Key Points
Npm1c and Nras-G12D co-mutation in mice leads to AML with a longer latency and a more mature phenotype than the Npm1c/Flt3-ITD combination.
Mutant Flt3 or Nras allele amplification is the dominant mode of progression in both Npm1c/Flt3-ITD and Npm1c/Nras-G12D murine AML.
Abstract
NPM1 mutations define the commonest subgroup of acute myeloid leukemia (AML) and frequently co-occur with FLT3 internal tandem duplications (ITD) or, less commonly, NRAS or KRAS mutations. Co-occurrence of mutant NPM1 with FLT3-ITD carries a significantly worse prognosis than NPM1-RAS combinations. To understand the molecular basis of these observations, we compare the effects of the 2 combinations on hematopoiesis and leukemogenesis in knock-in mice. Early effects of these mutations on hematopoiesis show that compound Npm1cA/+;NrasG12D/+ or Npm1cA;Flt3ITD share a number of features: Hox gene overexpression, enhanced self-renewal, expansion of hematopoietic progenitors, and myeloid differentiation bias. However, Npm1cA;Flt3ITD mutants displayed significantly higher peripheral leukocyte counts, early depletion of common lymphoid progenitors, and a monocytic bias in comparison with the granulocytic bias in Npm1cA/+;NrasG12D/+ mutants. Underlying this was a striking molecular synergy manifested as a dramatically altered gene expression profile in Npm1cA;Flt3ITD, but not Npm1cA/+;NrasG12D/+, progenitors compared with wild-type. Both double-mutant models developed high-penetrance AML, although latency was significantly longer with Npm1cA/+;NrasG12D/+. During AML evolution, both models acquired additional copies of the mutant Flt3 or Nras alleles, but only Npm1cA/+;NrasG12D/+ mice showed acquisition of other human AML mutations, including IDH1 R132Q. We also find, using primary Cas9-expressing AMLs, that Hoxa genes and selected interactors or downstream targets are required for survival of both types of double-mutant AML. Our results show that molecular complementarity underlies the higher frequency and significantly worse prognosis associated with NPM1c/FLT3-ITD vs NPM1/NRAS-G12D-mutant AML and functionally confirm the role of HOXA genes in NPM1c-driven AML.
Introduction
Advances in genomics have defined the somatic mutational landscape of acute myeloid leukemia (AML), leading to a detailed characterization of their prognostic significance and patterns of mutual co-occurrence or exclusivity.1,2 Mutations in NPM1, the gene for Nucleophosmin, characterize the most common subgroup of AML representing 25% to 30% of all cases, result in cytoplasmic dislocation of the protein (NPM1c), and are mutually exclusive of leukemogenic fusion genes.1-3 As is often the case for fusion genes, progression to AML after the acquisition of mutant NPM1 is contingent upon the gain of additional somatic mutations, such as those that activate STAT or RAS signaling.3,4 For reasons that are not clear, this transforming step favors acquisition of internal tandem duplications in FLT3 (FLT3-ITD) over other somatic mutations with similar effects such as those involving NRAS or KRAS.1-4 Furthermore, the NPM1c/FLT3-ITD combination is associated with a significantly worse prognosis than are combinations of NPM1c with mutant NRAS, KRAS, or related mutations.2
Although the adverse prognostic impact of NPM1/FLT3-ITD vs NPM1/RAS co-mutation influences clinical decisions in AML, its molecular basis and that of the frequent co-occurrence of NPM1c and FLT3-ITD in AML are unknown. Here, to investigate these phenomena, we compare the interaction of Npm1c with Flt3-ITD to its interaction with NrasG12D in knock-in mice. Individually, knock-in models of NPM1c, FLT3-ITD, and NRAS-G12D display enhanced myelopoiesis and progression to myeloproliferative disorders or AML in a significant proportion of animals.5-7 Also, we and others have previously shown that Npm1c and Flt3-ITD synergize to drive rapid-onset AML,8,9 however the interaction between Npm1c and mutant NrasG12D has not, to our knowledge, been previously investigated in knock-in mice.10 Our findings reveal that the combination of Npm1c and Flt3-ITD has an early profound effect on gene expression and hematopoiesis, whereas Npm1c and Nras-G12D display only modest molecular synergy and subtler cellular changes. Also, while both types of co-mutation drove AML in the majority of mice, the leukemias in Npm1c;Flt3-ITD mice were more aggressive and undifferentiated than were those that developed in Npm1c;Nras-G12D animals. At the genomic level, there was frequent amplification in both models of the mutant Flt3-ITD or Nras-G12D allele; however, additional somatic mutations in AML driver genes (eg, Idh1 and Ptpn11) were seen only in Npm1c;Nras-G12D AMLs. Our findings propose that the molecular synergy between Npm1c and Flt3-ITD underpin the co-occurrence patterns, phenotype, and prognosis of NPM1-mutant AML.
Materials and methods
Animal husbandry
Mx1-Cre+;Npm1flox-cA/+ were crossed with NrasLSL-G12D or Flt3ITD mice to generate triple- transgenic animals (Mx1-Cre;Npm1flox-cA/+;NrasLSL-G12D/+ and Mx1-Cre;Npm1flox-cA/+;Flt3ITD/+). To activate conditional alleles (Npm1cA and NrasG12D) in approximately 12- to 14-week-old Mx1-Cre;Npm1flox-cA/+;NrasLSL-G12D/+ mice, Mx1-Cre was induced by administration of pIpC. As has been described previously, Mx-1 Cre;Npm1flox-cA/+;Flt3ITD/+ mutants do not require pIpC induction of Mx1-Cre and recombination of the Npm1flox-cA allele.8 For preleukemic analyses Npm1cA/+;NrasG12D/+ were sacrificed 4 to 5 weeks post- pIpC, and Npm1cA/+;Flt3ITD/+ were sacrificed at 5 weeks of age. Genotyping for mutant alleles was performed as has been previously described.5-7 All animal procedures were carried out in accordance with the Home Office Animals (Scientific Procedures) Act 1986 Amendment Regulations (2012) under project license 80/2564.
Hematological measurements
Blood counts were performed on a VetABC analyzer (Horiba ABX).
Histopathology
Formalin-fixed, paraffin-embedded sections were stained with hematoxylin and eosin. Samples from leukemic mice were also stained with anti-CD3, anti-B220, anti-myeloperoxidase, anti-ERK1/2, or anti-phospho-Erk1/2 (pERK1/2). All material was examined by 2 experienced histopathologists (P.W. and M.A.) blinded to mouse genotypes.
Colony-forming assays and serial replating
Nucleated cells (3 × 104) from bone marrow (BM) aspirates of mutant and wild-type (WT) mice were suspended in cytokine-containing methylcellulose-based media (M3434, Stem Cell Technologies) and plated in duplicate wells of 6-well plates. Colony-forming units (CFUs) were counted 7 days later. For serial replating, 3 × 104 cells were reseeded and colonies counted after 7 days.
Flow cytometry and cell sorting
Single-cell suspensions of BM cells or splenocytes were incubated in 0.85% NH4Cl for 5 min to lyse erythrocytes. Cells were then suspended in Hank's Balanced Salt Solution, supplemented with 2% fetal calf serum and 10 μM N-2-hydroxyethylpiperazine-N′-2-ethanesulfonic acid. Progenitor populations were defined and stained as described in the supplemental Methods, available on the Blood Web site. Gated cellularity was calculated by multiplying the percentage of gated cells by the total number of nucleated cells from BM samples after erythrocyte depletion.
Viral transduction of BM progenitors and AML cell culture
Lineage-depleted (Lin−) BM aspirates, isolated from WT and Flt3ITD/+ mice, were transduced with murine stem cell virus (MSCV)–Hoxa9–green fluorescent protein (GFP) or MSCV-Nkx2-3–cyan fluorescent protein (CFP) retroviruses, or both, and expanded for 7 days in liquid culture (X-Vivo, Lonza; supplemented with 10 ng/mL interleukin-3 (IL-3), 10 ng/mL IL-6, and 50 ng/mL stem cell factor; Peprotech). CFP, GFP, or double-positive cells were fluorescence-activated cell sorted (FACS) and 2.5 × 104 cells replated in semisolid media, as was previously described. BM-derived AML cells from Rosa26-EF1a-Cas9 mice were cultured in vitro in the presence of cytokines. Disruption of individual candidate genes was performed by transduction with lentivirus expressing gene-specific guide RNA (gRNA) and blue fluorescent protein (BFP). The impact of gene disruption on AML cell growth was determined using competitive coculture of transduced (BFP+) vs nontransduced (BFP−) cells, as has been described previously11 (see Figure 6A; supplemental Methods).
