Key Points
Combined use of global and targeted analyses allows the cytogenetic classification of 88% of adult patients with Ph-negative BCP-ALL.
In GRAALL trials, t(4;11)/KMT2A-AFF1 and t(v;14q32)/IGH are the 2 cytogenetic anomalies significantly associated with a worse outcome.
Abstract
Multiple cytogenetic subgroups have been described in adult Philadelphia chromosome (Ph)-negative B-cell precursor (BCP) acute lymphoblastic leukemia (ALL), often comprising small numbers of patients. In this study, we aimed to reassess the prognostic value of cytogenetic abnormalities in a large series of 617 adult patients with Ph-negative BCP-ALL (median age, 38 years), treated in the intensified Group for Research on Adult Acute Lymphoblastic Leukemia (GRAALL)-2003/2005 trials. Combined data from karyotype, DNA index, fluorescence in situ hybridization, and polymerase chain reaction screening for relevant abnormalities were centrally reviewed and were informative in 542 cases (88%), allowing classification in 10 exclusive primary cytogenetic subgroups and in secondary subgroups, including complex and monosomal karyotypes. Prognostic analyses focused on cumulative incidence of failure (including primary refractoriness and relapse), event-free survival, and overall survival. Only 2 subgroups, namely t(4;11)/KMT2A-AFF1 and 14q32/IGH translocations, displayed a significantly worse outcome in this context, still observed after adjustment for age and after censoring patients who received allogeneic stem cell transplantation (SCT) in first remission at SCT time. A worse outcome was also observed in patients with low hypodiploidy/near triploidy, but this was likely related to their higher age and worse tolerance to therapy. The other cytogenetic abnormalities, including complex and monosomal karyotypes, had no prognostic value in these intensive protocols designed for adult patients up to the age of 60 years.
Introduction
B-cell precursor (BCP) acute lymphoblastic leukemia (ALL) is a genetically heterogeneous disease.1 The revised World Health Organization classification recognizes multiple BCP-ALL subtypes according to recurrent genetic abnormalities.2,3 These abnormalities have been established as independent prognostic factors, mainly in children.4-7 In adults, the t(9;22)(q34;q11)/BCR-ABL1 translocation (Philadelphia chromosome [Ph]), rarely observed in children, is the most prevalent subtype, occurring in about one third of all BCP-ALL cases and now differently treated with the addition of a tyrosine kinase inhibitor.8-14 The prognostic value of other subtypes is more difficult to establish because of their rarity.15-22 In the largest United Kingdom ALL (UKALL)/Eastern Cooperative Oncology Group (ECOG) study, t(4;11)(q21;q23), t(8;14)(q24;q32), low hypodiploidy/near triploidy (Ho-Tr) and a complex karyotype were associated with a shorter event-free survival (EFS), whereas patients with high hyperdiploidy (HeH) had a better outcome.21 The poor prognosis associated with complex karyotypes nevertheless remains a matter of debate.23,24 Among patients not entering any recurrent subtype, a Ph-like subgroup associated with a poor prognosis and harboring various kinase activating alterations has been recently identified, emphasizing the value of a precise cytogenetic assignment of all BCP-ALL patients.23,25
In this study, we aimed to reassess the prognostic value of the main cytogenetic abnormalities of Ph-negative BCP-ALL in a large series of adult patients treated in the intensive Group for Research on Adult Acute Lymphoblastic Leukemia (GRAALL) 2003 and 2005 trials.
Patients and methods
The GRAALL 2003 and 2005 trials
The GRAALL 2003 and 2005 trials were conducted in 70 centers in France, Belgium, and Switzerland. Protocols are detailed in a supplemental Protocol File (found online on the Blood Web site). Results of the GRAALL-2003 trial (ClinicalTrial.gov, NCT00222027) have already been reported.26 The subsequent GRAALL-2005 trial (ClinicalTrial.gov, NCT00327678) included the addition of a randomized evaluation of (1) hyperfractionated cyclophosphamide during induction and late intensification27 and (2) rituximab during all phases of therapy in CD20-positive BCP-ALL patients.28 Allogeneic stem cell transplantation (SCT) was offered in first remission to patients ages 55 years old or younger presenting at least 1 conventional ALL high-risk factor, as listed below. Informed consent was obtained from all patients at trial entry. Both trials were conducted in accordance with the Declaration of Helsinki and approved by research ethics committees. Between 2003 and 2011, 955 patients ages 15- to 59-years-old with newly diagnosed Ph-negative ALL were enrolled in these GRAALL 2003/2005 trials, including 617 BCP-ALL (149 GRAALL-2003, 468 GRAALL-2005) with a median age of 38 years.
BCP-ALL risk classification
Conventional high-risk factors included (a) white blood cell count (WBC) ≥30 × 109/L; (b) central nervous system (CNS) involvement; (c) KMT2A (MLL) gene rearrangement, defined as t(4;11)(q21;q23), KMT2A-AFF1, or both fusion transcripts, or another KMT2A rearrangement; (d) t(1;19)(q23;p13), TCF3-PBX1, or both fusion transcripts; (e) Ho-Tr on karyotype or DNA index analysis; (f) early resistance to steroid prephase, defined as a peripheral-blood (PB) blast cell count higher than 1.0 × 109/L after the prephase; (g) poor early bone marrow (BM) blast clearance, defined by BM morphological blast cell percentage higher than 5% after the first week of induction chemotherapy; (h) late complete remission (CR), requiring the planned salvage course to reach CR; and (i) immunoglobin T-cell receptor minimal residual disease level ≥ 10−2 after the first induction course in the GRAALL-2003 trial only. Two other factors were introduced in the GRAALL-2005 trial only: (a) complex karyotype, according to Moorman et al,21 and (b) CD10-negative pro-B immunophenotype.
Cytogenetic analysis and study population
Pretreatment diagnosis BM or PB samples were cultured and analyzed by standard cytogenetic methods at local laboratories. Fluorescence in situ hybridization (FISH) was performed on cytogenetic preparations using commercially available probes. Karyotypes were centrally reviewed in annual workshops and reported according to the International System of Cytogenetic Nomenclature.29 Absence of clonal abnormality was classified as a normal karyotype or karyotype failure, depending on the number of metaphases analyzed: at least 20, or less than 20, respectively. Reverse transcriptase polymerase chain reaction (RT-PCR) analyses were performed locally or referred to other laboratories of the group. All patients were screened for BCR-ABL1 and, if negative, for TCF3-PBX1 and KMT2A-AFF1. In patients younger than 25 years old, screening for the cryptic t(12;21)(p13;q22)/ETV6-RUNX1 rearrangement was performed by RT-PCR, FISH, or both. Whenever possible, DNA content analysis was performed to identify aneuploid cases.30
A primary cytogenetic classification was applied first, adapted from Moorman et al,21 taking into account primary abnormalities identified by karyotype, FISH, DNA index, or RT-PCR. Accordingly, 10 exclusive primary subgroups were considered: (1) t(4;11)(q21;q23)/KMT2A-AFF1; (2) other 11q23 abnormalities with rearranged KMT2A gene; (3) t(1;19)(q23;p13)/TCF3-PBX1; (4) Ho-Tr, including low hypodiploidy with 30 to 39 chromosomes and near triploidy with 60 to 78 chromosomes; (5) HeH with 51 to 65 chromosomes; (6) t(12;21)(p13;q22)/ETV6-RUNX1 translocation; (7) intrachromosomal amplification of chromosome 21 (iAMP21); (8) 14q32/IGH translocations; (9) other abnormalities; and (10) no identified abnormalities (normal karyotype and no abnormality identified by FISH, RT-PCR, or DNA content analyses). Next, a detailed analysis of various secondary anomalies, that is, whole chromosome losses and gains and classical ALL chromosomal deletions (del(6q), del(9p), del(12p), del(13q), del(17p)), was performed. Finally, taking into account primary and secondary abnormalities, complex karyotypes and monosomal karyotypes were identified. Complex karyotypes were defined as cases lacking specific primary abnormalities and presenting with at least 5 chromosomal abnormalities in the 40 to 50 chromosome range, according to Moorman et al.21 Monosomal karyotypes were defined, according to Breems et al,31 as cases presenting with 2 or more distinct autosomal monosomies or a single autosomal monosomy in the presence of a structural clonal abnormality (other than a marker or ring chromosome).
