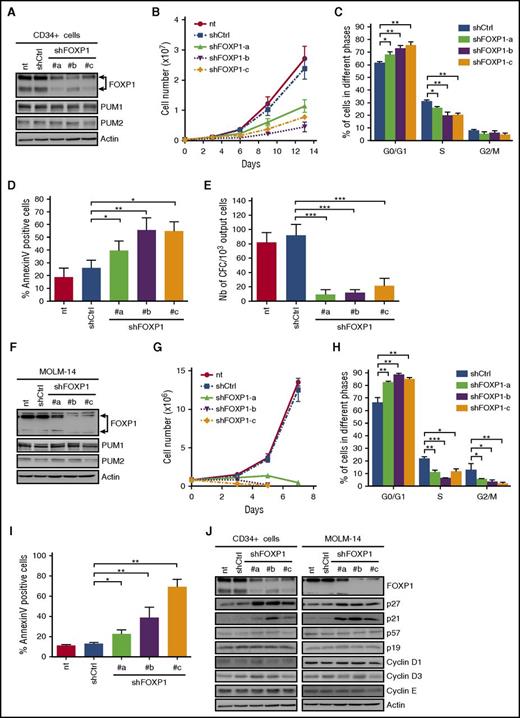
FOXP1 is 1 of the 4 FOXP subfamily members, which regulate a large number of genes involved in cell proliferation, differentiation, and development.22 Nothing is currently known on the relationship between FOXP1 and early hematopoiesis. To address the biological activities of FOXP1 in HSPCs, we identified 3 shRNAs efficiently targeting human FOXP1 (Figure 5A). Each of them inhibited CD34+ cell expansion (Figure 5B) by keeping cells in G0/G1 phase (Figure 5C), followed by enhanced cell death (Figure 5D), without impairing PUM1/2 expression (Figure 5A). Furthermore, shFOXP1-CD34+ cells displayed a severely reduced clonogenic potential (Figure 5E). As for shPUM1 or shPUM2, FOXP1 shRNAs inhibited AML cell growth (MOLM-14, MV4-11, UT7-GM) by keeping cells in G0/G1 phase and further induced apoptosis without impairing PUM1/2 expression (Figure 5F-I; supplemental Figure 5). To nail down how FOXP1 regulates HSPC growth, we analyzed expression of a panel of cyclins and cell cycle inhibitors involved in HSPC expansion. FOXP1 KD in HSPCs and AML cells enhanced expression of the 2 cell cycle inhibitors, p21CIP1 and p27KIP1, without modification of other cell cycle regulators (Figure 5J; supplemental Figure 5A,D).
FOXP1 contributes to human CD34+HSPC and leukemic cell growth. (A-E) Human cord blood CD34+ cells were transduced or not (nt) with lentiviral vectors encoding shRNAs targeting FOXP1 (shFOXP1#a, #b, #c; sequences detailed in supplemental Table 8) or luciferase as control (shCtrl). (A) Immunoblot analysis of the indicated proteins at day 3 posttransduction. Actin was used as loading control. (B) Cell expansion analysis along time posttransduction (n = 4). (C) Cell cycle analysis upon propidium iodide labeling at day 3 posttransduction (n = 3). (D) Annexin V labeling assessed at day 4 posttransduction (n = 4). (E) CFC assays at day 7 posttransduction (n = 3). (F-I) MOLM-14 leukemia cells were transduced or not (nt) with the indicated shRNA-encoding vectors. (F) Immunoblot analysis of the indicated proteins at day 3 posttransduction. Actin was used as loading control. (G) Cell expansion analysis along time posttransduction (n = 3). (H) Cell cycle analysis upon propidium iodide labeling at day 2 posttransduction (n = 3). (I) Annexin V labeling at day 3 posttransduction (n = 3). (J) Immunoblot analysis of the indicated proteins at day 3 (CD34+) and day 2 (MOLM-14) posttransduction. Actin was used as loading control. Data are expressed as mean ± SEM. *P < .05; **P < .01; ***P < .001 (Student t test).