Microarray and comparative genomic hybridization analysis
Mouse gene expression profiles (GEPs) were generated using the Illumina MouseWG-6 version 2 Expression BeadChip platform (Illumina). DNA copy number variation in leukemic samples was assessed with Mouse Genome Comparative Genomic Hybridization 244K Microarray (array comparative genomic hybridization [aCGH], Agilent Technologies). Full details of analysis are provided in the supplemental Methods. For mouse gene expression profiling, n = 4 to 10 (Lin−) or n = 3 to 5 (multipotent progenitor [MPP]).
AML exome sequencing and mutation calling
Whole exome sequencing (WES) of AML BM and control C57BL/6N or 129Sv tail DNA was performed using the Agilent SureSelect Mouse Exon Kit (Agilent Technologies) and paired-end sequencing on a HiSeq2000 sequencer (Illumina). Validation of mutations was performed using MiSeq sequencing (Illumina) of amplicon libraries, as was previously described (see supplemental Methods, supplemental Figure 1, and supplemental Tables 6 and 7 for primer sequences).12,13 Full details of analysis are provided in the supplemental Methods.
Datasets
Microarray data were deposited at Array Express (accession number E-MTAB-5356), and RNA sequencing (accession numbers ERS1732539 to ERS1732546, ERS812461, and ERS812462) as well as exome and Miseq sequencing (accession numbers PRJEB18526 and ERP020464) were deposited at EBI ENA.
Results
Mutant Npm1 cooperates with Nras-G12D and Flt3-ITD to increase self-renewal of hematopoietic progenitors and expand myelopoiesis
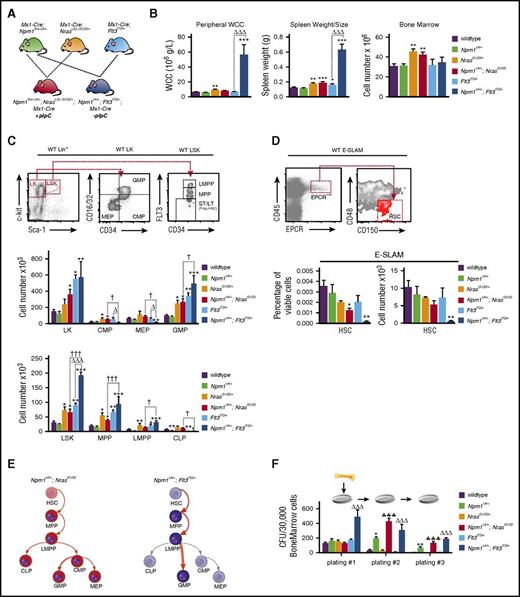
To understand the impact of the studied mutations, we analyzed hematopoietic cell compartments of Npm1cA/+;NrasG12D/+, Npm1cA/+;Flt3ITD/+, NrasG12D/+, Flt3ITD/+, and WT mice 4 to 6 weeks after activation of conditional mutations (Figure 1). In comparison with Flt3ITD/+ single mutants, Npm1cA/+;Flt3ITD/+ mice displayed higher WCCs (56 ± 13.4 vs 6.5 ± 0.5 × 106 g/L; P < .001) and spleen weights (0.63 g vs 0.16 g; P < .001), but not BM cellularity (Figure 1B). By contrast, both NrasG12D/+ and Npm1cA/+;NrasG12D/+ mutants exhibited subtler increases in spleen weight (WT: 0.12 g; NrasG12D/+: 0.18 g; Npm1cA/+;NrasG12D/+: 0.19 g; P < .01 and P < .001, respectively vs WT), but increased BM cellularity (WT: 28.1 ± 1.9 × 106; NrasG12D/+: 43.7 ± 2.6 × 106 and Npm1cA/+;NrasG12D/+: 41.3 ± 3.2 × 106; P < .01 for either comparison vs WT) (Figure 1B).
Mutant Npm1 cooperates with Nras-G12D and Flt3-ITD to enhance myeloid differentiation and enhance progenitor self-renewal. (A) Schema for Mx-1 Cre, Npm1flox-cA, NrasLSL-G12D, and Flt3ITD intercrosses. (B) NrasG12D/+ mice show a subtle and Npm1cA/+;Flt3ITD/+ mice a marked increase in WCC, in comparison with wild-type. Splenic sizes were significantly increased in all mutant genotypes except Npm1cA/+, with Npm1cA/+;Flt3ITD/+ showing the most striking phenotype. Bone marrow cellularity was increased only in the presence of the NrasG12D/+ allele. (C) FACS analysis at 4 to 5 weeks after mutation induction. Gating strategies depicted are from wild-type mice. There were significant differences in the stem and progenitor cell compartments of NrasG12D/+ and Flt3ITD/+, but not of Npm1cA/+ single-mutant, mice, as has been previously reported. In double-mutant mice, the Npm1cA/+;NrasG12D/+ combination was not significantly different to NrasG12D/+, in contrast to Npm1cA/+;Flt3ITD/+, which was markedly different from both Flt3ITD/+and Npm1cA/+ single mutants. (D) Using a cell surface phenotype independent of FLT3 staining, we found that CD45+/EPCR+/CD150+/CD48− HSCs were reduced slightly in Npm1cA/+;NrasG12D/+ and markedly in Npm1cA/+;Flt3ITD/+ mice. (E) Summary of hematopoietic effects of Npm1cA/+;NrasG12D/+ and Npm1cA/+;Flt3ITD/+ double mutations in mice. (F) Single Npm1cA/+- and double Npm1cA/+;NrasG12D/+- or Npm1cA/+;Flt3ITD/+-mutant hematopoietic progenitors show increased self-renewal potential in whole bone marrow serial replating assays (n = 4-8). Mean ± SEM are plotted. Significant values are reported for 1-way analysis of variance (Bonferroni adjusted). LT, long-term; Prog., progenitors; ST, short-term. *P < .05, **P < .01, ***P < .001, all vs wild-type; ΔP < .05, ΔΔΔP < .001, all vs Flt3ITD/+; ♣♣♣P < .001, all vs NrasG12D/+; †P < .05, ††P < .01, †††P < .001, all Npm1cA/+;NrasG12D/+ vs Npm1cA/+;Flt3ITD/+. WCC, white cell count.
Mutant Npm1 cooperates with Nras-G12D and Flt3-ITD to enhance myeloid differentiation and enhance progenitor self-renewal. (A) Schema for Mx-1 Cre, Npm1flox-cA, NrasLSL-G12D, and Flt3ITD intercrosses. (B) NrasG12D/+ mice show a subtle and Npm1cA/+;Flt3ITD/+ mice a marked increase in WCC, in comparison with wild-type. Splenic sizes were significantly increased in all mutant genotypes except Npm1cA/+, with Npm1cA/+;Flt3ITD/+ showing the most striking phenotype. Bone marrow cellularity was increased only in the presence of the NrasG12D/+ allele. (C) FACS analysis at 4 to 5 weeks after mutation induction. Gating strategies depicted are from wild-type mice. There were significant differences in the stem and progenitor cell compartments of NrasG12D/+ and Flt3ITD/+, but not of Npm1cA/+ single-mutant, mice, as has been previously reported. In double-mutant mice, the Npm1cA/+;NrasG12D/+ combination was not significantly different to NrasG12D/+, in contrast to Npm1cA/+;Flt3ITD/+, which was markedly different from both Flt3ITD/+and Npm1cA/+ single mutants. (D) Using a cell surface phenotype independent of FLT3 staining, we found that CD45+/EPCR+/CD150+/CD48− HSCs were reduced slightly in Npm1cA/+;NrasG12D/+ and markedly in Npm1cA/+;Flt3ITD/+ mice. (E) Summary of hematopoietic effects of Npm1cA/+;NrasG12D/+ and Npm1cA/+;Flt3ITD/+ double mutations in mice. (F) Single Npm1cA/+- and double Npm1cA/+;NrasG12D/+- or Npm1cA/+;Flt3ITD/+-mutant hematopoietic progenitors show increased self-renewal potential in whole bone marrow serial replating assays (n = 4-8). Mean ± SEM are plotted. Significant values are reported for 1-way analysis of variance (Bonferroni adjusted). LT, long-term; Prog., progenitors; ST, short-term. *P < .05, **P < .01, ***P < .001, all vs wild-type; ΔP < .05, ΔΔΔP < .001, all vs Flt3ITD/+; ♣♣♣P < .001, all vs NrasG12D/+; †P < .05, ††P < .01, †††P < .001, all Npm1cA/+;NrasG12D/+ vs Npm1cA/+;Flt3ITD/+. WCC, white cell count.