Statistical methods
The primary aim of the study was to investigate how cytogenetic features may interfere with resistance to therapy. With EFS or overall survival (OS) as endpoints, mortality not associated with primary refractoriness or relapse represents an obvious competing and confounding factor. We thus also considered cumulative incidence of failure (CIF), defined as the incidence of either primary ALL resistance or relapse after CR achievement. In CIF evaluation, deaths not associated with primary refractoriness or relapse were considered competing events. Because approximately one third of the patients (198 of 542) received allogeneic SCT in the first CR and numbers of transplanted patients might be relatively low in some cytogenetic subgroups, we primarily performed the analysis without censoring patients transplanted in the first CR. We paid particular attention to correlations between age and cytogenetic abnormalities and performed age adjustments when necessary. We used Cox models for EFS and OS and cause-specific Cox models for CIF comparisons. All statistical tests were 2-sided, with a type I error at 5%. STATA/IC 12.1 (STATA, College Station, TX) was used for all analyses. Hazard ratios (HRs) and cause-specific HRs were given with a 95% confidence interval (CI). The median actuarial follow-up was 5.7 years (6.0 and 5.5 years for the GRAALL 2003 and 2005 trials, respectively).
Results
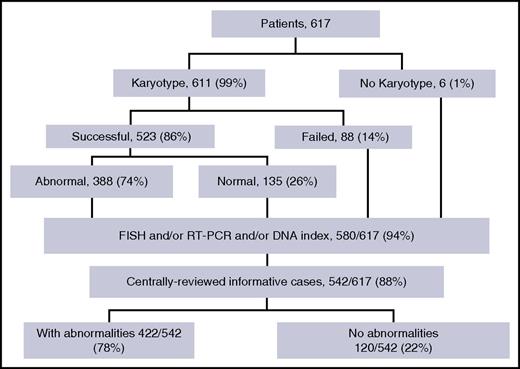
As is presented in the cytogenetics flowchart (Figure 1), karyotyping was undertaken in 611 (99%) of the 617 patients. A successful karyotype was obtained in 523 patients (86%), of whom 388 (74%) had an abnormal karyotype. Among the 6 cases for whom karyotype was not performed and the 88 cases with karyotype failure, 19 cases had informative abnormal data obtained by FISH (n = 9), RT-PCR (n = 6), or DNA content (n = 7) analyses. Therefore, a total of 542 out of 617 patients (88%) had informative cytogenetics. These 542 patients represent the study population, and abnormalities were finally identified in 422 of them (78%). No differences in patient characteristics or outcome were found between the 542 study patients and the remaining 75 patients (not shown). The main and detailed cytogenetic features found in the 542 study patients are shown in Table 1 and supplemental Table 1, respectively, whereas clinical patient characteristics and outcome by cytogenetic subgroup are given in Tables 2 and 3, respectively.
Main cytogenetic characteristics according to cytogenetic subgroups
| Cytogenetic subgroup . | Cases, N . | Abnormal karyotype, N (%) . | Median MNC . | Chromosome gains, N (%) . | Chromosome losses, N (%) . | Structural CA, N (%) . | Deletions,* N (%) . | Additional CA, N (%) . | Median CA, N (range) . | Complex karyotype, N (%) . | Monosomal karyotype, N (%) . |
|---|---|---|---|---|---|---|---|---|---|---|---|
| All patients | 542 | 388 (72) | 46 | 180 (33) | 102 (19) | 338 (62) | 133 (25) | 237 (44) | 1 (1-26) | 27 (5) | 82 (15) |
| Primary subgroups | |||||||||||
| t(4;11)(q21;q23) | 72 | 69 (96) | 46 | 25 (35) | 4 (6) | 72 (100) | 8 (11) | 30 (42) | 1 (1-13) | NA | 4 (6) |
| Other t(v;11q23) | 11 | 9 (82) | 46 | 4 (36) | 1 (9) | 11 (100) | 1 (9) | 5 (45) | 2 (1-11) | NA | 4 (6) |
| t(1;19)(q23;p13) | 28 | 26 (93) | 46 | 5 (18) | 5 (18) | 28 (100) | 13 (46) | 17 (61) | 3 (1-7) | NA | 2 (7) |
| Ho-Tr | 33 | 30 (91) | 36-69 | 6 (18) | 33 (100) | 15 (45) | 1 (3) | 21 (45) | NA | NA | 33 (100) |
| HeH | 36 | 32 (89) | 56 | 36 (100) | 5 (14) | 21 (58) | 9 (25) | 23 (64) | NA | NA | 4 (11) |
| ETV6-RUNX1 | 2 | 1 | 52 | 1 | 0 | 2 | 0 | 1 | 11 | NA | 0 |
| iAMP21 | 3 | 3 | 47 | 3 | 1 | 3 | 0 | 3 | 3 (2-10) | 1 | 1 |
| 14q32/IGH | 27 | 27 (100) | 46 | 16 (59) | 8 (30) | 27 (100) | 10 (37) | 22 (81) | 3 (1-20) | NA | 6 (22) |
| Other abnormalities | 210 | 191 (91) | 46 | 84 (40) | 46 (22) | 159 (76) | 91 (43) | 115 (55) | 2 (1-26) | 26 (12) | 31 (15) |
| Secondary subgroups | |||||||||||
| Complex karyotypes | 27 | 27 (100) | 46 | 23 (85) | 21 (78) | 26 (96) | 18 (67) | 27 (100) | 6 (5-12) | 27 (100) | 21 (78) |
| Monosomal karyotypes | 82† | 79 (96) | 46 | 47 (57) | 82 (100) | 61 (74) | 30 (37) | 79 (96) | 6 (2-26) | 21 (26) | 82 (100) |
| Cytogenetic subgroup . | Cases, N . | Abnormal karyotype, N (%) . | Median MNC . | Chromosome gains, N (%) . | Chromosome losses, N (%) . | Structural CA, N (%) . | Deletions,* N (%) . | Additional CA, N (%) . | Median CA, N (range) . | Complex karyotype, N (%) . | Monosomal karyotype, N (%) . |
|---|---|---|---|---|---|---|---|---|---|---|---|
| All patients | 542 | 388 (72) | 46 | 180 (33) | 102 (19) | 338 (62) | 133 (25) | 237 (44) | 1 (1-26) | 27 (5) | 82 (15) |
| Primary subgroups | |||||||||||
| t(4;11)(q21;q23) | 72 | 69 (96) | 46 | 25 (35) | 4 (6) | 72 (100) | 8 (11) | 30 (42) | 1 (1-13) | NA | 4 (6) |
| Other t(v;11q23) | 11 | 9 (82) | 46 | 4 (36) | 1 (9) | 11 (100) | 1 (9) | 5 (45) | 2 (1-11) | NA | 4 (6) |
| t(1;19)(q23;p13) | 28 | 26 (93) | 46 | 5 (18) | 5 (18) | 28 (100) | 13 (46) | 17 (61) | 3 (1-7) | NA | 2 (7) |
| Ho-Tr | 33 | 30 (91) | 36-69 | 6 (18) | 33 (100) | 15 (45) | 1 (3) | 21 (45) | NA | NA | 33 (100) |
| HeH | 36 | 32 (89) | 56 | 36 (100) | 5 (14) | 21 (58) | 9 (25) | 23 (64) | NA | NA | 4 (11) |
| ETV6-RUNX1 | 2 | 1 | 52 | 1 | 0 | 2 | 0 | 1 | 11 | NA | 0 |
| iAMP21 | 3 | 3 | 47 | 3 | 1 | 3 | 0 | 3 | 3 (2-10) | 1 | 1 |
| 14q32/IGH | 27 | 27 (100) | 46 | 16 (59) | 8 (30) | 27 (100) | 10 (37) | 22 (81) | 3 (1-20) | NA | 6 (22) |
| Other abnormalities | 210 | 191 (91) | 46 | 84 (40) | 46 (22) | 159 (76) | 91 (43) | 115 (55) | 2 (1-26) | 26 (12) | 31 (15) |
| Secondary subgroups | |||||||||||
| Complex karyotypes | 27 | 27 (100) | 46 | 23 (85) | 21 (78) | 26 (96) | 18 (67) | 27 (100) | 6 (5-12) | 27 (100) | 21 (78) |
| Monosomal karyotypes | 82† | 79 (96) | 46 | 47 (57) | 82 (100) | 61 (74) | 30 (37) | 79 (96) | 6 (2-26) | 21 (26) | 82 (100) |
CA, cytogenetic abnormality.