FOXP1 contributes to human CD34+HSPC and leukemic cell growth. (A-E) Human cord blood CD34+ cells were transduced or not (nt) with lentiviral vectors encoding shRNAs targeting FOXP1 (shFOXP1#a, #b, #c; sequences detailed in supplemental Table 8) or luciferase as control (shCtrl). (A) Immunoblot analysis of the indicated proteins at day 3 posttransduction. Actin was used as loading control. (B) Cell expansion analysis along time posttransduction (n = 4). (C) Cell cycle analysis upon propidium iodide labeling at day 3 posttransduction (n = 3). (D) Annexin V labeling assessed at day 4 posttransduction (n = 4). (E) CFC assays at day 7 posttransduction (n = 3). (F-I) MOLM-14 leukemia cells were transduced or not (nt) with the indicated shRNA-encoding vectors. (F) Immunoblot analysis of the indicated proteins at day 3 posttransduction. Actin was used as loading control. (G) Cell expansion analysis along time posttransduction (n = 3). (H) Cell cycle analysis upon propidium iodide labeling at day 2 posttransduction (n = 3). (I) Annexin V labeling at day 3 posttransduction (n = 3). (J) Immunoblot analysis of the indicated proteins at day 3 (CD34+) and day 2 (MOLM-14) posttransduction. Actin was used as loading control. Data are expressed as mean ± SEM. *P < .05; **P < .01; ***P < .001 (Student t test).
In order to definitively conclude on the FOXP1 and PUM1/2 protein-interrelated biological properties, we constitutively expressed FOXP1 in shPUM1- or shPUM2-transduced HSPCs. Exogenous FOXP1 reversed cell growth defects and apoptosis of PUM1 or PUM2 KD HSPCs (Figure 6A-C) and also improved the clonogenic potential of PUM KD HSPCs (Figure 6D). Furthermore, enforced FOXP1 expression counteracted shPUM1- or shPUM2-dependent growth defects, and cell apoptosis of myeloid leukemia cells (Figure 6E-G; supplemental Figure 6). Taken together, our results demonstrate that FOXP1 supports HSPC and leukemic cell expansion and mediates PUM1- and PUM2-dependent growth activities in both normal progenitors and leukemic cells.
FOXP1 rescues growth defects of PUM1 and PUM2 KD cells. (A-D) Cord blood CD34+ HSPCs were transduced first with pLenti-PGK-USIL DEST lentiviral vectors that encode (+) or not (−) FOXP1, and then with shCtrl (−), shPUM1 or shPUM2 vectors, and sorted at day 3 posttransduction. (A) Immunoblot analysis of the indicated proteins at day 3 posttransduction. Actin was used as loading control. (B) Relative cell expansion at day 10 posttransduction (n = 3). (C) Annexin V labeling at day 7 (shPUM1) and at day 10 (shPUM2) posttransduction. (D) CFC assay at day 7 posttransduction (n = 3). (E-G) MOLM-14 cells were transduced first with pINDUCER21 inducible lentiviral vectors that encode (+) or not (−) FOXP1, then with shCtrl (−), shPUM1, or shPUM2 vectors. The day after, FOXP1 expression was induced with doxycycline (0.05 µg/mL). (E) Immunoblot analysis of the indicated proteins at day 4 posttransduction. Actin was used as loading control. (F) Relative cell expansion of double-transduced cells at day 8 posttransduction (n = 4). (G) Annexin V labeling at day 6 posttransduction (n = 4). Data are expressed as mean ± SEM. Ns, not significant. *P < .05; **P < .01 (Student t test).
FOXP1 rescues growth defects of PUM1 and PUM2 KD cells. (A-D) Cord blood CD34+ HSPCs were transduced first with pLenti-PGK-USIL DEST lentiviral vectors that encode (+) or not (−) FOXP1, and then with shCtrl (−), shPUM1 or shPUM2 vectors, and sorted at day 3 posttransduction. (A) Immunoblot analysis of the indicated proteins at day 3 posttransduction. Actin was used as loading control. (B) Relative cell expansion at day 10 posttransduction (n = 3). (C) Annexin V labeling at day 7 (shPUM1) and at day 10 (shPUM2) posttransduction. (D) CFC assay at day 7 posttransduction (n = 3). (E-G) MOLM-14 cells were transduced first with pINDUCER21 inducible lentiviral vectors that encode (+) or not (−) FOXP1, then with shCtrl (−), shPUM1, or shPUM2 vectors. The day after, FOXP1 expression was induced with doxycycline (0.05 µg/mL). (E) Immunoblot analysis of the indicated proteins at day 4 posttransduction. Actin was used as loading control. (F) Relative cell expansion of double-transduced cells at day 8 posttransduction (n = 4). (G) Annexin V labeling at day 6 posttransduction (n = 4). Data are expressed as mean ± SEM. Ns, not significant. *P < .05; **P < .01 (Student t test).
PUM1/2 upregulate FOXP1 expression via two 3′UTR PBE
To further explore the mechanism by which PUM1/2 positively regulate FOXP1 levels, we screened FOXP1 mRNA sequence for potential PBEs. The presence of 2 canonical PBEs (5′UGUAXAUA-3′)23 were uncovered at positions 3005 (PBE1) and 3877 (PBE2) from the beginning of the 4.5-kb-long FOXP1 3′UTR (Figure 7A). Using oligoRNA pull-down assays with MOLM-14 (Figure 7B) and UT7-GM cell lysates (supplemental Figure 7A), we found interactions between PUM1/2 and the 2 PBEs identified (PBE1 and PBE2) on FOXP1 mRNA. Competition assays in the presence of excess of WT or mutated (MUT) PBE1 or PBE2 confirmed the specificity of these interactions. We further assessed PUM protein/FOXP1 mRNA interactions using RNA immunoprecipitation assays. FOXP1 transcripts were strongly enriched in PUM1 and PUM2 IPs prepared from untreated MOLM-14 cells, in contrast to control IPs (Figure 7C) and to GAPDH and HPRT transcripts (supplemental Figure 7B). FOXP1 mRNA was similarly enriched in PUM-IPs prepared from prefixed crosslinked MOLM-14 cells, thus precluding any cell lysis artifacts (supplemental Figure 7B). As expected, less FOXP1 transcripts were associated with PUM-IPs prepared from PUM1 or PUM2 KD cells, in accordance with lower PUM1 or PUM2 amounts (Figure 7C). Similar observations were obtained using UT7-GM cells (supplemental Figure 7C). Overall, our data indicate that PUM1 and PUM2 bind to endogenous FOXP1 mRNA of myeloid cells.
PUM1 and PUM2 positively regulate FOXP1 mRNA expression via two 3′UTR located PBE. (A) Schematic representation of human FOXP1 mRNA with the 2 PUM-binding elements (PBE), PBE1 and PBE2, boxed (red), and regions A, B, C delineated. The numbers indicate positions in 3′UTR, starting from translation stop codon. (B) RNA pull-down assay. MOLM-14 cell lysates were incubated with WT1 or MUT1 biotin-labeled FOXP1-PBE1 probes, and none or ×5 unlabeled PBE1 (WT1 or MUT1) or PBE2 (WT2 or MUT2) competitor riboprobes, as indicated. Pulled‐down PUM proteins were analyzed by immunoblotting. (C) RNA immunoprecipitation assays. Lysates from shRNA-transduced MOLM-14 cells were immunoprecipitated (IP) using control (IgG), PUM1 or PUM2 antibody, as indicated. Upper panel: Immunoblot analysis of the indicated proteins. Lower panel: RT-qPCR analysis of FOXP1 mRNA present in the IPs. The results are expressed as percent of input (n = 3). (D) Luciferase assay. MOLM-14 cells were transfected with control (siCtrl), PUM1 (siPUM1), or PUM2 (siPUM2) siRNA, and with the indicated luciferase reporter construct harboring the FL FOXP1-3′UTR. Relative luciferase activity was monitored 24 hours later and expressed as the ratio between Renilla luciferase (Rluc) activity and Firefly luciferase (Fluc) activity, used as transfection control. Data are expressed relative to expression in FOXP1-3′UTR FL and siCtrl transfected cells (n = 4). (E) Luciferase assays were performed as in panel D, with luciferase constructs harboring the indicated regions of the FOXP1-3′UTR (A-C) with WT, or doubly MUT1/2 PBEs. Data are expressed relative to expression in WT-FOXP1-3′UTR-C and siCtrl transfected cells (n = 4). (F) Luciferase assay. MOLM-14 cells were transfected with FOXP1-3′UTR-C luciferase reporter constructs harboring WT PBE1 and PBE2, mutated PBE1 (MUT1), mutated PBE2 (MUT2), or doubly mutated PBE1 and PBE2 (MUT1/2). Relative luciferase activity was monitored 24 hours later, expressed as the ratio Rluc/Fluc, and compared relative to expression in WT-FOXP1-3′UTR-C transfected cells (n = 4). Data are expressed as mean ± SEM. *P < .05; **P < .01; ***P < .001 (Student t test).
PUM1 and PUM2 positively regulate FOXP1 mRNA expression via two 3′UTR located PBE. (A) Schematic representation of human FOXP1 mRNA with the 2 PUM-binding elements (PBE), PBE1 and PBE2, boxed (red), and regions A, B, C delineated. The numbers indicate positions in 3′UTR, starting from translation stop codon. (B) RNA pull-down assay. MOLM-14 cell lysates were incubated with WT1 or MUT1 biotin-labeled FOXP1-PBE1 probes, and none or ×5 unlabeled PBE1 (WT1 or MUT1) or PBE2 (WT2 or MUT2) competitor riboprobes, as indicated. Pulled‐down PUM proteins were analyzed by immunoblotting. (C) RNA immunoprecipitation assays. Lysates from shRNA-transduced MOLM-14 cells were immunoprecipitated (IP) using control (IgG), PUM1 or PUM2 antibody, as indicated. Upper panel: Immunoblot analysis of the indicated proteins. Lower panel: RT-qPCR analysis of FOXP1 mRNA present in the IPs. The results are expressed as percent of input (n = 3). (D) Luciferase assay. MOLM-14 cells were transfected with control (siCtrl), PUM1 (siPUM1), or PUM2 (siPUM2) siRNA, and with the indicated luciferase reporter construct harboring the FL FOXP1-3′UTR. Relative luciferase activity was monitored 24 hours later and expressed as the ratio between Renilla luciferase (Rluc) activity and Firefly luciferase (Fluc) activity, used as transfection control. Data are expressed relative to expression in FOXP1-3′UTR FL and siCtrl transfected cells (n = 4). (E) Luciferase assays were performed as in panel D, with luciferase constructs harboring the indicated regions of the FOXP1-3′UTR (A-C) with WT, or doubly MUT1/2 PBEs. Data are expressed relative to expression in WT-FOXP1-3′UTR-C and siCtrl transfected cells (n = 4). (F) Luciferase assay. MOLM-14 cells were transfected with FOXP1-3′UTR-C luciferase reporter constructs harboring WT PBE1 and PBE2, mutated PBE1 (MUT1), mutated PBE2 (MUT2), or doubly mutated PBE1 and PBE2 (MUT1/2). Relative luciferase activity was monitored 24 hours later, expressed as the ratio Rluc/Fluc, and compared relative to expression in WT-FOXP1-3′UTR-C transfected cells (n = 4). Data are expressed as mean ± SEM. *P < .05; **P < .01; ***P < .001 (Student t test).
To investigate how PUM1/2 positively regulate FOXP1 transcripts, we focused on FOXP1-3′UTR carrying the 2 PBEs. Full-length (FL) 4.5-kb-long FOXP1-3′UTR was cloned downstream from Luciferase reporter to determine whether this FOXP1 region mediates PUM-dependent positive regulation of gene expression. Figure 7D shows that siRNA-mediated PUM1 or PUM2 KD consistently decreased luciferase expression from FL FOXP1-3′UTR reporter. FOXP1-3′UTR sequence was further split into 3 shorter fragments, A, B, and C, to delineate PUM-responsive region. Neither A nor B fragment mediated luciferase repression induced upon PUM1/2 KD. Outstandingly, only fragment C, containing the 2 canonical PBEs, responded to PUM1/2 expression changes and mediated siPUM-dependent luciferase repression (Figure 7E). Furthermore, mutation of each of the 2 PBEs decreased FOXP1-3′UTR-C activity; combined mutations showed an additive effect (Figure 7F). Finally, mutation of both PBEs rendered FOXP1-3′UTR-C no longer responsive to PUM1 or PUM2 KD (Figure 7E), confirming the requirement of the 2 PBEs in PUM-dependent regulation. Altogether, these data indicate that FOXP1 mRNA is a target of PUM1 and PUM2 factors, which enhance its expression by binding to two 3′UTR-located PBEs.
Discussion
We previously identified PUM1 and PUM2 as genes whose expression is upregulated by Dll4/Notch signaling pathway and homeoprotein HOXB4, 2 well-known regulators of HSPC self-renewal.6,8,11,12 Here, we identify a novel molecular link, where the RBP PUM1/2 integrate both self-renewal signals to FOXP1 transcription factor activity to control HSPC expansion. Moreover, we reveal that this PUM1/2-FOXP1 node contributes to leukemic cell expansion. Finally, we show that PUM1/2, as opposed to their well-known repressive functions, rather promote FOXP1 expression directly through binding to two 3′UTR-PBEs.
We first demonstrate that PUM1 and PUM2 RBPs are both required to maintain human and murine HSPC growth in vitro and also in vivo: loss of either PUM1 or PUM2 induces loss of HSPC survival, expansion, and clonogenic properties in vitro and renders them unable to reconstitute hematopoiesis in vivo after myeloablation. PUM loss further inhibits HOXB4 growth-promoting activity (data not shown). Contrary to what we have observed, PUM1- or PUM2-deficient mice remain viable and do not exhibit obvious alterations of native hematopoiesis.24,25 This observation could be due to compensatory mechanisms that occur in primitive hematopoiesis, which may mask PUM activities in adult HSPCs, as observed in some other knockout mouse models.26 Our results indicate that the 2 mammalian PUM exhibit similar biological activities and suggest that global PUM expression level is limiting. Indeed, we have observed that murine HSPCs displayed a stronger reduction in cell expansion, and a complete inhibition of the CFC potential upon PUM1 and PUM2 double KD, as compared with single KD. Similarities in PUM1/2 activities might be explained by their highly conserved amino-acid sequences (83% similarity), with 91% homology in their RNA-binding domains.18 Unexpectedly, overexpression of PUM1/2 was similarly deleterious for HSPC and leukemic cell survival (data not shown), probably reflecting PUM major contribution to chromosome instability, as recently reported.27,28 These observations indicate that PUM levels are subjected to fine-tuning to control myeloid cell growth.
PUM factors are ubiquitously expressed. Their multiple biological activities depend on association with differentially expressed partners,29 on posttranslational modifications that can be modulated by growth factors,30 on PUM stability itself,13 and also on the presence of RNA sponge that binds and sequesters PUM proteins.27,28 Our data indicate that PUM1 and PUM2 gene expression levels are also regulated along cell differentiation, being maximal in undifferentiated progenitors.
It has previously been reported that PUM2 KD reduces cell proliferation of human cultured adipose-derived stem cells.17 Conversely, PUM1 favors growth of human primary fibroblasts by cooperating with miR221/miR222 to repress p27KIP1 cell cycle inhibitor through binding to 2 PBEs in p27KIP1-3′UTR.30 We similarly observed that PUM1/2 through FOXP1 inhibit p27KIP1 expression in myeloid cells, indicating that cell cycle control through p27KIP1 regulation is a widespread PUM activity in mammalian cells.
We identified the forkhead transcription factor, FOXP1, as a novel target of PUM1 and PUM2, and above all, as a new key regulator of HSPC expansion, able to mediate PUM1/2 growth-promoting activity. FOXP1 is expressed in virtually all cells, yet to variable levels. Like all forkhead factors, FOXP1 is a multifaceted transcription factor responsible for fine-tuning the spatial and temporal expression of a broad range of genes both during development and in adult tissues.22 In hematopoietic cells, FOXP1 contributes to differentiation of immature B cells and plasmocytes31,32 ; it enforces naive T-cell quiescence by limiting cell cycle progression.33,34 It also inhibits differentiation of monocytes to macrophages.35 Whereas FOXP1 contribution to HSPC expansion was not foreseen, other forkhead factors, namely FOXOs, are well-known critical regulators of hematopoietic stem cell quiescence and inhibit cell cycle progression.36,37 They moreover display antileukemic activities.38 FOXP1, like certain other FOX proteins, might thus compete for binding the same DNA motifs of some FOXO target promoters as described in a few contexts.22,33,39
As opposed to HSPCs, FOXP1 maintains hair follicle stem cell quiescence,40 whereas it sustains ESC self-renewal and contributes to efficient reprogramming of somatic cells into induced pluripotent stem cells.41 The reasons behind these paradoxical observations are unclear, but may be linked to expression of dedicated spliced variants of FOXP1 observed in some cellular contexts,42-45 as exemplified in ESCs.41 Transcription-dependent overexpression of FOXP1 has been observed following recurrent chromosome translocations of FOXP1 locus and linked to lymphoma development.46 MicroRNA-dependent posttranscriptional deregulation of FOXP1 is also reported in various cancers.47-49 Extra alternatively spliced FOXP1 mRNA isoforms represent an additional mechanism for FOXP1 overexpression and/or differential activities that correlate with lymphomagenesis.42 Our data reveal that RBPs such as PUMs also tune FOXP1 growth-promoting and oncogenic activities.
To date, the best-characterized function of PUM proteins is as posttranscriptional repressors.13,50 PUF proteins inhibit mRNA expression by recruiting the Ccr4-Pop2-NOT deadenylase complex or by impeding interactions of translation initiation factors with the 5′mRNA cap, by changing ribonucleoprotein structure, and by limiting translation elongation and termination. Also, PUM binding to RNA can induce local conformational changes that favor association with microRNA, as demonstrated for p27KIP1 and E2F3.30,51 Contrary to this canonical role of PUMs as repressors, here PUM1/2 promote FOXP1 expression through binding to 2 PBEs in FOXP1-3′UTR. An activator function of PUM1/2 has been occasionally reported in Xenopus and Caenorhabditis elegans,52,53 linked to extension of the polyA tail54 or polyA-dependent translational activation.55 We observed that the 2 PBEs present in FOXP1-3′UTR work in an additive manner to mediate PUM-dependent FOXP1 regulation, thus strengthening the key role of PUM on FOXP1 mRNA regulation. Whether PUM1/2 activity impinges both FOXP1 mRNA stability and FOXP1 mRNA translation await further investigation.
In conclusion, our data identify the RBPs PUM, and the posttranscriptional processes they control, as new regulators of HSPC expansion. PUM factors are positioned at the crossroad of Notch pathway and the homeoprotein HOXB4 from 1 side, and FOXP1 on the other side to tune HSPC growth. As FOXP1 is a transcription factor, our data indicate that PUM1/2 have the capacity to change the transcription networks of uncommitted cells to sustain their expansion, and modify their transition between cell proliferation and differentiation, and possibly to contribute to leukemia cell growth. It provides new targets to amplify HSPC in vitro for stem cell–based therapies.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors acknowledge the 3P5 proteomic facility for proteome analysis (Université Paris Descartes, Sorbonne Paris Cité, Institut Cochin, Paris), and the Animal Facility for handling mice, CYBIO for cell sorting, and GENOM'IC of Institut Cochin/INSERM U1016 for help in RT-qPCR analysis. The authors thank H. Tucker (Austin, TX) for providing them with FOXP1 antibody; A. Yacia, B. Izac, G. Dieffenbach, A. Artus, M. Le-Gall, and I. Leray for technical assistance; and O. Kosmider for his help for AML data analysis. The authors acknowledge the Obstetric Unit from Orsay Hospital, Centre Hospitalier de Marne-la-Vallée, and Cell Therapy Center from Saint-Louis Hospital (Paris, France) for providing them with cord blood samples. The authors thank the FILOtheque (BB-0033-00073; Tumour Bank of the FILO group, Cochin Hospital, Paris), and Saint-Louis Hospital’s Tumor Biobank for managing patient biological samples.
This work was supported by grants from INSERM, Centre National de la Recherche Scientifique, Paris Descartes University, Region Ile de France to 3P5, Ligue Nationale Contre le Cancer (RS13/75-84 and R14077KK), and Association pour la Recherche Contre le Cancer (ECL2010R01075).
A.H. and F.M. were fellows from the French Research Minister, A.B. from the “Ligue Nationale Contre le Cancer,” F.M. from “Fondation pour la Recherche Médicale,” A.H. and A.M.-N. from “Société Française d'Hématologie,” and C.N. from “Association de la Recherche Contre le Cancer.”
The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Authorship
Contribution: E.L., I.D.-F., I.V., and S.F. designed the research and analyzed the data; C.N., A.H., F.M., A.M.-N., A.B., and F.G. performed the experiments; E.L. and I.D.-F. wrote the original draft; E.L., I.D.-F., I.V., F.P., S.F., and C.N. wrote, reviewed, and edited the paper; C.N., A.H., F.M., I.V., and A.M.-N. created the figures; and E.L., I.D.-F., and S.F. oversaw funding.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
The current affiliation for I.V. is INSERM U1035, Équipe CSHNL, Université de Bordeaux, Bordeaux, France.
Correspondence: Evelyne Lauret, U1016 INSERM, Institut Cochin, 22 rue Méchain 75014 Paris, France; e-mail: evelyne.lauret@inserm.fr; and Isabelle Dusanter-Fourt, U1016 INSERM, Institut Cochin, 22 rue Méchain 75014 Paris, France; e-mail: isabelle.dusanter@inserm.fr.
References
Author notes
C.N., A.H., and F.M. contributed equally to this study.
I.V., I.D.-F., and E.L. are joint senior authors.



![Figure 4. PUM1 and PUM2 regulate FOXP1 expression in CD34+ HSPCs and leukemic cells. (A) Immunoblot analysis of the indicated proteins in CD34+ HSPCs transduced or not (nt) with the indicated shRNA-encoding vectors at day 4 post-transduction. Actin was used as loading control. (B) RT-qPCR analysis of hFOXP1 mRNA in shRNA-transduced CD34+ cells at day 4 post-transduction. Results are normalized to GAPDH mRNA expression, and expressed relative to shCtrl-expressing cells (n = 3). (C) RT-qPCR analysis of hPUM1, hPUM2 and hFOXP1 transcripts in the indicated human stem/progenitor subpopulations (see Figure 1A). Results are normalized to GAPDH expression, and expressed relative to expression in HSC (n = 3). (D) Immunoblot analysis of the indicated proteins in shRNA-transduced MOLM-14 cells at day 3 post-transduction. Actin was used as loading control. (E) RT-qPCR analysis of hFOXP1 mRNA in shRNA-transduced MOLM-14 cells at day 3 post-transduction. Results are normalized to GAPDH expression and expressed relative to shCtrl-expressing cells (n = 3). (F) RT-qPCR analysis of FOXP1 expression in AML patient samples grouped according to FAB classification, and in CD34+ cells from healthy donors. Each symbol represents data from a single patient. CD34+: 11 samples; AML0: 4 samples; AML1: 9 samples; AML2: 17 samples; AML3: 6 samples; AML4: 6 samples; AML5: 4 samples. Results are normalized to GAPDH, HPRT and Cyclophilin A mRNA levels. (G) Correlation between PUM1 or PUM2 and FOXP1 mRNA expressions in AML samples. r = Pearson correlation’s coefficient. Data are expressed as mean ± SEM. ns: not significant, * P < .05; **P < .01; ***P < .001 [Mann-Whitney test in (F); Student t test in (B, C, E)].](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/129/18/10.1182_blood-2016-10-747436/7/m_blood747436f4.jpeg?Expires=1767715238&Signature=YYMgqVlbg2MVZKHlaaUjfjuz5QqvLq3pdx5mTOKKZpaZt99EJSYVZ~vWowUgCsxHZI655VC-f-DPkHKUyP8zo0e4EfkN9h3qJvtPn-ht2sreHzmxbtDmwDXYIgxlxRHUdl5KjQVHCwF-SI2N-Gx3HbFhkw0ZUatBiaFSPL04lJbVsc4jSJm3z3yj2yuj9UuYFZa3ihJJ62isfF4NnaMwQyyd0KZKWCHdH0YKgw3RxEGPM6M9gVRyR5vGAis9SjMmpcWaJhE8UAjD6UxnMvJzrX~VXin-3nZKI1~OG8c3zB0c-cj9c8bzG0uEHQSDZk-BkvSCBrbanVCKp0FTD-qReA__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)