Expanded myelopoiesis and myeloproliferation were previously documented in single NrasG12D/+ and Flt3ITD/+-mutant mice.5,6 Mutant Npm1 augmented these phenotypes with increases in total Mac-1+ splenocytes (from 27% to 50% for NrasG12D/+ and 57% to 73% for Flt3ITD/+). Notably, these cells were predominantly granulocytic (Mac-1+/Gr-1+) in Npm1cA/+;NrasG12D/+ and predominantly monocytic (Mac-1+/Gr-1−) in Npm1cA/+;Flt3ITD/+ mice (supplemental Figure 1A).
NrasG12D/+ mice have been shown to have increased hematopoietic stem (HSC) and progenitor cell numbers, due to increased proliferation and self-renewal of the HSC and MPP compartments.14,15 Our results confirm these data, demonstrating significant increases in total myeloid progenitors, that is, granulocyte-macrophage (GMP) and common-myeloid progenitors (CMP). Total numbers of Lin−/Sca1+/Kit+ early progenitors (LSK) and MPPs are also increased in both Npm1cA/+;NrasG12D/+ and NrasG12D/+ BM cells (Figure 1C; supplemental Figure 2A). However, NrasG12D/+ progenitor cell composition was largely unaltered by the addition of mutant NPM1. Concordant with previous studies, hematopoiesis in Flt3ITD/+ mice was characterized by increased numbers of total myeloid progenitors (Lin−/Kit+ [LK], P < .05, and GMPs, P < .01) and early progenitor populations (LSK, MPP, and lymphoid primed multipotent progenitor [LMPP], P < .01, P < .01, and P < .05, respectively) (Figure 1C; supplemental Figure 2A).16,17 Of note, there were detectable decreases in the size of the common lymphoid progenitor (CLP) population in Flt3ITD/+ and Npm1cA/+;Flt3ITD/+ mice (Figure 1C) (in part due to the reduction in Il-7Rα-positive cells) (supplemental Figure 2B). Npm1cA/+;Flt3ITD/+ mice also exhibited robust increases in numbers of LK, LSK, MPP, and LMPP populations, above what was observed with Flt3ITD/+, when compared with WT. In direct comparison with Flt3ITD/+ mutants, numbers of CMP and megakaryocyte-erythroid progenitor (MEP) progenitors in Npm1cA/+; Flt3ITD/+ mice were reduced (from 55 × 103 to 16 × 103, P < .05, and from 61 × 103 to 17 × 103, P < .05), yet GMPs, proposed as direct descendants of CMPs,18 are significantly increased. This demonstrates that Flt3ITD/+-mutant myelopoiesis is dramatically altered by the addition of Npm1cA/+. In direct comparison with Npm1cA/+;NrasG12D/+, Npm1cA/+;Flt3ITD/+ mice showed increased LMPP and GMP populations with reduced numbers of CLP (Figure 1E).
To assess the effects on the earliest detectable HSC, we opted to perform E-SLAM staining (CD45+/endothelial protein C receptor-positive [EPCR+]/CD48−/CD150+).19 Importantly, this does not rely on cell surface expression of FLT3, and reveals that the percentage of E-SLAM detectable HSCs is decreased in Npm1cA/+;NrasG12D/+ mice and further so in Npm1cA/+;Flt3ITD/+mutants (Figure 1D). Finally, using serial replating of BM cells in semisolid media, we show that Npm1cA/+ co-mutation markedly increased self-renewal of Flt3ITD/+ (as was shown previously8 ) and of NrasG12D/+ cells (Figure 1F).
An Npm1cA/+ transcriptional signature persists in double-mutant hematopoietic progenitors
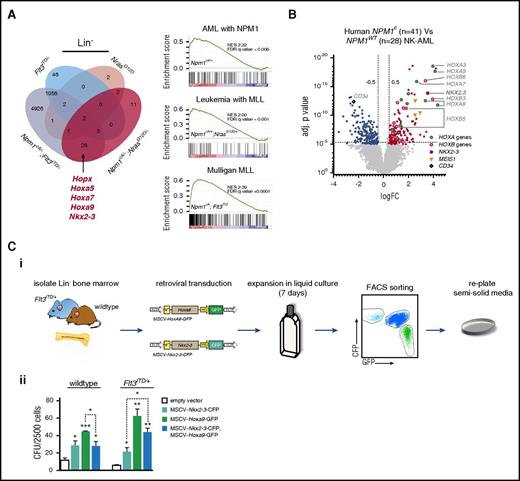
To examine their combined effects on transcription, we performed comparative global gene expression profiling of lineage negative (Lin−) BM cells using microarrays. Npm1cA/+;NrasG12D/+ and Npm1cA/+;Flt3ITD/+ cells displayed a dramatically altered GEP in comparison with single NrasG12D/+ or Flt3ITD/+ mutants (Figure 2A; supplemental Figure 3B). Previously, we showed that mouse Npm1cA/+ Lin− cells overexpressed several homeobox (Hox) genes (in particular Hoxa5, Hoxa7, Hoxa9, and 2 other homeobox genes, Hopx and Nkx2-3).7 Here, we show that this signature, absent from NrasG12D/+ or Flt3ITD/+ single-mutant mice, persists in compound Npm1cA/+;NrasG12D/+ and Npm1cA/+;Flt3ITD/+ Lin− progenitors (Figure 2A; supplemental Figure 3A-C). Gene Set Enrichment Analysis of Npm1cA/+ single- and compound-mutant cell differentially expressed genes showed significant enrichment for genes upregulated in NPM1-mutant and MLL-fusion gene positive human leukemias (Figure 2A).
Impact of Npm1cA/+on the transcriptome of NrasG12D/+- and Flt3ITD/+-mutant hematopoietic progenitors. (A) Overlap of differentially expressed mRNAs reveals that Npm1cA/+ has a dramatic impact on Lin− progenitor GEPs when combined with Flt3ITD/+ but only a modest impact when combined with NrasG12D/+. Nonetheless, the characteristic hallmarks of Npm1cA/+, namely overexpression of Hoxa genes and of the homeobox genes Hopx and Nkx2-3, are retained in both types of double-mutant progenitors. Gene Set Enrichment Analysis reveals enrichment of differentially expressed genes from these models in human AMLs harboring mutant NPM1 or MLL gene fusions. (B) Comparison of human NPM1-mutant (NPM1c) vs NPM1-wildtype (NPM1WT) normal karyotype AML also shows marked overexpression of HOXA and HOXB genes as well as of NKX2.3, raising the possibility that the latter may mediate some of the effect of NPM1c. (C) Effects of Nkx2-3 and Hoxa9 overexpression on mouse hematopoietic progenitors. (i) Lin− bone marrow progenitors from wild-type and Flt3ITD/+ mice were transduced with MSCV-Nkx2.3-CFP or MSCV-Hoxa9-GFP constructs or both, maintained in liquid culture for 7 days, FACS sorted for CFP and GFP single- and double-transfected cells, and plated in semisolid media. (ii) Colony assays of 2,500 transduced cells show that both MSCV-Hoxa9 and MSCV-Nkx2-3 conferred an increase in self-renewal of both wild-type and Flt3ITD/+ cells. However, double MSCV-Hoxa9/MSCV-Nkx2-3 transfected cells showed no further changes in self-renewal when compared with MSCV-Hoxa9 alone. Mean ± SEM (n = 3). *P < .05; **P < .01; ***P < .001; Student t test. (D) Sorting strategy for LSK/CD34+/Flt3+/CD48+ progenitor cells and overlap of differentially expressed genes (Illumina MouseWG-6 version 2 Expression BeadChip) for NrasG12D/+ vs (i) Npm1cA/+;NrasG12D/+ and (ii) Flt3ITD/+ vs Npm1cA/+;Flt3ITD/+ MPP datasets. (E) Heat maps of normalized Hox gene expression in (i) MPP and (ii) Lin− progenitor populations reveal that, unlike Lin− cells, Npm1cA/+-mutant (single or double) MPPs have similar patterns of Hox gene expression to Npm1-WT MPPs (normalized average expression values are used to generate heat map values). (F) Differentially expressed genes in Npm1cA/+;Flt3ITD/+ MPPs vs WT controls. Adj., adjusted; FC, fold change; NK, normal karyotype.