Classical ALL deletions: del(6q), del(9p), del(12p), del(13q), and del(17p).
Including 3 Ho-Tr cases identified by DNA index and FISH analyses.
Main clinical characteristics according to cytogenetic subgroups
| Cytogenetic subgroup . | Patients, N (%) . | Males, N (%) . | Median age, y (range) . | Age 15-24 y, N (%) . | Age 25-44 y, N (%) . | Age 45-59 y, N (%) . | Median WBC 109/L (range) . | WBC >30 × 109/L, N (%) . | CNS+, N (%) . | CD10-negative, N (%) . |
|---|---|---|---|---|---|---|---|---|---|---|
| All patients | 542 | 302 (56) | 38 (15-59) | 151 (28) | 206 (38) | 185 (34) | 8.21 (0.4-474) | 142 (26) | 24 (4) | 165 (32) |
| Primary subgroups† | ||||||||||
| t(4;11)(q21;q23) | 72 (13) | 34 (47) | 41 (18-59) | 15 | 36 | 21 | 113.3 (3.8-474)** | 55 (76)** | 1 | 62 (96)** |
| Other t(v;11q23) | 11 (2) | 7 (64) | 31 (21-53) | 3 | 5 | 3 | 32.9 (1.6-142)** | 7 (64)** | 2 | 9 (82)** |
| t(1;19)(q23;p13) | 28 (5) | 11 (39) | 36 (18-59) | 7 | 14 | 7 | 13 (2.6-396)* | 5 (18) | 4 | 0 (0)** |
| Ho-Tr | 33 (6) | 18 (55) | 53 (22-59)** | 1 | 4 | 28 (85)** | 6.4 (0.8-134) | 1 (3) | 0 | 17 (55)** |
| HeH | 36 (7) | 25 (69) | 30 (17-59) | 15 | 10 | 11 | 2.8 (0.7-62)** | 1 (3)* | 1 | 5 (14) |
| ETV6-RUNX1 | 2 (0.4) | 2 | 20 (19-20) | 2 | 0 | 0 | 8.5 (6.9-10) | 0 (0) | 0 | 0 (0) |
| iAMP21 | 3 (0.5) | 3 | 20 (18-20)* | 3 | 0 | 0 | 3.7 (1.7-25) | 0 (0) | 0 | 0 (0) |
| 14q32/IGH | 27 (5) | 16 (59) | 43 (18-59) | 5 | 11 | 11 | 7.5 (1.3-262) | 4 (15) | 2 | 3 (12) |
| Other abnormalities | 210 (39) | 119 (57) | 34 (15-59) | 63 | 79 | 68 | 6.9 (0.4-343) | 50 (24) | 9 | 40 (20) |
| No identified abnormalities | 120 (22) | 68 (57) | 37 (16-59) | 37 | 47 | 36 | 5.6 (0.5-342) | 19 (16) | 5 | 29 (25) |
| Secondary abnormalities‡ | ||||||||||
| del(6q) | 23/492 (5) | 12 (52) | 40 (18-58) | 5 | 9 | 9 | 12.9 (1.5-396) | 9 (39) | 3 | 5 (22) |
| del(7p) | 29/494 (6) | 19 (66) | 42 (18-59) | 6 | 11 | 12 | 8.7 (1.2-438) | 10 (34) | 1 | 6 (22) |
| Monosomy 7 | 53/510 (10) | 30 (60) | 49 (18-59)** | 7 | 11 | 35 (66)** | 5.2 (0.7-45)** | 2 (4)** | 0 | 19 (38) |
| del(9p) | 67/493 (14) | 37 (55) | 35 (18-59) | 22 | 21 | 24 | 7.9 (0.7-396) | 16 (24) | 5 | 6 (9)** |
| Monosomy 9 | 39/511 (8) | 22 (56) | 53 (18-59)** | 4 | 7 | 28 (74)** | 8.2 (0.9-222) | 4 (10)* | 0 | 17 (46) |
| del(13q) | 15/494 (3) | 10 (66) | 42 (19-58) | 4 | 5 | 6 | 7.3 (1-159) | 3 (20) | 2 | 2 (13) |
| Monosomy 13 | 32/506 (6) | 16 (50) | 51 (19-59)** | 3 | 5 | 24 (75)** | 6.7 (0.7-142)* | 2 (6)** | 0 | 13 (43) |
| del(17p) | 11/493 (2) | 6 (55) | 33 (19-49) | 3 | 4 | 4 | 6.6 (1.2-124) | 3 (27) | 0 | 4 (40) |
| Monosomy 17 | 36/508 (7) | 21 (58) | 53 (19-59)** | 2 | 5 | 29 (81)** | 6.5 (0.8-57)** | 1 (3)** | 0 | 13 (68) |
| Complex karyotype | 27/527 (5) | 19 (70) | 33 (18-59) | 10 | 9 | 8 | 6.6 (1.1-222) | 2 (7)* | 0 | 5 (19) |
| Monosomal karyotype | 82/518 (16) | 47 (57) | 48 (18-59)** | 15 | 17 | 50 (61)** | 6.7 (0.7-230)** | 8 (10)** | 0* | 27 (35) |
| Cytogenetic subgroup . | Patients, N (%) . | Males, N (%) . | Median age, y (range) . | Age 15-24 y, N (%) . | Age 25-44 y, N (%) . | Age 45-59 y, N (%) . | Median WBC 109/L (range) . | WBC >30 × 109/L, N (%) . | CNS+, N (%) . | CD10-negative, N (%) . |
|---|---|---|---|---|---|---|---|---|---|---|
| All patients | 542 | 302 (56) | 38 (15-59) | 151 (28) | 206 (38) | 185 (34) | 8.21 (0.4-474) | 142 (26) | 24 (4) | 165 (32) |
| Primary subgroups† | ||||||||||
| t(4;11)(q21;q23) | 72 (13) | 34 (47) | 41 (18-59) | 15 | 36 | 21 | 113.3 (3.8-474)** | 55 (76)** | 1 | 62 (96)** |
| Other t(v;11q23) | 11 (2) | 7 (64) | 31 (21-53) | 3 | 5 | 3 | 32.9 (1.6-142)** | 7 (64)** | 2 | 9 (82)** |
| t(1;19)(q23;p13) | 28 (5) | 11 (39) | 36 (18-59) | 7 | 14 | 7 | 13 (2.6-396)* | 5 (18) | 4 | 0 (0)** |
| Ho-Tr | 33 (6) | 18 (55) | 53 (22-59)** | 1 | 4 | 28 (85)** | 6.4 (0.8-134) | 1 (3) | 0 | 17 (55)** |
| HeH | 36 (7) | 25 (69) | 30 (17-59) | 15 | 10 | 11 | 2.8 (0.7-62)** | 1 (3)* | 1 | 5 (14) |
| ETV6-RUNX1 | 2 (0.4) | 2 | 20 (19-20) | 2 | 0 | 0 | 8.5 (6.9-10) | 0 (0) | 0 | 0 (0) |
| iAMP21 | 3 (0.5) | 3 | 20 (18-20)* | 3 | 0 | 0 | 3.7 (1.7-25) | 0 (0) | 0 | 0 (0) |
| 14q32/IGH | 27 (5) | 16 (59) | 43 (18-59) | 5 | 11 | 11 | 7.5 (1.3-262) | 4 (15) | 2 | 3 (12) |
| Other abnormalities | 210 (39) | 119 (57) | 34 (15-59) | 63 | 79 | 68 | 6.9 (0.4-343) | 50 (24) | 9 | 40 (20) |
| No identified abnormalities | 120 (22) | 68 (57) | 37 (16-59) | 37 | 47 | 36 | 5.6 (0.5-342) | 19 (16) | 5 | 29 (25) |
| Secondary abnormalities‡ | ||||||||||
| del(6q) | 23/492 (5) | 12 (52) | 40 (18-58) | 5 | 9 | 9 | 12.9 (1.5-396) | 9 (39) | 3 | 5 (22) |
| del(7p) | 29/494 (6) | 19 (66) | 42 (18-59) | 6 | 11 | 12 | 8.7 (1.2-438) | 10 (34) | 1 | 6 (22) |
| Monosomy 7 | 53/510 (10) | 30 (60) | 49 (18-59)** | 7 | 11 | 35 (66)** | 5.2 (0.7-45)** | 2 (4)** | 0 | 19 (38) |
| del(9p) | 67/493 (14) | 37 (55) | 35 (18-59) | 22 | 21 | 24 | 7.9 (0.7-396) | 16 (24) | 5 | 6 (9)** |