Impact of Npm1cA/+on the transcriptome of NrasG12D/+- and Flt3ITD/+-mutant hematopoietic progenitors. (A) Overlap of differentially expressed mRNAs reveals that Npm1cA/+ has a dramatic impact on Lin− progenitor GEPs when combined with Flt3ITD/+ but only a modest impact when combined with NrasG12D/+. Nonetheless, the characteristic hallmarks of Npm1cA/+, namely overexpression of Hoxa genes and of the homeobox genes Hopx and Nkx2-3, are retained in both types of double-mutant progenitors. Gene Set Enrichment Analysis reveals enrichment of differentially expressed genes from these models in human AMLs harboring mutant NPM1 or MLL gene fusions. (B) Comparison of human NPM1-mutant (NPM1c) vs NPM1-wildtype (NPM1WT) normal karyotype AML also shows marked overexpression of HOXA and HOXB genes as well as of NKX2.3, raising the possibility that the latter may mediate some of the effect of NPM1c. (C) Effects of Nkx2-3 and Hoxa9 overexpression on mouse hematopoietic progenitors. (i) Lin− bone marrow progenitors from wild-type and Flt3ITD/+ mice were transduced with MSCV-Nkx2.3-CFP or MSCV-Hoxa9-GFP constructs or both, maintained in liquid culture for 7 days, FACS sorted for CFP and GFP single- and double-transfected cells, and plated in semisolid media. (ii) Colony assays of 2,500 transduced cells show that both MSCV-Hoxa9 and MSCV-Nkx2-3 conferred an increase in self-renewal of both wild-type and Flt3ITD/+ cells. However, double MSCV-Hoxa9/MSCV-Nkx2-3 transfected cells showed no further changes in self-renewal when compared with MSCV-Hoxa9 alone. Mean ± SEM (n = 3). *P < .05; **P < .01; ***P < .001; Student t test. (D) Sorting strategy for LSK/CD34+/Flt3+/CD48+ progenitor cells and overlap of differentially expressed genes (Illumina MouseWG-6 version 2 Expression BeadChip) for NrasG12D/+ vs (i) Npm1cA/+;NrasG12D/+ and (ii) Flt3ITD/+ vs Npm1cA/+;Flt3ITD/+ MPP datasets. (E) Heat maps of normalized Hox gene expression in (i) MPP and (ii) Lin− progenitor populations reveal that, unlike Lin− cells, Npm1cA/+-mutant (single or double) MPPs have similar patterns of Hox gene expression to Npm1-WT MPPs (normalized average expression values are used to generate heat map values). (F) Differentially expressed genes in Npm1cA/+;Flt3ITD/+ MPPs vs WT controls. Adj., adjusted; FC, fold change; NK, normal karyotype.
Overexpression of the homeobox gene NKX2.3 in human NPM1-mutant AML
Using the human The Cancer Genome Atlas (TCGA) AML dataset, we compared GEPs of NPM1-mutant (NPM1c+ve) to NPM1-wild-type (NPM1wt) AML.1 In agreement with previously published analyses, both HOXA and HOXB genes were significantly overexpressed in NPM1c+ve AML (Figure 2B).20 We noted that NKX2-3 was also overexpressed, in keeping with our findings in Npm1cA/+ mice (Figure 2A). Recently, NKX2-3 overexpression was shown to be the most effective discriminant of MLL-MLLT4 (MLL-AF6)-driven AML from AMLs driven by other MLL-fusion genes.21 Although overexpression of Hox genes such as Hoxa9 has been shown to impart increased self-renewal and proliferation of hematopoietic progenitors, the effects of Nkx2-3 overexpression are unknown.22 To study this, we performed retroviral gene transfer of fluorescently tagged Nkx2-3-CFP and Hoxa9-GFP into wild-type and Flt3ITD/+ Lin− cells. Cells were subsequently sorted and plated in semisolid methylcellulose media for colony-formation assays (Figure 2Ci). We find that overexpression of Nkx2-3 increases clonogenic potential, albeit to a lesser extent to Hoxa9 overexpression, in both wild-type and Flt3ITD/+ progenitors. Notably, this was not augmented in combined transfected cells (Figure 2Cii).
Hoxa gene expression is unaltered in Npm1-mutant early multipotent progenitors
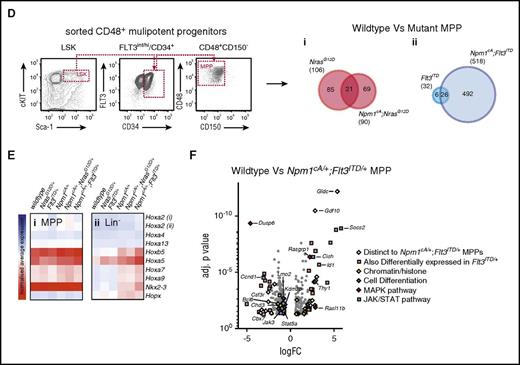
To mitigate the impact of the studied driver mutations on cell surface phenotypes, we performed transcriptome analysis on a homogeneous population of early progenitors, purified LSK MPPs (Figure 2D). Hox gene expression was not significantly altered in this population in any of the Npm1cA/+ models when compared with wild-type or single NrasG12D/+ and Flt3ITD/+ mutants (Figure 2E; supplemental Figure 3C). These results are in agreement with observations that Hox gene expression in human NPM1c AML blasts is comparable to that seen in WT human HSCs and myeloid progenitors.20 Because we do not observe statistically significant expansion in total (Lin−) progenitors in single Npm1cA/+ mice (Figure 1C), these data propose that unlike HSCs, the observed pattern of Hox overexpression in these progenitors is a molecular consequence of NPM1c rather than a change in cellular composition. This concurs with our published observations that the Hox signature is detectable even in CD19-positive B cells.7
MPPs from single NrasG12D/+ or Flt3ITD/+ and the respective Npm1cA/+ compound-mutant MPPs also had distinct transcriptional changes. In comparison with WT, both NrasG12D/+ and Npm1cA/+;NrasG12D/+ MPPs displayed small numbers of differentially expressed genes, yet only ∼20% of these were shared (Figure 2Di). Gene Set Enrichment Analysis did not uncover significant overlap with any preestablished expression signatures (data not shown). In contrast, the “addition” of Npm1cA/+ to Flt3ITD/+ in MPPs led to differential expression of a large number of additional genes, while also retaining most of the transcriptional changes attributable to Flt3ITD/+ (Figure 2Dii; supplemental Table 2), demonstrating the powerful synergy between Npm1cA/+ and Flt3ITD/+. Pathway analysis of genes differentially expressed in Npm1cA/+;Flt3ITD/+ MPPs revealed enrichment of genes in the JAK-STAT pathway (supplemental Figure 3E; supplemental Table 4), including the negative regulators Cish and Socs2 (Figure 2F). A number of genes encoding proteins involved in MAPK signaling were also deregulated, as were genes involved in chromatin regulation/organization and hematopoietic/myeloid differentiation (Figure 2F; supplemental Figure 3D). Many of the genes in our Npm1cA/+;Flt3ITD/+ dataset were also found deregulated in a recently published Tet2−/−;Flt3ITD/+ mouse model of AML (supplemental Figure 3F; supplemental Table 6) which serves to verify our mouse dataset technically but also reveals a distinguishing expression signature of FLT3-ITD, which includes Socs2, Id1, Csfr3r, and Bcl11a.17 In contrast, a lack of correlation between deregulated gene sets of Npm1cA/+;Flt3ITD/+, and Npm1cA/+;NrasG12D/+ MPPs (supplemental Figure 3D) emphasizes the molecular distinction between these compound mutants.
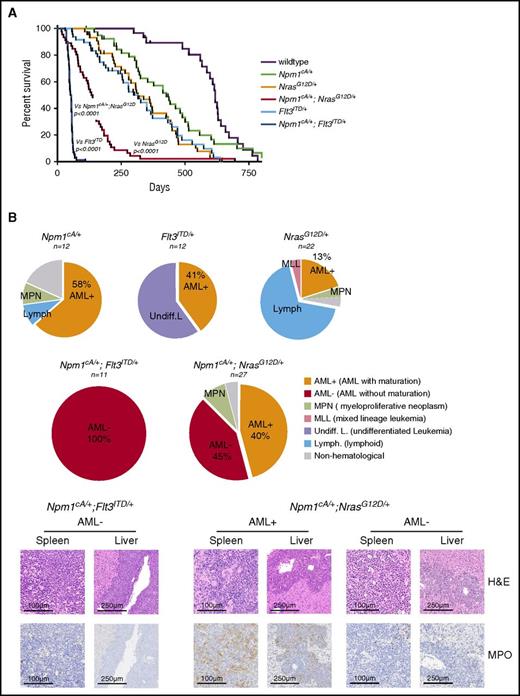
Npm1cA/+ and NrasG12D collaborate to promote high-penetrance AML
To understand the leukemogenic potential of combined Npm1cA/+ and NrasG12D mutations, we aged combined and single-mutant cohorts. Compound Npm1cA/+;NrasG12D/+ and Npm1cA/+;Flt3ITD/+ mice had significantly reduced survival (median 138 and 52.5 days, respectively) when compared with wild-type (618 days), Npm1cA/+ (427 days), NrasG12D/+ (315 days), and Flt3ITD/+ (also 315 days) (Figure 3A; supplemental Figure 4A). No difference in the survival of NrasG12D/+ and Flt3ITD/+-mutant mice was observed (P = .85; see supplemental Figure 4A for all comparisons). At time of sacrifice, blood counts and tissues were collected and subjected to histopathological analysis. Aged Npm1cA/+;NrasG12D/+ and Npm1cA/+;Flt3ITD/+ mice exhibited characteristic AML pathological findings at a much higher frequency than did single-mutant mice. These included significantly higher WCC, reduced platelet numbers, and substantial organ infiltration with leukemic cells (supplemental Figure 4B-D). Histological analysis verified the increased AML incidence from 41% (Flt3ITD/+) to 100% in Npm1cA/+;Flt3ITD/+ samples and from 13% (NrasG12D/+) to 85% in Npm1cA/+;NrasG12D/+ samples (45% AML with maturation, AML+ and 40% AML without maturation, AML− as defined by the Bethesda classification23 ) (Figure 3B).
Npm1cAand NrasG12Dcooperate to drive high-penetrance AML. (A) Kaplan- Meier survival curves of wild-type (n = 23), Npm1cA/+ (n = 34), NrasG12D/+ (n = 40), Flt3ITD/+ (n = 39), Npm1cA/+;NrasG12D/+ (n = 46), and Npm1cA/+;Flt3ITD+/ (n = 40). Double mutant (Npm1cA/+;NrasG12D/+ and Npm1cA/+;Flt3ITD/+) mice had a significantly shortened survival when compared with single mutants, and Npm1cA/+;Flt3ITD had significantly shorter survival than Npm1cA/+;NrasG12D/+ mice. (B) Results of independent histopathological analysis of aged moribund mice. Incidence of AML in compound Npm1cA/+;NrasG12D/+ and Npm1cA/+;Flt3ITD/+ mice is increased in comparison with Npm1cA/+, NrasG12D/+, and Flt3ITD/+ mice. Examples of complete effacement of splenic tissue and infiltration of myeloid blast cells in liver tissue from Npm1cA/+;NrasG12D/+ and Npm1cA/+;Flt3ITD+/ AMLs are presented. Reduced myeloperoxidase (MPO) staining in diseased tissues is observed in samples categorized as AML without maturation (AML−) in comparison with those categorized as AML with maturation (AML+). H&E, haematoxylin and eosin.
Npm1cAand NrasG12Dcooperate to drive high-penetrance AML. (A) Kaplan- Meier survival curves of wild-type (n = 23), Npm1cA/+ (n = 34), NrasG12D/+ (n = 40), Flt3ITD/+ (n = 39), Npm1cA/+;NrasG12D/+ (n = 46), and Npm1cA/+;Flt3ITD+/ (n = 40). Double mutant (Npm1cA/+;NrasG12D/+ and Npm1cA/+;Flt3ITD/+) mice had a significantly shortened survival when compared with single mutants, and Npm1cA/+;Flt3ITD had significantly shorter survival than Npm1cA/+;NrasG12D/+ mice. (B) Results of independent histopathological analysis of aged moribund mice. Incidence of AML in compound Npm1cA/+;NrasG12D/+ and Npm1cA/+;Flt3ITD/+ mice is increased in comparison with Npm1cA/+, NrasG12D/+, and Flt3ITD/+ mice. Examples of complete effacement of splenic tissue and infiltration of myeloid blast cells in liver tissue from Npm1cA/+;NrasG12D/+ and Npm1cA/+;Flt3ITD+/ AMLs are presented. Reduced myeloperoxidase (MPO) staining in diseased tissues is observed in samples categorized as AML without maturation (AML−) in comparison with those categorized as AML with maturation (AML+). H&E, haematoxylin and eosin.
Additional somatic mutations are required for progression to AML in Npm1cA/+;NrasG12D/+ mice
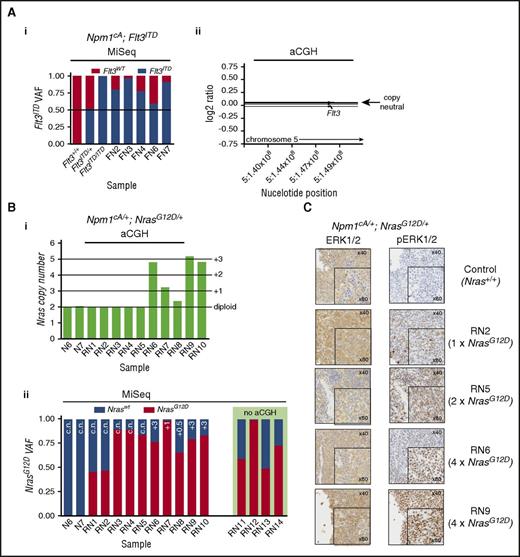
Npm1cA/+;Flt3ITD/+ mice succumb to AML significantly more rapidly than do Npm1cA/+ and Npm1cA/+;NrasG12D/+ mice. We hypothesized that the slower onset of AML in the latter 2 genotypes may be due to the requirement for additional cooperating mutations. To test this, we performed aCGH and WES of AMLs from Npm1cA/+, Npm1cA/+;Flt3ITD/+, and Npm1cA/+;NrasG12D/+ mice. We first confirmed the frequent development of loss-of-heterozygosity (LOH) at the Flt3 locus in Npm1cA/+;Flt3ITD/+ AMLs8,24 and verified this by quantifying Flt3ITD variant allele fractions using polymerase chain reaction (PCR)–MiSeq (Figure 4Ai). aCGH showed that LOH was copy-neutral and due to uniparental disomy of Flt3ITD (supplemental Figure 4Aii). Interestingly, aCGH of Npm1cA/+;NrasG12D/+ samples revealed amplification of chr3 in 5 of 10 samples tested (Figure 4Bi). This was exclusive to Npm1cA/+;NrasG12D/+ AMLs and mapped to a minimally amplified region (chr3: 102743581-103470550) containing Nras (supplemental Table 10). We confirmed these NrasG12Dcopy gains using PCR-MiSeq and found copy-neutral LOH for NrasG12D in 3 of 10 AMLs. In addition, we found copy-neutral LOH in 3 of 4 Npm1cA/+;NrasG12D/+ AMLs not studied by aCGH. In summary, increased NrasG12D dosage was detected in 11 of 14 Npm1cA/+;NrasG12D/+ AMLs (Figure 4Bii), and this correlated with levels of RAS pathway activation as measured by phosphorylated extracellular signal-regulated kinase 1/2 (pERK1/2) staining (Figure 4C).