| Monosomy 9 | 39/511 (8) | 22 (56) | 53 (18-59)** | 4 | 7 | 28 (74)** | 8.2 (0.9-222) | 4 (10)* | 0 | 17 (46) |
| del(13q) | 15/494 (3) | 10 (66) | 42 (19-58) | 4 | 5 | 6 | 7.3 (1-159) | 3 (20) | 2 | 2 (13) |
| Monosomy 13 | 32/506 (6) | 16 (50) | 51 (19-59)** | 3 | 5 | 24 (75)** | 6.7 (0.7-142)* | 2 (6)** | 0 | 13 (43) |
| del(17p) | 11/493 (2) | 6 (55) | 33 (19-49) | 3 | 4 | 4 | 6.6 (1.2-124) | 3 (27) | 0 | 4 (40) |
| Monosomy 17 | 36/508 (7) | 21 (58) | 53 (19-59)** | 2 | 5 | 29 (81)** | 6.5 (0.8-57)** | 1 (3)** | 0 | 13 (68) |
| Complex karyotype | 27/527 (5) | 19 (70) | 33 (18-59) | 10 | 9 | 8 | 6.6 (1.1-222) | 2 (7)* | 0 | 5 (19) |
| Monosomal karyotype | 82/518 (16) | 47 (57) | 48 (18-59)** | 15 | 17 | 50 (61)** | 6.7 (0.7-230)** | 8 (10)** | 0* | 27 (35) |
*P < .05; **P < .01.
Comparisons were performed using the subgroup of patients with no identified abnormality as control.
Comparisons were performed in the entire populations of evaluable patients for each secondary abnormality.
Patient outcome according to cytogenetic subgroups
| . | . | CIF . | EFS . | OS . | ||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Cytogenetic subgroup . | Patients . | 5-y estimates (95% CI) . | SHR (95% CI) . | P . | 5-y estimates (95% CI) . | HR (95% CI) . | P . | 5-y estimates (95% CI) . | HR (95% CI) . | P . |
| All patients | 542 | |||||||||
| Primary subgroups† | N (%) | |||||||||
| t(4;11)(q21;q23) | 72 (13) | 37.6% (28-50) | 1.13 (1.01-1.27) | .040 | 42.8% (31-54) | 1.12 (1.01-1.24) | .027 | 45.7% (34-57) | 1.17 (1.05-1.30) | .005 |
| Other t(v;11q23) | 11 (2) | 18.2% (5-55) | 0.90 (0.68-1.20) | .48 | 63.6% (30-85) | 0.96 (0.79-1.18) | .73 | 72.7% (37-90) | 0.93 (0.74-1.18) | .57 |
| t(1;19)(q23;p13) | 28 (5) | 28.6% (15-49) | 1.13 (0.91-1.40) | .28 | 53.6% (34-70) | 1.05 (0.86-1.29) | .63 | 57.1% (37-73) | 1.09 (0.88-1.35) | .43 |
| Ho-Tr | 33 (6) | 36.4% (23-55) | 1.45 (0.79-2.64) | .23 | 35.8% (20-52) | 1.81 (1.10-2.98) | .020 | 38.6% (22-55) | 2.06 (1.21-3.51) | .007 |
| HeH | 36 (7) | 22.9% (12-41) | 1.00 (0.71-1.40) | .99 | 63.0% (45-77) | 0.88 (0.65-1.20) | .42 | 72.0% (54-84) | 0.93 (0.68-1.28) | .67 |
| ETV6-RUNX1 | 2 (0.3) | — | NT | — | — | NT | — | — | NT | — |
| iAMP21 | 3 (0.5) | — | NT | — | — | NT | — | — | NT | — |
| 14q32/IGH | 27 (5) | 41.2% (25-62) | 1.06 (0.98-1.15) | .12 | 19.7% (7-37) | 1.13 (1.06-1.20) | <.001 | 30.6% (14-49) | 1.13 (1.06-1.21) | <.001 |
| Other abnormalities | 210 (39) | 31.8% (26-37) | 1.02 (0.98-1.06) | .28 | 48.2% (41-55) | 1.02 (0.99-1.06) | .16 | 53.0% (46-60) | 1.03 (0.99-1.07) | .07 |
| No identified abnormalities | 120 (22) | 22.6% (16-31) | Control | — | 60.3% (51-69) | Control | — | 63.2% (54-71) | Control | — |
| Secondary abnormalities‡ | N/N evaluable (%) | |||||||||
| del(6q) | 23/492 (5) | — | 0.96 (0.47-1.95) | .91 | — | 0.82 (0.44-1.55) | .55 | — | 0.94 (0.50-1.77) | .84 |
| del(7p) | 29/494 (6) | — | 0.84 (0.43-1.64) | .60 | — | 0.89 (0.51-1.56) | .69 | — | 1.02 (0.58-1.79) | .94 |
| Monosomy 7 | 53/510 (10) | — | 1.00 (0.63-1.57) | .99 | — | 1.18 (0.81-1.73) | .39 | — | 1.23 (0.82-1.83) | .32 |
| del(9p) | 67/493 (14) | — | 1.10 (0.73-1.66) | .65 | — | 1.05 (0.74-1.51) | .78 | — | 0.86 (0.57-1.28) | .46 |
| Monosomy 9 | 39/511 (8) | — | 1.07 (0.65-1.76) | .80 | — | 1.48 (0.99-2.23) | .056 | — | 1.62 (1.06-2.47) | .026 |
| del(13q) | 15/494 (3) | — | 0.51 (0.16-1.60) | .25 | — | 0.53 (0.22-1.29) | .17 | — | 0.50 (0.18-1.34) | .17 |
| Monosomy 13 | 32/506 (6) | — | 0.99 (0.56-1.74) | .97 | — | 1.38 (0.87-2.18) | .16 | — | 1.55 (0.97-2.48) | .07 |
| del(17p) | 11/493 (2) | — | 1.98 (0.88-4.47) | .10 | — | 1.71 (0.81-3.63) | .16 | — | 1.79 (0;84-3.81) | .13 |
| Monosomy 17 | 36/508 (7) | — | 1.01 (0.60-1.69) | .97 | — | 1.53 (0.01-2.31) | .043 | — | 1.69 (1.10-2.61) | .016 |
| Complex karyotype | 27/527 (5) | 18.5% (8-39) | 0.53 (0.24-1.20) | .13 | 59.0% (38-75) | 0.72 (0.39-1.31) | .28 | 62.7% (42-78) | 0.74 (0.39-1.39) | .35 |
| Monosomal karyotype | 82/518 (16) | 33.3% (24-45) | 1.22 (0.85-1.75) | .27 | 40.4% (30-51) | 1.36 (0.996-1.85) | .053 | 45.7% (35-56) | 1.38 (0.995-1.92) | .054 |
| . | . | CIF . | EFS . | OS . | ||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Cytogenetic subgroup . | Patients . | 5-y estimates (95% CI) . | SHR (95% CI) . | P . | 5-y estimates (95% CI) . | HR (95% CI) . | P . | 5-y estimates (95% CI) . | HR (95% CI) . | P . |
| All patients | 542 | |||||||||
| Primary subgroups† | N (%) | |||||||||
| t(4;11)(q21;q23) | 72 (13) | 37.6% (28-50) | 1.13 (1.01-1.27) | .040 | 42.8% (31-54) | 1.12 (1.01-1.24) | .027 | 45.7% (34-57) | 1.17 (1.05-1.30) | .005 |
| Other t(v;11q23) | 11 (2) | 18.2% (5-55) | 0.90 (0.68-1.20) | .48 | 63.6% (30-85) | 0.96 (0.79-1.18) | .73 | 72.7% (37-90) | 0.93 (0.74-1.18) | .57 |
| t(1;19)(q23;p13) | 28 (5) | 28.6% (15-49) | 1.13 (0.91-1.40) | .28 | 53.6% (34-70) | 1.05 (0.86-1.29) | .63 | 57.1% (37-73) | 1.09 (0.88-1.35) | .43 |