Leukemic progression in double-mutant mice involves increased NrasG12Dor Flt3ITDallele dosage. (A) Increase in Flt3ITD allele burden in AMLs from Npm1cA;Flt3ITD mice through loss of heterozygosity for the locus. (i) Flt3ITD amplicon sequencing (MiSeq) of leukemic bone marrow or spleen DNA (FN2-FN7). Tail DNA amplified from 2-week-old Flt3+/+, Flt3ITD/+, and Flt3ITD/ITD mice were used as control. (ii) Normalized log2 ratio plots show normal copy number for the Flt3 locus in 7 of 7 Npm1cA;Flt3ITD murine AMLs (FN-AMLs) tested. (Bi) Summary of aCGH showing copy number gain at the Nras locus in AMLs RN6-10. (Bii) Allele fractions for Nraswt vs NrasG12D show that copy number gains in RN6-10 involved NrasG12D and that an additional 3 cases (RN3-5) show copy-neutral (c.n.) loss of heterozygosity. In addition, 2 more RN AMLs show gains in mutant NRAS when measuring Nraswt vs NrasG12D allele fractions (aCGH was not performed on these). Results of 2 Npm1cA/+ samples are also shown for comparison purposes (N6, N7). (C) Increased gene dosage of NrasG12D correlates with increased levels of phosphorylated RAS effectors pERK1/2. FN2,3,4,6,7 = NPM1cA/+;Flt3ITD/+ AMLs, RN1-14 = Npm1cA/+;NrasG12D/+ AMLs. VAF, variant allele fraction.
Leukemic progression in double-mutant mice involves increased NrasG12Dor Flt3ITDallele dosage. (A) Increase in Flt3ITD allele burden in AMLs from Npm1cA;Flt3ITD mice through loss of heterozygosity for the locus. (i) Flt3ITD amplicon sequencing (MiSeq) of leukemic bone marrow or spleen DNA (FN2-FN7). Tail DNA amplified from 2-week-old Flt3+/+, Flt3ITD/+, and Flt3ITD/ITD mice were used as control. (ii) Normalized log2 ratio plots show normal copy number for the Flt3 locus in 7 of 7 Npm1cA;Flt3ITD murine AMLs (FN-AMLs) tested. (Bi) Summary of aCGH showing copy number gain at the Nras locus in AMLs RN6-10. (Bii) Allele fractions for Nraswt vs NrasG12D show that copy number gains in RN6-10 involved NrasG12D and that an additional 3 cases (RN3-5) show copy-neutral (c.n.) loss of heterozygosity. In addition, 2 more RN AMLs show gains in mutant NRAS when measuring Nraswt vs NrasG12D allele fractions (aCGH was not performed on these). Results of 2 Npm1cA/+ samples are also shown for comparison purposes (N6, N7). (C) Increased gene dosage of NrasG12D correlates with increased levels of phosphorylated RAS effectors pERK1/2. FN2,3,4,6,7 = NPM1cA/+;Flt3ITD/+ AMLs, RN1-14 = Npm1cA/+;NrasG12D/+ AMLs. VAF, variant allele fraction.
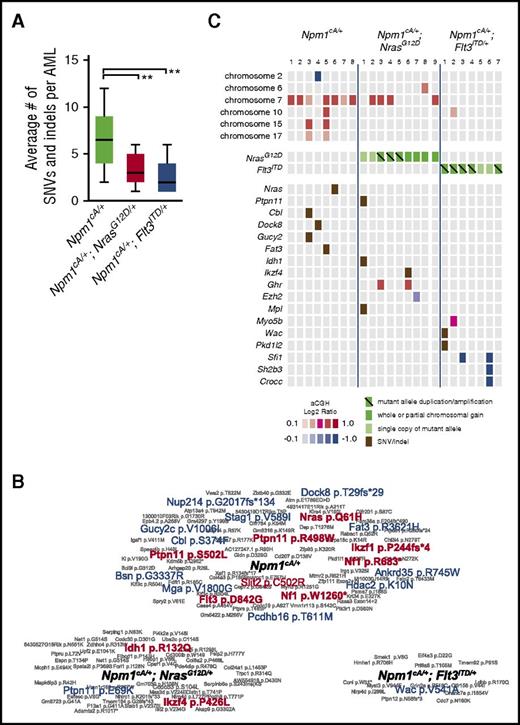
WES showed that the average number of single nucleotide variants (SNVs) and small insertions/deletions (indels) per AML sample correlated with mouse genotype (Figure 5A). Npm1cA/+ AMLs spontaneously acquired mutations in genes involved in RAS signaling (Nras-p.Q61H, Cbl-p.S374F, Ptpn11-p.S502L, Nf1-p.W1260*, and Nf1-R683*) confirming this genetic interaction. Likewise, we detected a spontaneous tyrosine kinase domain mutation in Flt3 (Flt3-p.D842G), confirming the importance of FLT3 mutations in progression of NPM1-mutant AML (Figure 5B-C; supplemental Table 9). Interestingly, a single Npm1cA/+;NrasG12D/+ AML harbored an Idh1-p.R132Q mutation and mirroring the R132H/R132C mutations commonly seen in human AML,1 and IDH1-R132Q itself that was reported in human chondrosarcoma.25 aCGH also revealed complete or partial gain of a minimally amplified region on chr7 in 7 of 8 Npm1cA/+ and 4 of 9 Npm1cA/+;NrasG12D/+ AMLs containing genes implicated in leukemogenesis, including Nup98, Wee1, and Eed, (supplemental Figure 5C).7,26-28 Single-copy loss of a region containing the epigenetic modifiers Wt1, Asxl1, Dnmt3a (1/8 Npm1cA/+), and a focal deletion of Ezh2 (1/9 Npm1cA/+; NrasG12D/+) were also detected (Figure 5C; supplemental Figure 5C).
Somatic mutations in Npm1cA/+, Npm1cA/+;NrasG12D/+, and Npm1cA;Flt3ITDAMLs. (A) Exome sequencing identifies an increased number of SNVs and small indels in Npm1cA/+, in comparison with Npm1cA/+;NrasG12D/+ (RN-AML) and Npm1cA;Flt3ITD (FN-AML) AML samples. Npm1cA/+ 6.8 ± 0.9, Npm1cA/+;NrasG12D/+ 3.3 ± 0.5, and Npm1cA/+;Flt3ITD/+ 2.6 ± 0.7 (mean ± SEM) (**P < .01 vs Npm1cA/+, 1-way analysis of variance, Bonferroni adjusted). Total AMLs sequenced; Npm1cA/+ (n = 12), Npm1cA/+;NrasG12D/+ (n = 14) and Npm1cA;Flt3ITD (n = 7). (B) Summary of SNVs/indels detected in AMLs from each genotype, as indicated. Genes mutated in the TCGA AML dataset1 are depicted in red (exact mutation reported) or in blue (different mutations reported). (C) Co-occurrence of SNVs and copy number variants (CNVs) that have been formally detected in the TCGA AML dataset1 or identified as common insertion sites in our previously published Npm1cA/+ Sleeping Beauty Transposon screen.7 Mutant allele copy gains, chromosome gains, and losses are depicted separately. For copy number variation, color-coded boxes are based on log2 ratios (aCGH) and are not representative of CNV size. For a complete overview of all CNV and SNV co-occurrences, see supplemental Figure 6.