| Ho-Tr | 33 (6) | 36.4% (23-55) | 1.45 (0.79-2.64) | .23 | 35.8% (20-52) | 1.81 (1.10-2.98) | .020 | 38.6% (22-55) | 2.06 (1.21-3.51) | .007 |
| HeH | 36 (7) | 22.9% (12-41) | 1.00 (0.71-1.40) | .99 | 63.0% (45-77) | 0.88 (0.65-1.20) | .42 | 72.0% (54-84) | 0.93 (0.68-1.28) | .67 |
| ETV6-RUNX1 | 2 (0.3) | — | NT | — | — | NT | — | — | NT | — |
| iAMP21 | 3 (0.5) | — | NT | — | — | NT | — | — | NT | — |
| 14q32/IGH | 27 (5) | 41.2% (25-62) | 1.06 (0.98-1.15) | .12 | 19.7% (7-37) | 1.13 (1.06-1.20) | <.001 | 30.6% (14-49) | 1.13 (1.06-1.21) | <.001 |
| Other abnormalities | 210 (39) | 31.8% (26-37) | 1.02 (0.98-1.06) | .28 | 48.2% (41-55) | 1.02 (0.99-1.06) | .16 | 53.0% (46-60) | 1.03 (0.99-1.07) | .07 |
| No identified abnormalities | 120 (22) | 22.6% (16-31) | Control | — | 60.3% (51-69) | Control | — | 63.2% (54-71) | Control | — |
| Secondary abnormalities‡ | N/N evaluable (%) | |||||||||
| del(6q) | 23/492 (5) | — | 0.96 (0.47-1.95) | .91 | — | 0.82 (0.44-1.55) | .55 | — | 0.94 (0.50-1.77) | .84 |
| del(7p) | 29/494 (6) | — | 0.84 (0.43-1.64) | .60 | — | 0.89 (0.51-1.56) | .69 | — | 1.02 (0.58-1.79) | .94 |
| Monosomy 7 | 53/510 (10) | — | 1.00 (0.63-1.57) | .99 | — | 1.18 (0.81-1.73) | .39 | — | 1.23 (0.82-1.83) | .32 |
| del(9p) | 67/493 (14) | — | 1.10 (0.73-1.66) | .65 | — | 1.05 (0.74-1.51) | .78 | — | 0.86 (0.57-1.28) | .46 |
| Monosomy 9 | 39/511 (8) | — | 1.07 (0.65-1.76) | .80 | — | 1.48 (0.99-2.23) | .056 | — | 1.62 (1.06-2.47) | .026 |
| del(13q) | 15/494 (3) | — | 0.51 (0.16-1.60) | .25 | — | 0.53 (0.22-1.29) | .17 | — | 0.50 (0.18-1.34) | .17 |
| Monosomy 13 | 32/506 (6) | — | 0.99 (0.56-1.74) | .97 | — | 1.38 (0.87-2.18) | .16 | — | 1.55 (0.97-2.48) | .07 |
| del(17p) | 11/493 (2) | — | 1.98 (0.88-4.47) | .10 | — | 1.71 (0.81-3.63) | .16 | — | 1.79 (0;84-3.81) | .13 |
| Monosomy 17 | 36/508 (7) | — | 1.01 (0.60-1.69) | .97 | — | 1.53 (0.01-2.31) | .043 | — | 1.69 (1.10-2.61) | .016 |
| Complex karyotype | 27/527 (5) | 18.5% (8-39) | 0.53 (0.24-1.20) | .13 | 59.0% (38-75) | 0.72 (0.39-1.31) | .28 | 62.7% (42-78) | 0.74 (0.39-1.39) | .35 |
| Monosomal karyotype | 82/518 (16) | 33.3% (24-45) | 1.22 (0.85-1.75) | .27 | 40.4% (30-51) | 1.36 (0.996-1.85) | .053 | 45.7% (35-56) | 1.38 (0.995-1.92) | .054 |
NT, not tested; SHR, cause-specific hazard ratios.
Comparisons were performed using the subgroup of patients with no identified abnormality as control.
Comparisons were performed in the entire populations of evaluable patients for each secondary abnormality.
Primary cytogenetic classification
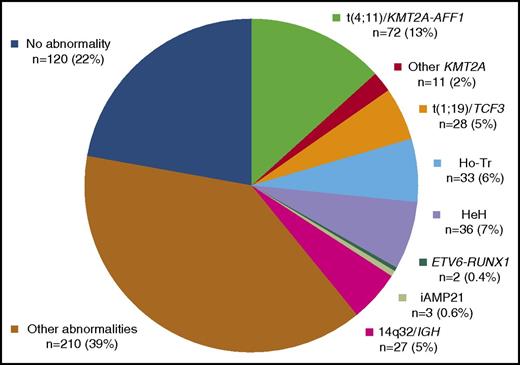
Distribution of the 542 study patients into the 10 primary cytogenetic subgroups is shown in Figure 2. Detailed age distributions are given in supplemental Figure 1.
t(4;11)(q21;q23)/KMT2A-AFF1 translocation was the most prevalent primary subgroup (n = 72, 13%). Cases were mainly diagnosed by conventional cytogenetics (n = 69). Three cases with karyotype failure were diagnosed by RT-PCR and confirmed by KMT2A FISH analyses in 1 tested case. The median modal number of chromosomes (MNC) was 46 (range, 46-102). Few additional clonal abnormalities (ACAs) were present, the most prevalent one being a gain of chromosome X. As was expected, these patients had significantly higher WBC, and their leukemic blasts for the most part did not express CD10 (Table 2).
Other 11q23 abnormalities with a rearranged KMT2A gene were found in 11 patients (2%), mostly identified by karyotype (n = 10), and 1 KMT2A-AFF1 negative case with a normal karyotype was diagnosed by FISH on interphase nuclei. The most frequently identified chromosome partner was 19p13 (n = 6), more precisely 19p13.3 (ie, location of the MLLT1/ENL gene). Other identified chromosome partners were 5q31, 6p21, 11q25, and 17q23 (1 case each). The median MNC was 46 (range, 46-59), and few ACA were present. Again, these patients had higher WBC and mostly lacked CD10 antigen expression.
t(1;19)(q23;p13)/TCF3-PBX1 translocation was diagnosed in 28 patients (5%), mostly identified by karyotype (n = 26). TCF3 involvement was confirmed by RT-PCR (n = 19), FISH (n = 7), or both (n = 2) in the 24 cases tested. The 2 remaining cases were diagnosed by RT-PCR after karyotype failure. The median MNC was 46 (range, 41-91). The unbalanced der(19)t(1;19) form of this translocation was prevalent (n = 20), leading to trisomy 1q. ACAs were present in 17 cases, with a median of 2 ACAs per case, mainly as classical deletions (n = 13); the most prevalent of these was a deletion of 9p (n = 9), mainly resulting from an isochromosome 9q (n = 5). Interestingly, trisomy 1q was observed in 22 out of 26 cases, either due to the unbalanced der(19)t(1;19) or due to another unbalanced 1q translocation (n = 6); this led to tetrasomy 1q in 5 cases. A higher rate of CNS involvement was observed in this subgroup, and all cases expressed the CD10 antigen.
Low hypodiploidy/near triploidy (Ho-Tr) was identified in 33 patients (6%). The median MNCs were 36 (range, 33-40) and 69 (range, 58-75) for the hypodiploid and the hypotriploid clones, respectively. In the 4 cases falling outside the classical 30 to 39 and 60 to 78 ranges, the diagnosis was established according to the chromosomal profile, the presence of a second clone (either hypodiploid or hypotriploid), or both on karyotype, FISH, or DNA content analyses (supplemental Table 1). Thirty cases were diagnosed by karyotype as an isolated hypodiploid (n = 9) or hypotriploid (n = 11) clone or a combination of both (n = 10). DNA content analysis allowed the identification of 3 cases with a normal karyotype, because they presented with a hypodiploid DNA content peak (range, 0.67-0.78), associated in 2 cases with an additional hypotriploid DNA content peak (1.35 and 1.54, respectively) corresponding to the duplication of the hypodiploid clone. Furthermore, characteristic chromosome losses such as monosomy 7 were confirmed by FISH in these 2 cases. Considering that the near-triploid clone is instead a low-hypotetraploid clone resulting from the duplication of a hypodiploid clone, as was recently proposed,32 all patients had monosomy 7, either as 1 copy in the hypodiploid clone or as 2 copies in the hypotetraploid clone. Other prevalent monosomies were monosomies 3, 13, 9, 15, 16, 4, and 17 by decreasing order of frequency (supplemental Table 2 and supplemental Figure 2). Additional structural abnormalities were identified in 15 out of 28 evaluable Ho-Tr cases, but classical ALL deletions such as del(9p) were rarely observed. Interestingly, patients with Ho-Tr ALL were significantly older (median age, 53 years) and tended to have lower WBC, and in more than half of the cases, their blasts did not express CD10.
High hyperdiploidy (HeH) was diagnosed in 36 patients (7%), mainly by karyotype (n = 32), with a median MNC of 56 (range, 51-63). Four cases were identified by DNA content analysis (range, 1.21 to 1.4). The 32 cases with abnormal karyotypes could be subclassified into 2 subgroups according to differences in the profile of chromosomal gains (supplemental Table 2 and supplemental Figure 3): (1) a typical HeH subgroup (n = 15), with the characteristic childhood BCP-ALL HeH profile, harboring gains of chromosomes 4, 10, 17, or 18; and (2) an atypical HeH subgroup (n = 17), mainly lacking these “typical” gains and harboring “atypical” HeH gains such as gains of chromosomes 1, 3, 5, 11, 12, 13, 15, or 22. A triple combination of trisomies 4, 10, and 17 was found only in typical HeH cases (n = 8), all presenting with trisomy 18 and high MNC (median, 58; range, 54-59). Both subgroups harbored prevalent gains of chromosomes X and 21. Atypical cases did not differ from typical cases in median MNC (56 vs 57), but in 3 of 17 cases the HeH clone resulted from the clonal evolution of a non-HeH clone. Atypical cases tended to have more frequent structural ACAs (14 of 17 vs 7 of 14). Interestingly, these 2 subgroups also seemed to differ in the gender distribution and median age. Typical HeH was predominantly observed in younger males (median age, 29 years), whereas patients with atypical HeH were older (median age, 45 years).
t(12;21)(p13;q22)/ETV6-RUNX1 translocation was diagnosed after a total of 232 patients were screened by RT-PCR, FISH, or both for the ETV6-RUNX1 rearrangement, including 106 of the 180 patients below the age of 25 years. Only 2 positive cases were found (age, 19 and 20 years), confirming the rarity of this entity in adult patients.
Intrachromosomal amplification of chromosome 21 (iAMP21) were identified after a total of 105 patients were screened by FISH for the iAMP21 abnormality, including 61 of the 180 patients below the age of 25 years. Three patients, all younger than 21 years, were diagnosed with iAMP21, a rare high-risk entity mostly found in older children.33 They presented with an abnormal chromosome 21 on karyotype and amplified RUNX1 FISH signals on this abnormal chromosome 21. As is described in pediatric cases, they also had low WBC.
14q32/IGH translocations were diagnosed in 27 patients (5%), mostly by karyotype (n = 26) confirmed by IGH FISH in 4 patients. The remaining case with del9p and incomplete karyotype was diagnosed by IGH FISH. The median MNC was 46 (range, 44-60), and most cases (n = 22) presented with ACAs at a high rate (median, 3). The main 14q32 partners were 18q21 (n = 5), 8q11 (n = 4), 19q13 (n = 4), and 8q24 (n = 2), likely corresponding to the partner genes BCL2, MYC, CEBPD, and CEBPA, respectively. Interestingly, 4 cases presented with a karyotype evocative of a “double-hit” lymphoma,34 that is, a 14q32 translocation involving 18q21 (n = 2), 6p21 (n = 1), or an unidentified partner (n = 1) associated with another translocation involving 8q24/MYC in a context of numerous ACAs (median, 7; range, 3-19) and high MNC (median, 48; range, 47-60). These 4 patients tended to be older (median, 51 years; range 43-57) than the whole 14q32 subgroup.
Other various cytogenetic abnormalities were identified in 210 patients (39%), mostly by karyotype (n = 191). Other cases, with karyotype failure (n = 8) or normal karyotypes (n = 11), were diagnosed by FISH (n = 11), DNA content analysis (n = 8), or both. The median MNC was 46 (range, 43-94). Five cases had near tetraploid karyotypes (87-94 chromosomes), which were clonal evolutions of a hypodiploid clone in 2 cases. The median number of cytogenetic abnormalities was 2 (range, 1-26). Among cases with chromosomal gains (n = 84), trisomy 21 and gain of a chromosome X were prevalent. Chromosomal losses were less frequent (n = 46), most often monosomy 7 (n = 19). Structural abnormalities were identified in most cases (n = 159), mainly as classical ALL deletions (n = 91).
The last subgroup of patients, with no identified abnormality, comprised 120 patients (22%).
Secondary cytogenetic classification
Deletions.
Overall, 133 patients (25%) had at least 1 of the main partial chromosome deletions described in ALL, including del(9p) (n = 67), del(12p) (n = 33), del(7p) (n = 29), del(6q) (n = 23), del(13q) (n = 15), and del(17p) (n = 11). These deletions were particularly frequent in the t(1;19)/TCF3-PBX1 and the 14q32/IGH subgroups, as well as in the subgroup with other various abnormalities. No specific clinical characteristics could be assigned to any of these secondary subgroups.
Chromosome losses.
These losses were found in 102 patients (19%), obviously mainly in the Ho-Tr subgroup. After exclusion of Ho-Tr cases (n = 33), monosomy 7 remained the most frequent monosomy (n = 22), followed by monosomies 9 (n = 13) and 17 (n = 11). Overall, patients with monosomies 9, 13, and 17 were older (Table 2), although this was not observed in the case of monosomy 7.
Chromosome gains.
Gains were observed in 180 patients (33%). Among the non-HeH patients, chromosome gains were markedly found in the 14q32/IGH subgroup (n = 16, 59%), and the most frequent gains in non-HeH patients involved chromosomes X (n = 39), 21 (n = 24, mainly as trisomy 21, n = 22), 8 (n = 13), and 5 (n = 13).
Complex karyotypes.
Complex karyotypes were found in 27 patients (5%), 26 belonging to the primary subgroup with other various abnormalities and the remaining 1 to the iAMP21 subgroup. In these cases, the median MNC was 46 (range, 42-49) and the median number of chromosomal abnormalities was 6 (range, 5-12). Chromosome gains (mainly of structurally abnormal chromosomes) were observed in 23 cases. Chromosome losses were observed in 21 cases, mainly involving chromosomes 7 and 9, with all of these cases being classified also as monosomal karyotypes. Among structural abnormalities (n = 26), unbalanced translocations were prevalent (n = 20), classical ALL deletions being observed in 18 cases. No specific clinical characteristics were found for these patients, except that they rarely had high WBC.
Monosomal karyotype.
The monosomal karyotype was identified in 82 patients, either due to loss of at least 2 autosomes (n = 57), including all the Ho-Tr cases, or due to loss of only 1 autosome but presence of a structural abnormality (n = 25). The median MNC was 46 (range, 33-94). Monosomy 7 was found in more than half of the monosomal cases (n = 47). After exclusion of Ho-Tr cases, patients with a monosomal karyotype did not differ from other patients in terms of age or WBC.
Correlations with patient outcome
Among the 617 study patients, 563 (91.2%) achieved CR. At 5 years, EFS was estimated at 50.2% (95% CI, 46.1-54.1) and OS at 55.0% (95% CI, 50.1-58.9).
Among the primary cytogenetic subgroups, the CR rate was 88.9% in the t(4;11)/KMT2A-AFF1 subgroup; 100% in the other t(v;11q23)/KMT2A subgroup; 85.7% in the t(1;19)/TCF3-PBX1 subgroup; 90.9%, in the Ho-Tr subgroup; 91.7% in the HeH subgroup; 81.5% in the 14q32/IGH subgroup; 93.3% in the subgroup of patients with other various abnormalities; and 90.8% in patients with no identified abnormality. Table 3 provides specific HRs and HRs for CIF, EFS, and OS for the primary and secondary cytogenetic subgroups, as well as 5-year estimates for the main abnormalities, and Figure 3 illustrates EFS and OS by primary subgroups. Of note, among the HeH subgroup, a trend for a worse outcome was found in the atypical HeH subtype (supplemental Figure 4). Because some abnormalities were considered as high-risk factors orienting patients toward allogeneic SCT in first CR if a donor, we repeated the analysis after censoring the 198 patients transplanted in first CR at SCT time. As is indicated in supplemental Table 3, results were globally similar.
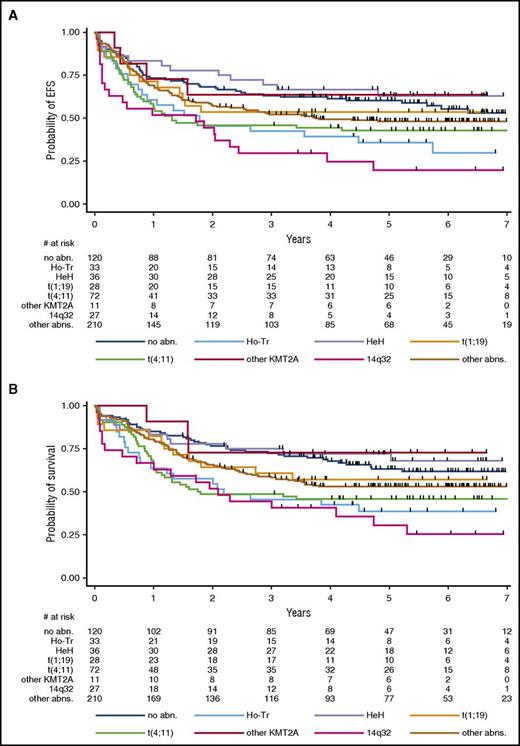
Outcome by primary cytogenetic subgroup. (A) Event-free survival. Five-year EFS estimates are given in Table 3, which also provides hazard ratios and P values. When compared with the control group without identified abnormalities, EFS was significantly shorter in the 3 t(4;11)/KMT2A-AFF1 (P = .027), Ho-Tr (P = .020), and 14q32/IGH (P < .001) subgroups. (B) Overall survival. Five-year OS estimates are given in Table 3, which also provides hazard ratios and P values. When compared with the control group without identified abnormalities, OS was significantly shorter in the 3 t(4;11)/KMT2A-AFF1 (P = .005), Ho-Tr (P = .007), and 14q32/IGH (P < .001) subgroups.
Outcome by primary cytogenetic subgroup. (A) Event-free survival. Five-year EFS estimates are given in Table 3, which also provides hazard ratios and P values. When compared with the control group without identified abnormalities, EFS was significantly shorter in the 3 t(4;11)/KMT2A-AFF1 (P = .027), Ho-Tr (P = .020), and 14q32/IGH (P < .001) subgroups. (B) Overall survival. Five-year OS estimates are given in Table 3, which also provides hazard ratios and P values. When compared with the control group without identified abnormalities, OS was significantly shorter in the 3 t(4;11)/KMT2A-AFF1 (P = .005), Ho-Tr (P = .007), and 14q32/IGH (P < .001) subgroups.
The t(4;11)/KMT2A-AFF1 and 14q32/IGH translocation subgroups were associated with a worse outcome. Patients with Ho-Tr also displayed worse EFS and OS. As is mentioned above, patients with Ho-Tr were significantly older, as were those with monosomy 9 or 17, or those with a monosomal karyotype in whom an impact on EFS and OS without higher CIF was also observed (Table 3). Notably, the prognostic impacts of Ho-Tr, monosomy 9, monosomy 17, and monosomal karyotype did not remain significant after adjustment for age (not shown), suggesting that a higher incidence of deaths unrelated to ALL resistance or recurrence may have played a role in worsening EFS and OS in these patients. For instance, in older patients ages 45 to 59 years old, EFS was not significantly shorter in the 28 Ho-Tr cases in comparison with the 157 non-Ho-Tr cases. Conversely, the worse EFS and OS associated with t(4;11)/KMT2A-AFF1 and 14q32/IGH translocations (Figure 4) were still observed after adjusting for age (HR, 1.12 [1.01-1.24] and 1.12 [1.05-1.20]; P = .031 and .001 for EFS; and HR, 1.16 [1.04-1.30] and 1.12 [1.04-1.20]; P = .007 and .001 for OS, respectively). Finally, it is of note that complex karyotypes did not have a poor prognostic value in this GRAALL cohort.
Overall survival in the t(4;11)/KMT2A-AFF1 and 14q32/IGH subgroups. (A) Without censoring patients who received allogeneic SCT in first CR. At 5 years, OS was estimated at 45.8% (95% CI, 34-57) in the t(4;11)/KMT2A-AFF1 subgroup and at 30.6% (95% CI, 14-49) in the 14q32/IGH subgroup, in comparison with 63.2% (95% CI, 54-71) in the subgroup with no identified cytogenetic abnormality (HR, 1.17 [95% CI, 1.05-1.30] and 1.13 [95% CI, 1.06-1.21]; P = .005 and <.001, respectively). (B) Censoring patients who received allogeneic SCT in first CR at SCT time. At 5 years, OS was estimated at 27.6% (95% CI, 12-46) in the t(4;11)/KMT2A-AFF1 subgroup and at 20.2% (95% CI, 4-45) in the 14q32/IGH subgroup, in comparison with 65.1% (95% CI, 53-75) in the subgroup with no identified cytogenetic abnormality (HR, 1.32 [95% CI, 1.14-1.53] and 1.16 [95% CI, 1.07-1.26]; P < .001 and <.001, respectively).
Overall survival in the t(4;11)/KMT2A-AFF1 and 14q32/IGH subgroups. (A) Without censoring patients who received allogeneic SCT in first CR. At 5 years, OS was estimated at 45.8% (95% CI, 34-57) in the t(4;11)/KMT2A-AFF1 subgroup and at 30.6% (95% CI, 14-49) in the 14q32/IGH subgroup, in comparison with 63.2% (95% CI, 54-71) in the subgroup with no identified cytogenetic abnormality (HR, 1.17 [95% CI, 1.05-1.30] and 1.13 [95% CI, 1.06-1.21]; P = .005 and <.001, respectively). (B) Censoring patients who received allogeneic SCT in first CR at SCT time. At 5 years, OS was estimated at 27.6% (95% CI, 12-46) in the t(4;11)/KMT2A-AFF1 subgroup and at 20.2% (95% CI, 4-45) in the 14q32/IGH subgroup, in comparison with 65.1% (95% CI, 53-75) in the subgroup with no identified cytogenetic abnormality (HR, 1.32 [95% CI, 1.14-1.53] and 1.16 [95% CI, 1.07-1.26]; P < .001 and <.001, respectively).
Discussion
In this study, we analyzed the cytogenetic features of a large multicenter cohort of 617 adult patients with Ph-negative BCP-ALL treated in intensive adult ALL protocols with a median follow-up of more than 5 years. After central review, the proportion of cases with informative cytogenetics was very high (88%). The 2 t(4;11)/KMT2A-AFF1 and 14q32/IGH translocation subgroups were the only ones associated with a worse outcome independent of age.
The t(4;11)/KMT2A-AFF1 translocation was the most prevalent primary abnormality, accounting for 13% of cases. We confirmed its association with known clinical characteristics and its independent value as an indicator of poor prognosis,21,35,36 suggesting the need for novel targeted therapies in this specific subgroup.37-39 Although other KMT2A cases also presented with higher WBC and CD10 negativity, we found that in contrast to what is observed in children,4,5 these adult cases did not seem to share the poor prognosis observed for t(4;11)/KMT2A-AFF1 cases. Conversely, and as reported in numerous adult series,21,40,41 we confirmed that in patients treated with modern intensified protocols, the t(1;19)/TCF3-PBX1 translocation was no longer associated with a poor prognosis.
We also confirmed the unfavorable prognosis associated with 14q32/IGH translocations, as has been reported previously.42,43 It should be noted that this subgroup is actually genetically heterogeneous, with various oncogenes that can be transcriptionally dysregulated by juxtaposition to IGH regulatory elements. In this subgroup, we identified a small subset of older patients harboring 8q24/MYC and 18q21/BCL2 abnormalities, potentially representing the leukemic counterpart of double-hit lymphomas (high-grade mature lymphomas),3,34,44 as has been recently reported in rare cases of adolescents and young adults with BCP-ALL.44 Interestingly, a higher rate of 14q32/IGH cases has been reported through a systematic IGH FISH approach, which can also detect cryptic IGH translocations such as the IGH-CRLF2 rearrangement.43 Although IGH translocations are mainly primary cytogenetic abnormalities,45 the association of IKZF1 gene deletion and Ph-like profile could participate in the poor prognosis of these cases.42,46-48
The relationship between prognosis and patient age is of special interest. For instance, the worse prognosis associated with Ho-Tr cases was likely related to an older age. It should be noted that this relationship with older age has been previously reported,23,25,49,50 even if it was not observed in the UKALL/ECOG trial.21 Conversely, we did not observe the favorable outcome reported for the HeH subgroup as well as the secondary del(9p) subgroup, which in the case of the UKALL/ECOG study could have been related to their younger age.21 Indeed, relatively few young patients were included in our study, because adolescents under the age of 18 were treated in pediatric trials.51 Furthermore, about half of our HeH patients presented with an older age and an atypical pattern of chromosomal gains, especially fewer “good prognosis” trisomies described in children such as trisomy 4, 10, 17, and 1852-55 and a higher incidence of trisomy 5 that has been associated with a worse outcome in childhood56 and Ph-positive BCP-ALL.57 This latter observation emphasizes the need for an accurate cytogenetic analysis of HeH cases52-57 and warrants further differential genomic characterization.58,59
In contrast to the large studies published to date,21,23 we failed to identify any unfavorable prognostic value associated with complex karyotypes, even after censoring patients allografted in first CR. This lack of poor prognosis has also been observed in another risk-adapted trial,24 suggesting that these differences could be related to treatment. On the other hand, conversely to other studies,24,60 we observed marked trends for shorter EFS and OS in patients with monosomal karyotypes, which became significant after SCT censoring, suggesting that SCT might benefit these patients. Monosomy 7, resulting in the entire loss of one IKZF1 allele and previously reported as a poor risk factor in childhood ALL61 and in a smaller series of adult ALL,22 had no prognostic impact in our study.
In conclusion, through the combined use of global and targeted cytogenetic analyses and centralized data reviewing, as has been recommended,62,63 we report here a very high rate of informative cytogenetic results allowing the accurate classification of patients with BCP-ALL and the identification of prognostically relevant patient subgroups. The large “B-other” subgroup of cases with no specific primary cytogenetic abnormality certainly warrants further genomic analyses to identify poor prognosis markers such as an IKZF1 deletion43,64,66,67 as well as Ph-like cases that could eventually benefit from tyrosine kinase inhibitor therapy.46-48,67-72
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors are very grateful to Christine Harrison and Anne Hagemeijer and to Nicole Dastugue, Agnès Chassevent, Berna Beverloo, Anita Rijneveld, and Anthony Moorman for helpful support and discussions.
Authorship
Contribution: M.L.-P., L.B., V.L., N.I., and H.D. conceived and designed the study; V.L. provided administrative support; all of the authors provided study materials or patients; M.L.-P., L.B., and V.L. collected and assembled data; M.L-P., L.B., and H.D. analyzed and interpreted data; M.L.-P. and H.D. wrote the manuscript; and all authors approved the final manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
A complete list of the members of the Group for Research on Adult Acute Lymphoblastic Leukemia (GRAALL) appears in the online appendix.
Correspondence: Hervé Dombret, Institut Universitaire d’Hématologie, Hôpital Saint-Louis, 1 Avenue Claude Vellefaux, 75475 Paris Cedex 10, France; e-mail: herve.dombret@aphp.fr.
References
Author notes
M.L.-P. and L.B. contributed equally to this study.

![Figure 4. Overall survival in the t(4;11)/KMT2A-AFF1 and 14q32/IGH subgroups. (A) Without censoring patients who received allogeneic SCT in first CR. At 5 years, OS was estimated at 45.8% (95% CI, 34-57) in the t(4;11)/KMT2A-AFF1 subgroup and at 30.6% (95% CI, 14-49) in the 14q32/IGH subgroup, in comparison with 63.2% (95% CI, 54-71) in the subgroup with no identified cytogenetic abnormality (HR, 1.17 [95% CI, 1.05-1.30] and 1.13 [95% CI, 1.06-1.21]; P = .005 and <.001, respectively). (B) Censoring patients who received allogeneic SCT in first CR at SCT time. At 5 years, OS was estimated at 27.6% (95% CI, 12-46) in the t(4;11)/KMT2A-AFF1 subgroup and at 20.2% (95% CI, 4-45) in the 14q32/IGH subgroup, in comparison with 65.1% (95% CI, 53-75) in the subgroup with no identified cytogenetic abnormality (HR, 1.32 [95% CI, 1.14-1.53] and 1.16 [95% CI, 1.07-1.26]; P < .001 and <.001, respectively).](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/130/16/10.1182_blood-2017-05-783852/4/m_blood783852f4.jpeg?Expires=1767713206&Signature=VnpsrAA~ubFI9Iwj9K76XRjfqQS0RRLvwkohCVBCw9kKaHViBBCCc4b5yDOpk1rV0J8cgmfZlJwNjTcjSEydrIlLbh6b-uHCfTXm4jCNEeriewRqwQw0cJmSolfqgbwSVD8xMHuAL7GTW719H1kLGIrRW53xXyD0lsy1qItSRR9aVIDiZkvciG0p-Gngfew9buTQ7XW7ZX8Z76FA~0eRXt5W8sIeeZMhmR2HR55Gb8sC4EznrebjPijydsYLi0tjniouLK8MJdQVIJVaKPW9e6M0IB3kH-bTbqXAL-i53Dx1bEtq4B887KUvEYDAs6DsTUqD1ZrQWfer2wYSNstyng__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
This feature is available to Subscribers Only
Sign In or Create an Account Close Modal