Somatic mutations in Npm1cA/+, Npm1cA/+;NrasG12D/+, and Npm1cA;Flt3ITDAMLs. (A) Exome sequencing identifies an increased number of SNVs and small indels in Npm1cA/+, in comparison with Npm1cA/+;NrasG12D/+ (RN-AML) and Npm1cA;Flt3ITD (FN-AML) AML samples. Npm1cA/+ 6.8 ± 0.9, Npm1cA/+;NrasG12D/+ 3.3 ± 0.5, and Npm1cA/+;Flt3ITD/+ 2.6 ± 0.7 (mean ± SEM) (**P < .01 vs Npm1cA/+, 1-way analysis of variance, Bonferroni adjusted). Total AMLs sequenced; Npm1cA/+ (n = 12), Npm1cA/+;NrasG12D/+ (n = 14) and Npm1cA;Flt3ITD (n = 7). (B) Summary of SNVs/indels detected in AMLs from each genotype, as indicated. Genes mutated in the TCGA AML dataset1 are depicted in red (exact mutation reported) or in blue (different mutations reported). (C) Co-occurrence of SNVs and copy number variants (CNVs) that have been formally detected in the TCGA AML dataset1 or identified as common insertion sites in our previously published Npm1cA/+ Sleeping Beauty Transposon screen.7 Mutant allele copy gains, chromosome gains, and losses are depicted separately. For copy number variation, color-coded boxes are based on log2 ratios (aCGH) and are not representative of CNV size. For a complete overview of all CNV and SNV co-occurrences, see supplemental Figure 6.
MLL, Hox genes and their partners are required for the survival of Npm1cA-driven AML cells
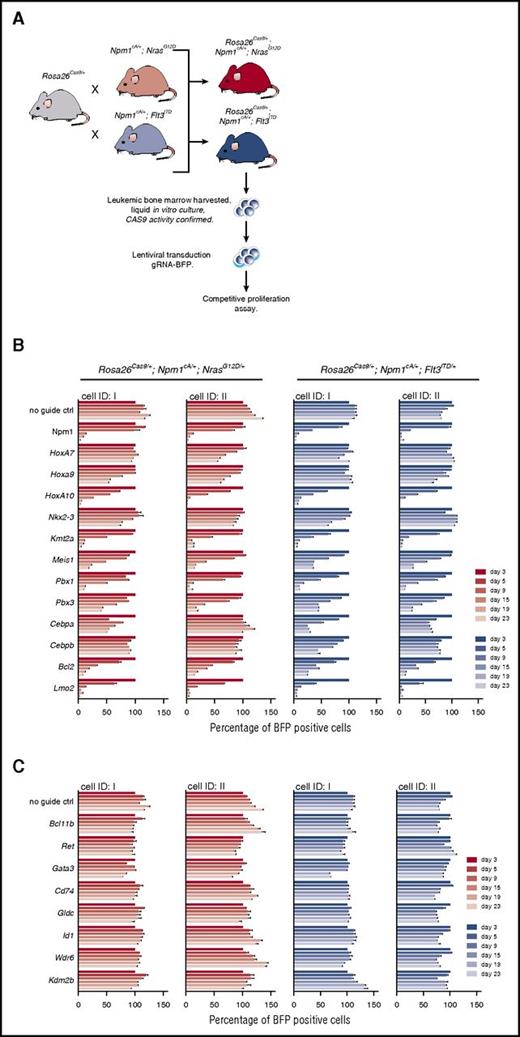
To assess their contribution to AML maintenance in Npm1cA/+;NrasG12D/+ and Npm1cA;Flt3ITD mice, we used CRISPR-Cas9 to disrupt selected deregulated genes identified by our preleukemic GEP studies. For this, we bred our mice with Rosa26-EF1a-Cas9 animals11 to generate Rosa26Cas9/+;Npm1cA/+;NrasG12D/+ and Rosa26Cas9/+;Npm1cA/+;Flt3ITD/+ mice. Competitive coculture of gRNA transduced and nontransduced BM cells from these mice revealed that Hoxa10 and to a lesser degree Hoxa9, but not Hoxa7, are required for Npm1cA/+;NrasG12D/+ and Npm1cA;Flt3ITD/+ AML maintenance (Figure 6B). In contrast, all 3 Hoxa genes were required for growth of AMLs generated by retroviral MLL-AF9 transformation of Flt3ITD/+ BM cells (supplemental Figure 7C).11,29,30 Notably, although Nkx2-3 overexpression enhanced colony-forming ability of wild-type and Flt3ITD/+ BM (Figure 2C), disruption of endogenous Nkx2-3 did not significantly affect proliferation of Npm1cA/+;NrasG12D/+ or Npm1cA;Flt3ITD/+ AMLs in vitro. Other genes whose disruption reduced proliferation of Npm1cA-driven AMLs included the Mll (Kmt2a) gene, recently shown to be a therapeutic target in this AML type31 ; Hoxa9/10 partners or cofactors including Meis1, Pbx1, and Pbx3; and the HOXA9 targets Bcl2 and Lmo2.32-34 A number of genes with altered expression in mutant preleukemic MPP cells were not required for survival of AML cells in vitro (Figure 6C). However, we cannot exclude a potential role for these in leukemia initiation.
MLL, Hox genes, and their partners are required for the survival of Npm1cA-driven AML cells. (A) Schematic depicting the derivation and liquid culture of Rosa26-EF1a-Cas9 expressing AML cell lines. CRISPR-Cas9–based assessment of individual genes aberrantly expressed in Npm1cA/+;NrasG12D/+ and Npm1cA;Flt3ITD mice. CAS9 activity of these mouse AML cell lines was validated, as described previously (supplemental Figure 7A).11 Individual Rosa26-EF1a-Cas9–expressing cell lines were derived from 2 mice (ID I and II) of each genotype. In vitro competitive assays were performed over a 23-day period using AML cell lines transduced with lentivirus expressing gRNAs for the indicated gene, and the BFP-positive fraction compared with the nontransduced population. Results were normalized to day 3 for each gRNA. Results from AML cell lines transduced with guide RNAs targeting Hoxa-related (B) and non-Hoxa–related (C) genes. gRNA sequences were selected from a previously published library11 and are detailed in supplemental Table 15. Guides against the pan-essential Npm1 gene are used as a control. Ctrl, control; ID, identity of mouse of origin.
MLL, Hox genes, and their partners are required for the survival of Npm1cA-driven AML cells. (A) Schematic depicting the derivation and liquid culture of Rosa26-EF1a-Cas9 expressing AML cell lines. CRISPR-Cas9–based assessment of individual genes aberrantly expressed in Npm1cA/+;NrasG12D/+ and Npm1cA;Flt3ITD mice. CAS9 activity of these mouse AML cell lines was validated, as described previously (supplemental Figure 7A).11 Individual Rosa26-EF1a-Cas9–expressing cell lines were derived from 2 mice (ID I and II) of each genotype. In vitro competitive assays were performed over a 23-day period using AML cell lines transduced with lentivirus expressing gRNAs for the indicated gene, and the BFP-positive fraction compared with the nontransduced population. Results were normalized to day 3 for each gRNA. Results from AML cell lines transduced with guide RNAs targeting Hoxa-related (B) and non-Hoxa–related (C) genes. gRNA sequences were selected from a previously published library11 and are detailed in supplemental Table 15. Guides against the pan-essential Npm1 gene are used as a control. Ctrl, control; ID, identity of mouse of origin.
We also wanted to investigate potential differences in JAK/STAT vs RAS signaling in our AMLs in a similar way. FLT3-ITD leads to constitutive activation of JAK/STAT signaling, driving growth and transformation of hematopoietic cells.35-37 In keeping with this, our transcriptome analysis revealed that genes involved in JAK/STAT signaling (Stat5a, Cish, Socs2) were differentially expressed in Npm1cA;Flt3ITD but not in Npm1cA;NrasG12D Lin− progenitors. Nevertheless, CRISPR-targeting of Jak2 and Stat5a/b genes inhibited the growth of both Npm1cA;Flt3ITD/+ and Npm1cA/+;NrasG12D/+ AML cells (supplemental Figure 8B). We confirmed by RNA-seq that this was due to activation of a JAK/STAT program in Npm1cA/+;NrasG12D/+ AML cells (supplemental Figure 9). In this light we conclude that the cytokines required for culturing primary murine AML cells in vitro (IL-3, IL-6, and stem cell factor), known to activate the JAK/STAT pathway, preclude the assessment of signaling genes in AML growth and proliferation.
Discussion
Although the mutational drivers of AML and their patterns of co-occurrence are well understood, the molecular basis for the frequency and prognostic impact of these patterns remains unknown. Of particular clinical relevance are the co-occurrence patterns of mutant NPM1 mutations, which characterize the most common AML subtype.1,2 Co-mutation of NPM1 with FLT3-ITD is both significantly more frequent and carries a worse prognosis than co-mutation with RAS genes.1,2 To understand the basis of this observation, we investigated the interactions of these mutations in bespoke experimental models (Figure 1A). Analysis of the short-term impact of these mutations on hematopoiesis confirmed that single Npm1cA/+-mutant mice have normal BM cellularity, WCC, and splenic weight.7 As was described before, single Flt3ITD/+ and NrasG12D/+ had moderate but significant increases in splenic size, whereas NrasG12D/+ had raised WCC and BM cellularity.5,6 Introduction of Npm1cA/+ into the NrasG12D/+ background did not alter these parameters significantly, yet the Npm1cA/+;Flt3ITD/+ co-mutation led to a dramatic rise in WCC and splenic size (Figure 1B). At the cellular level, the Npm1cA/+;NrasG12D/+ combination did not change progenitor and stem cell numbers when compared with NrasG12D/+ alone. In contrast, when compared with Flt3ITD/+ mutants, Npm1cA/+;Flt3ITD/+ mice displayed reductions in CMP and MEP and increases in LSK progenitors. Furthermore, Npm1cA/+;Flt3ITD/+ mice showed a profound reduction in phenotypic HSCs (Figure 1C-E).
The differential impact of Npm1cA/+ on Flt3ITD/+ vs NrasG12D/+ was reflected in marked differences in GEPs between double-mutant mice. The Npm1cA/+;NrasG12D/+ model displayed only minimal differences to single NrasG12D/+, whereas Npm1cA/+;Flt3ITD/+ Lin− progenitors had profoundly different GEPs to Flt3ITD/+. From these and complementary analyses of human NPM1c AML, we identify NKX2-3 as a marker of this type of AML. Expression of NKX2-3 distinguishes MLL-AF6 and MLL-ENL from other forms of MLL-mutant leukemia,21,38 highlighting the mechanistic links between NPM1c- and MLL-fusion genes. Here, we show that although potent overexpression of Nkx2.3 by lentivirus may have an impact on self-renewal, genetic disruption of the endogenous Nkx2.3 did not inhibit AML cell growth (Figure 6).
We went on to age double-mutant mice and report that, as with Npm1cA/+;Flt3ITD/+ animals, Npm1cA/+;NrasG12D/+ mice also develop highly penetrant AML, albeit with a much longer latency and a more mature phenotype overall. As single-mutant Flt3ITD/+ and NrasG12D/+ mice had similar survival (Figure 3A), this indicates that the interaction with Npm1cA was central to this difference. To understand the genetic events involved in leukemic progression, we performed exome sequencing and copy number analysis of Npm1cA/+;Flt3ITD/+ and Npm1cA/+;NrasG12D/+ AMLs. Interestingly, the most common somatic event during AML progression was an increase in NrasG12D/+- or Flt3ITD/+-mutant allele burden, through copy-neutral LOH or copy number gain. In human AML, copy-neutral LOH is common for FLT3-ITD, but less so for mutant NRAS; for example, in a recent study we identified only one such LOH event among 13 RAS-mutant human AMLs.13 Nevertheless, in keeping with our findings, studies using the NrasG12D/+ model, in combination with retroviral insertional mutagenesis, resulted in high-penetrance AML with frequent LOH for Nras-G12D when combined with overexpression of oncogenes such as Evi1.6,39 The different incidence of LOH for mutant RAS between murine and human AML may operate through the fact that, in comparison with the acquisition of other oncogenic mutations (eg, Idh1-R132Q in our study), LOH for Nras-G12D may be more expedient in mice given the large numbers of Npm1cA/+/NrasG12D/+ preleukemic HSCs. Other possible reasons may relate to the differences in human-mouse synteny and also the fact that mice are inbred, potentially making recombination events more likely. Notwithstanding mouse-human differences in LOH frequencies, our data provide strong evidence that increased mutant Flt3 and Ras gene dosage are important for leukemic transformation/progression.
Finally, to investigate their role in Npm1c AML, we use CRISPR-Cas9 to disrupt selected genes in Cas9-expressing primary mouse leukemia cells. Using this approach, we confirmed the requirement for the Hoxa9/10 functional gene network in Npm1c AML maintenance. Interestingly, although it is widely appreciated that overexpression of Hoxa9 stimulates leukemic transformation,22,29,33 in our model, disruption of Hoxa10 had a more detrimental impact on AML cell survival, mirroring our recent genome-wide essentiality screen in the NPM1c-harboring OCI-AML3 cell line.11
Our study describes the first faithful mouse model of the interaction of Npm1c with Nras-G12D, the preferred form of oncogenic NRAS in human AML.2 Both NPM1c models share a number of salient characteristics, which are imparted by mutant Npm1, such as homeobox gene overexpression and increased self-renewal of hematopoietic progenitors. However, we demonstrate that the co-occurrence of Npm1c/Flt3-ITD is significantly more leukemogenic and leads to strikingly different molecular and cellular consequences in comparison with Npm1c/Nras-G12D, providing a mechanistic explanation for the higher frequency and worse prognosis of NPM1c/FLT3-ITD AML. Furthermore, through the generation of Cas9-expressing AML models, we also present a versatile approach for the study of genetic interactions in primary mouse leukemias using CRISPR. Although our non-Cas9–expressing Npm1c/Flt3-ITD model was helpful in recent studies of new anti-AML therapies,31 these Cas9-expressing models can be used to study both genetic and pharmacological interactions in parallel and to perform targeted mechanistic studies.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank Gary Gilliland for the Flt3ITD mouse, Kevin Shannon and Tyler Jacks for the NrasG12D mouse, and Mark Dawson for the MLL-AF9 retroviral construct. The authors thank Servicio Santander Supercomputación for their support.
O.M.D., J.L.C., and G.S.V. are funded by a Wellcome Trust Senior Fellowship in Clinical Science (WT095663MA). A.M. was funded by the Kay Kendall Leukaemia Fund project grant (KKL634). C.S.G. was funded by a Bloodwise Clinical Research Training Fellowship. I.V. is funded by Spanish Ministerio de Economía y Competitividad subprograma Ramón y Cajal.
Authorship
Contribution: O.M.D., J.L.C., A.M., C.S.G., C.L., P.G., and G.S.V. performed mouse experiments; O.M.D. and G.S.V. analyzed results; P.W. and M.A. performed histopathological analysis of mouse samples; O.M.D., N.C., R.M.A., and M.S.V. performed transcriptome analysis; I.V. performed analysis of next-generation sequencing; O.M.D, S.P., K.T., and K.Y. performed or supervised CRISPR-Cas9 experiments; O.M.D. and G.S.V. designed the study; and O.M.D. and G.S.V. wrote the paper with the help of R.R., P.W., M.A., and A.B.
Conflict-of-interest disclosure: G.S.V. is a consultant for and holds stock in Kymab, Ltd., and receives an educational grant from Celgene. The remaining authors declare no competing financial interests.
Correspondence: George S. Vassiliou, Wellcome Trust Sanger Institute, Wellcome Trust Genome Campus, Hinxton, Cambridge CB10 1SA, United Kingdom; e-mail: gsv20@sanger.ac.uk.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal