Abstract
Clonal hematopoiesis occurs normally, especially with aging, and in the setting of disease, not only in myeloid cancers but in bone marrow failure as well. In cancer, malignant clones are characterized by recurrent somatic mutations in specific sets of genes, but the direct relationship of such mutations to leukemogenesis, when they occur in cells of an apparently healthy older individual or after recovery from immune aplastic anemia, is uncertain. Here we emphasize a view of clonal evolution that stresses natural selection over deterministic ontogeny, and we stress the selective role of the environment of the marrow and organism. Clonal hematopoieses after chemotherapy, in marrow failure, and with aging serve as models. We caution against the overinterpretation of clinical results of genomic testing in the absence of a better understanding of clonal selection and evolution.
Introduction
Clones, subclones, malignant clones; clonal architecture, clonal expansion, clonal progression, clonal selection; clonal evolution; clonal detection…. The Star Wars allusions are cute, but the terminology is confusing. Confusing to the patient and physician struggling to evaluate increasingly routine laboratory assays of mutations, allele frequencies, and pathogenicity and also to basic science experimentalists attempting to understand normal and malignant hematopoieses. Many excellent reviews and interpretative essays have addressed clones and their significance.1-5 Here our approach is from the field of bone marrow failure, especially of observations of patients and their clones over many years; there are analogies to studies of aging in the normal population. Our perspective will offer a counter to prevailing interest in early detection of potential disease and threatened interventions, the equating of mutations with pathology, and a view of clonal evolution in the marrow that emphasizes frightening terminations (a leukemic cell) rather than powerful and effective natural selection against such cells. We will argue against bias that arises when pathophysiology is equated with physiology, when disease states are extrapolated to normal homeostasis; the fallacy of induction, especially generalization from special cases; and the need to emphasize ignorance so as to ultimately increase understanding.
Define clone
To start, we make some attempt at defining terms and providing framework. Clones, per Wikipedia and others, are identical cell populations. Clonality in the vernacular of the hematology/oncology clinic refers to a uniform population of malignant cells, as in the leukemias. Historically, long before next-generation sequencing (NGS), myeloid and lymphoid neoplasms were recognized as clonal, as inferred from X chromosome genes,6 microsatellites,7 and other gross markers of cell origin; in animal models, cancers can be shown to originate by limiting dilution from injection of a single cell.8
However, clonality also applies to normal hematopoiesis. Inferences from large animals and humans suggest that normal hematopoiesis is supported by only a few hundreds to thousands of stem cells in a young adult at steady state.9-13 Normal hematopoietic clones can be readily visualized: in colony-forming assays, in macroscopic spleen colonies in the mouse,14 and in 3-dimensional cultures of primitive progenitors.15 Clones of normal cells in humans can also be inferred from markers, such as mitochondrial DNA sequence,16 measurement of acquired somatic variants,17,18 and with other methods to determine natural mosaicism.19
Furthermore, “identical” is a loose term; we know from single-cell studies that cell populations are extremely heterogeneous,20-22 and perhaps no cell is identical to its neighbor, whether in a tissue culture flask or in a tissue. Cells that undergo replication invariably acquire mutations over time,23 and even the resting genome is susceptible to permanent alterations, as by deamination of cytosines.24 By middle age, normal hematopoietic progenitor cells have acquired so-called passenger mutations, random background variants that do not affect cellular fitness, on the order of 10 to 20 per cell in coding regions of the genome.25 Cells vary also in their epigenomes, highly specific regions of DNA methylation and histone acetylation that alter the accessibility of DNA to regulatory molecular machines26 ; in their metabolic states27 ; and in topographic polarization.28 NGS has allowed deep and detailed examination of cell variability. NGS has shown that leukemias, and cancers in general, are mixed populations of cells in which subclones have acquired critical mutations.29,30 Thus, our use of the term clone in various iterations is more descriptive than prescriptive.
Clonal evolution and evolution
Of course, clonal evolution (progression, selection) and not the stable existence of tiny populations of even abnormal cells is the practical concern. Evolution can mean gradual development, with increasing complexity and diversity implied. Indeed, the application of the fundamentals of Darwinian evolution to hematopoiesis emphasizes ontogeny: progression, development, and maturation to an end.31 For both normal and abnormal hematopoieses, hematologists have reconstructed pathways backward, from blood cells to their marrow precursors originally by morphology, and now from cancer cells to their parental cells by sequencing of mutations in our own era.32 However, the popular view of evolution, as the acquisition of characteristics culminating in an existing species, obscures both the dominant role of selection, selection by the environment of the organism, and the high prevalence of extinction events (Figure 1).33 The critical, dynamic role of environmental factors is much more difficult to surmise than are the favorable (or unfavorable) features of the winning species (or cell). The fossil record of animals, or the detection of genetic changes in a cancer, can be measured, but the catastrophic or subtle macro- or microenvironmental events that have shaped selection usually must be imputed indirectly or indeed are subject to speculation.
Depictions of Darwinian evolution (after Gould33). The classic phylogenetic tree (A) entails both adaptation (B) and elimination (C). Emphasis is commonly on the former, ontogeny, as in the display of precursors of current forms of life, with the development of modern man the paragon (D). Natural selection, however, produces many extinct species; the enormous variety and abundance of dinosaur species are underrepresented in the family tree of reptiles, and the diversity of surviving species of this class is limited (E). As Gould emphasized, our focus is on the living end product, not the lost dead ends. A physician’s parents proudly frame the baby picture with stethoscope (F), but most childhood fantasies are not fulfilled and indeed forgotten. These views of evolution apply not only to species and individual organisms but also to diverse cells and cell populations within the bone marrow.
Depictions of Darwinian evolution (after Gould33). The classic phylogenetic tree (A) entails both adaptation (B) and elimination (C). Emphasis is commonly on the former, ontogeny, as in the display of precursors of current forms of life, with the development of modern man the paragon (D). Natural selection, however, produces many extinct species; the enormous variety and abundance of dinosaur species are underrepresented in the family tree of reptiles, and the diversity of surviving species of this class is limited (E). As Gould emphasized, our focus is on the living end product, not the lost dead ends. A physician’s parents proudly frame the baby picture with stethoscope (F), but most childhood fantasies are not fulfilled and indeed forgotten. These views of evolution apply not only to species and individual organisms but also to diverse cells and cell populations within the bone marrow.
Models of leukemogenesis often depict clonal evolution as a stepwise process,32,34 in which oncogenic mutations are serially acquired, and the environmental component of natural selection is omitted. The emphasis on cell-intrinsic factors implies that the acquisition of favorable characteristics that increase cell proliferation, resistance to cell death, and propensity to differentiate is sufficient for clonal dominance. However, adaptation is more complex; there are costs to new characteristics, to the organism or to the cell (eg, in energy expenditures and structural alterations), and acquired features must always be advantageous relative to survival within a specific environment. Mutations that favor a leukemic cell in a dysregulated tumor microenvironment may not necessarily improve fitness in healthy bone marrow; indeed, that these polymorphisms are rarely seen in the germ line argues against their being optimal under normal conditions.35 Natural selection does not simply result from changes in a cell, and a shifting environment, whether sudden or gradual, will alter the fitness of existing heterogeneous cell populations and favor clonal selection.
Preleukemia vs normal variation
Approximately 50 genes are recurrently mutated in myelodysplastic syndrome (MDS) and acute myeloid leukemia (AML).36-39 Is the presence of such a mutation in an otherwise healthy individual preleukemia? Steensma et al32 coined CHIP, clonal hematopoiesis of indeterminate potential, in an attempt to categorize such a laboratory finding and distinguish it from true myeloid neoplasia. Explicit analogies to conditions with low rates of malignant progression, such as monoclonal gammopathy of unknown significance or monoclonal B-cell lymphocytosis, were intended to alleviate concerns that detection of CHIP signified that an individual was destined to develop MDS/AML.
In seminal publications, clonal hematopoiesis was detected as mutations in genes such as DNMT3A, TET2, or ASXL1 and as small clones in a sizable percentage of so-called normal persons in the fifth and sixth decades of life.17,18,40 Furthermore, the presence of clonal hematopoiesis was associated with a small age-adjusted risk of developing a hematologic malignancy as well as increased risk of overall mortality.17,18 Hematologic malignancy was defined broadly in these reports and included not only myeloid neoplasms but also lymphomas and monoclonal gammopathy of unknown significance. Although graphical representations depict CHIP as a first hit in progression to MDS or leukemia,32,34 there is limited evidence to support these models. In 1 study of 3342 individuals, there were 16 participants who subsequently developed a hematologic malignancy, of whom only 5 had a detectable candidate gene mutation; only 2 individuals were diagnosed with MDS/AML, 1 of whom had a prior history of non-Hodgkin lymphoma (so perhaps not truly healthy).18 Of more than 12 000 individuals screened in the Swedish report of aging-associated CHIP, 37 without a prior history of malignancy developed a hematologic cancer; a candidate gene mutation was detected in 10 of these 37, but only 5 of them had MDS/AML.17 In both studies, a majority of hematologic malignancies developed without antecedent detectable CHIP. Additionally, as with any population-based survey, only association and not causality could be inferred from any relationship between CHIP and disease risk.
The initial reports of clonal hematopoiesis in aging were limited in distinguishing true mutations at a low variant allele fraction (VAF) from artifact; the proposed definition of CHIP therefore used a minimum VAF threshold of 2.5%.32 Later investigations employed error-corrected sequencing techniques, which enabled ultrasensitive detection of somatic variants.41 When deeper sequencing was extended to younger persons, remarkably almost all individuals in their 50s had at least 1 small clone, most commonly harboring a DNMT3A or TET2 mutation. Clone sizes remained stable and small (<1% VAF) over a 10-year period, suggesting that acquisition of a mutation alone was insufficient to drive continuous clonal expansion.42 It is also notable that clonal hematopoiesis frequently occurs in the absence of a detectable MDS/AML candidate gene mutation. In a recent novel approach, a consortium of physicians, engineers, and anthropologists performed whole-genome sequencing of blood of >11 000 Icelanders, using barcoding to detect mosaic somatic mutations. Confirming previous large-scale work from Sweden,17 the prevalence of clonal hematopoiesis approached 50% in individuals entering the ninth decade of life.18,43 However, although clonal hematopoiesis was associated with shorter survival in the Icelanders, the risk of adverse outcomes (ie, death or hematologic malignancy) did not correlate specifically with MDS/AML candidate gene driver mutations. In a majority of cases, clonal hematopoiesis occurred in the absence of an MDS/AML-associated gene mutation. Simulations suggested that clonal hematopoiesis could arise stochastically by neutral drift within a diminished stem-cell pool.43 CHIP seems to be a biomarker that globally reflects an individual’s lifetime exposure to a genotoxic environment and/or susceptibility to cellular damage, as evidenced by the strong association between CHIP and cardiovascular disease,18,44 but the direct connection between CHIP and leukemogenesis, at least in the case of aging, is unclear. (Accelerated telomere attrition also has many negative medical associations45-48 and may similarly be a biomarker of cumulative unrepaired DNA damage.)
That small clones driven by somatic mutations in AML/MDS genes seem to be a near-universal feature of aging argues that acquisition of mutations is usually not the rate-limiting step in the initiation of malignant clonal hematopoiesis. Indeed, many current data from normal aging studies can be interpreted as not providing support for MDS/AML mutations as major risk factors in themselves for the development of myeloid malignancy. The ubiquity in normal tissues of somatic mutations in cancer-associated genes is not limited to hematopoietic cells.49,50 Far from rare, somatic mutations in cancer-associated genes seem to be a common feature of aging, and clonal hematopoiesis, with or without a candidate driver mutation, a biomarker for adverse aging-associated outcomes.
As we have described, normal hematopoiesis is clonal, with blood cells originating from a limited number of stem and progenitor cells and production shifting over time among various populations of parental cells. Experimentally distinguishing among normal human cells is not easy; even mouse experiments using genetically modified or barcoded stem cells to track clonal succession have yielded controversial conclusions, which of course may not be predictive of human marrow-cell behavior.9,51-55 Nevertheless, from simple inference of declining marrow cellularity with aging,56 poor hematologic tolerance to chemotherapy in the elderly,57 and the superior behavior of grafts from young vs old donors,58 it is likely that the number of functional stem cells is reduced with chronologic aging. The observation of a single highly mutated clone that had entirely supported hematopoiesis in a supercentenarian is perhaps a salutary example of the most common outcome of clonal hematopoiesis in aging, which is no clinical consequence.59 Small, even heavily mutated clones likely are clinically innocuous in a large majority of individuals.
Clonal hematopoiesis in bone marrow failure
Further evidence that clones containing MDS/AML gene mutations are not strongly linked to leukemogenesis comes from studies of bone marrow failure. Paroxysmal nocturnal hemoglobinuria (PNH) provides historical and powerful evidence of the benignity of clonal hematopoiesis. The etiology of PNH is an acquired mutation in PIGA; clonality is easily detectable in the clinical flow cytometry laboratory using sensitive methods to quantify missing cell-surface membrane proteins. PNH is strongly associated with marrow failure, deficient clones are present in more than half of patients with aplastic anemia (AA), and patients with hemolytic PNH commonly have deficiencies in hematopoiesis.60 Remarkably, tiny clonal populations of stem cells with a PNH phenotype and PIGA somatic mutation can be commonly detected in normal individuals.61 However, clonal expansion of PIGA-mutated cells to PNH is exceedingly rare.62 NGS of PNH clones, some of which likely had sustained hematopoiesis in individual patients for many years, disclosed a multitude of somatic mutations in candidate genes such as TET2, JAK2, and ASXL1. The emergence of subclones suggests serial acquisition of mutations that improves clonal fitness.63 Nevertheless, progression to leukemia in patients with PNH and from PNH clones in patients with marrow failure is rare.64-66
In acquired AA, which in most patients improves with immunosuppressive therapies, the disease evolves in a significant minority of patients, including hematologic responders, to MDS and AML, sometimes years after successful treatment.67 AA represents an opportunity to examine early events in leukemogenesis, especially when serial samples have been obtained before development of myeloid malignancy. Recent studies in bone marrow failure syndromes have been cited as supporting a connection between MDS/AML and clonal hematopoiesis, but again, the direct relationship between the 2 is not clear. From targeted sequencing of 150 cases of idiopathic bone marrow failure at King’s College, the presence of a somatic mutation was reported to be strongly predictive of the risk of clonal evolution.68 Two thirds of patients who experienced clonal evolution to MDS/AML had evidence of mutated clones at an earlier time point, and more than half of the patients with a somatic mutation in ASXL1 experienced progression to MDS. Our own larger study did not confirm these results.69 In this collaborative work, the presence of any single acquired somatic mutation was surprisingly not an independent risk factor for evolution. Clonal hematopoiesis involving a small cassette of mutated genes (BCOR1, DMNT3A, ASXL1) was seen in both evolvers and nonevolvers; machine learning was needed to identify “favorable” vs “unfavorable” mutations. Patients who responded to immunosuppressive therapy and did not experience evolution to MDS were more likely to have somatic clones with mutations in a set of favorable genes (BCOR and PIGA). Conversely, patients who developed MDS/AML were more likely to have mutations in a set of unfavorable genes (DNMT3A, ASXL1, RUNX1, and others). The relationship between unfavorable mutations and survival was more apparent than that between unfavorable mutations and progression to MDS/AML, and most patients with an unfavorable mutation have not developed MDS/AML. In the National Institutes of Health cohort, of the 17 patients with a somatic mutation in ASXL1, only 4 experienced clonal evolution to MDS, and only 3 of the 18 patients with somatic mutations in DNMT3A developed MDS.69
Serial tracking of somatic clones in AA further illustrates that clonal hematopoiesis is a complex and nondeterministic process. At the time of diagnosis, most clones were detected at a low VAF. Over years of follow-up, clones grow, shrink, or remain stable, with little connection to a patient’s blood counts. Clones bearing either ASXL1 or DNMT3A mutations tended to expand over time, and clones bearing mutations in BCOR and PIGA were more stable, but these trends were not universal. In some patients, small clones with unfavorable mutations were stable or diminished in size with observation.69
Nor were somatically mutated clones strongly associated with malignant transformation, historically labeled clonal evolution, the most serious late event in AA. Clonal evolution is typically diagnosed on the appearance of monosomy 7 and clinically often followed by refractory cytopenias, MDS, and AML. Our efforts to associate 7− with somatic mutations in MDS/AML genes were not successful. In 13 cases of AA that had progressed to 7− in which samples of marrow and blood had been collected annually, clonal hematopoesis was common, but most clones did not have candidate gene mutations, and in others, small mutated clones were not of obvious clinical significance.70 In larger cohorts, telomere length and attrition have been the major risk factors for development of 7− and evolution to MDS/AML.47 Telomere length correlated with evolution in patients treated with the combination of eltrombopag and standard immunosuppression, most of whom did not have recurrently mutated clones when 7− was detected.71 In refractory AA treated with eltrombopag alone, progression usually occurred without evidence of either the presence of a mutated clone or expansion of a clone, and conversely, mutated clones were detected in other patients in stable hematologic recovery (T. Winker and J.N.C., unpublished data).
Clonal dynamics in MDS compared with AA
Somatically mutated clones are prevalent in MDS.38 Mutations in epigenetic regulators and RNA splicing genes are early (founder, type 1) events, whereas high-risk cytogenetic abnormalities and mutations in transcription factors and growth signaling pathway genes occur later and presumably promote malignant transformation to AML.72,73 Studies that have serially tracked somatic clones in patients with MDS reveal several patterns. Expansion of a clone or subclone after acquisition of specific mutations (in FLT3 or the RAS family) may precede leukemic transformation in a pattern of linear evolution. In other cases, clones may expand and achieve clonal dominance for months or years only to be overtaken by an another subclone or even a novel clone (clone sweeping).72,73 Thus, as in AA, somatic clones in MDS can stably support hematopoiesis for years without changes in blood counts or evolution. In general, clinical characteristics, such as age and karyotype, have been far more predictive of patient outcome than have mutations, and genomic experts and attending hematologists have struggled to establish a role for deep-sequencing results.74-76 Some recent data do suggest a stronger relationship between the presence of somatically mutated clones and a true development of MDS.77,78 Deep sequencing for recurrently mutated MDS/AML genes was performed in a large number of cytopenic patients referred to a large European clinic. On average, anemia, thrombocytopenia, and neutropenia were mild, and mutated clones (mainly in TET2, DNMT3A, and ASXL1) were at low VAF. Strikingly, the presence of ≥1 clone in the setting of a cytopenia was almost 100% predictive of developing a frank myeloid malignancy.78 At present, hematologists face a conundrum: somatic clones, mutated in the same genes and at similar allele frequencies as in the healthy aged (in whom they are not particularly related to myeloid neoplasms), seem to be prognostic of MDS and AML when a mild cytopenia is also present.
Clonal expansion and the environment
Cells are more complicated than their genes, and organs are more than a collection of cells. Whatever their multifold intrinsic properties, cells exist in a complex environment of other cells, proteins, and chemicals. High-throughput sequencing is a powerful method to detect and track clonal hematopoiesis, but sequence alone does not fully determine the fate of a patient or a cell. DNA sequence is an intrinsic cell property, as are telomeres, epigenetic modifications, topographic polarization, and accumulated oxidative damage to mitochondria and cytoplasmic proteins, to name only a few examples of fundamental processes affecting cell fitness that change with normal aging and in disease.79 Sequencing cannot measure cell-extrinsic factors, but both the occurrence of mutations and the growth of mutated clones must reflect environment, and they may be imputed to characterize selective environmental influences. Experimental models have established that alterations in the bone marrow niche in both the mouse and human can perturb normal hematopoiesis as well as promote malignant transformation.27,80 In the aging mouse, alterations in bone marrow niche vascularization and changes to local cytokine signaling shift murine hematopoesis.81,82 Similarly, physicochemical toxicity perturbs hematopoiesis by altering both the stem-/progenitor-cell compartments and stroma.83-88 Acute and chronic inflammations alter normal hematopoiesis and remain underappreciated forces in the shaping of clonal architecture.89,90
Heterogeneity of cell-extrinsic factors may explain why specific somatic mutations and their clones arise under different clinical conditions. Peculiarly, somatic mutations that are not associated with aging (BCOR/BCORL1, PIGA) are commonly observed in AA.69 CHIP in healthy aging is predominantly driven by DNMT3A mutations, but CHIP occurring with a nonmyeloid malignancy is most frequently associated with mutations in TP53 or TET2.91,92 Even the selective pressures that drive clonal hematopoiesis in aging seem to vary with increasing age. Somatic mutations in DNMT3A or TET2 increase steadily after the fifth decade of life, and mutations in spliceosome genes are largely detected in the seventh decade and beyond.40 These patterns do not seem random, and they indirectly suggest that variation in cell-extrinsic factors, with aging and disease, shape clonal architecture.
Imputing environment from mutations and clonal architecture
We use 3 examples of altered hematopoietic homeostasis associated with clonal expansion of mutated cells. All involve some element of bone marrow failure and regeneration, but they vary clinically, in probability of subsequent malignant transformation and in kinetics over time, in addition to how well they have been characterized (Figures 2 and 3).
Clonal hematopoiesis and the environment. The bone marrow contains large numbers of cells (difficult to depict graphically), which by adulthood have stochastically accumulated mutations, most presumably without biological consequence (these cells and clones are shown with variable gray shading). By chance alone, mutations will also occur in genes that do affect function; a cell mutated in the PIGA gene is shown in normal adult bone marrow, because there is experimental evidence that such cells can be isolated from healthy donors.61 Whether such mutated cells expand to clones is highly dependent on extrinsic conditions. Chemotherapy is the most dramatic example of an environmental perturbation, shrinking (temporarily) the stem and progenitor compartment, effecting genotoxicity, and producing immediate regenerative stress. Selective pressure of the chemotherapy environment favors cells resistant to apoptotic signals, as has been demonstrated for TP53-mutated clones.91,92,97 In immune bone marrow failure, cells not recognized or resistant to immune destruction would be favored: PIGA-mutated cells and cells that have partially lost HLA genes. Additionally, an environment enriched for hematopoietic growth factors and other proliferative signals would allow passive accumulation of cells that normally respond poorly to such stimuli. The aging environment is not yet fully defined, but clonal hematopoiesis likely reflects a diminished stem-cell pool and a lifetime of intermittent inflammation and concurrent diseases, toxic exposures, and many other factors (Figure 3). HSC, hematopoietic stem cell.
Clonal hematopoiesis and the environment. The bone marrow contains large numbers of cells (difficult to depict graphically), which by adulthood have stochastically accumulated mutations, most presumably without biological consequence (these cells and clones are shown with variable gray shading). By chance alone, mutations will also occur in genes that do affect function; a cell mutated in the PIGA gene is shown in normal adult bone marrow, because there is experimental evidence that such cells can be isolated from healthy donors.61 Whether such mutated cells expand to clones is highly dependent on extrinsic conditions. Chemotherapy is the most dramatic example of an environmental perturbation, shrinking (temporarily) the stem and progenitor compartment, effecting genotoxicity, and producing immediate regenerative stress. Selective pressure of the chemotherapy environment favors cells resistant to apoptotic signals, as has been demonstrated for TP53-mutated clones.91,92,97 In immune bone marrow failure, cells not recognized or resistant to immune destruction would be favored: PIGA-mutated cells and cells that have partially lost HLA genes. Additionally, an environment enriched for hematopoietic growth factors and other proliferative signals would allow passive accumulation of cells that normally respond poorly to such stimuli. The aging environment is not yet fully defined, but clonal hematopoiesis likely reflects a diminished stem-cell pool and a lifetime of intermittent inflammation and concurrent diseases, toxic exposures, and many other factors (Figure 3). HSC, hematopoietic stem cell.
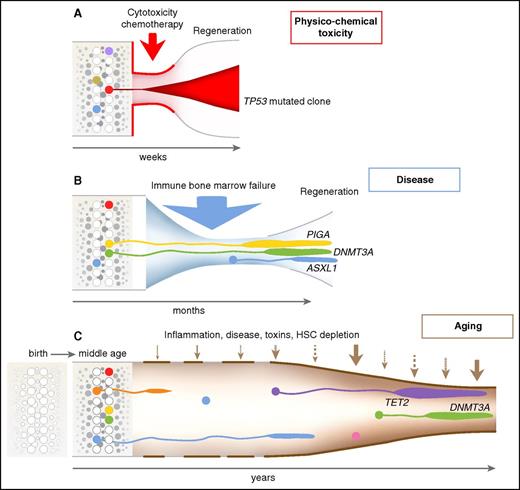
Dynamics of clonal selection. Time and the environment are emphasized in this visualization of hematopoiesis, following the classic diagrams of clonal evolution in myelodysplasia and AML.37 (A) Cytotoxic chemotherapy produces an acute reduction in hematopoiesis because of strong cell toxicity, in which survival of a cell resistant to apoptotic signals is strongly favored. (B) In immune bone marrow failure, as in AA and hypoplastic MDS, cells resistant to immune attack will be favored, as will cells that thrive in a regenerative environment depleted of normal progenitors but rich in hematopoietic growth factors. (C) Many poorly defined factors likely influence clonal selection with aging. Remarkably, of genes identified as recurrently mutated in myeloid malignancies, limited cassettes are mutated under these environmental stresses: TP53 after chemotherapy; DNMT3A, ASXL1, and BCOR (and PIGA, not a leukemia gene) in aplastic anemia; and TET2, DNMT3A, and ASXL1 in CHIP. HSC, hematopoietic stem cell.
Dynamics of clonal selection. Time and the environment are emphasized in this visualization of hematopoiesis, following the classic diagrams of clonal evolution in myelodysplasia and AML.37 (A) Cytotoxic chemotherapy produces an acute reduction in hematopoiesis because of strong cell toxicity, in which survival of a cell resistant to apoptotic signals is strongly favored. (B) In immune bone marrow failure, as in AA and hypoplastic MDS, cells resistant to immune attack will be favored, as will cells that thrive in a regenerative environment depleted of normal progenitors but rich in hematopoietic growth factors. (C) Many poorly defined factors likely influence clonal selection with aging. Remarkably, of genes identified as recurrently mutated in myeloid malignancies, limited cassettes are mutated under these environmental stresses: TP53 after chemotherapy; DNMT3A, ASXL1, and BCOR (and PIGA, not a leukemia gene) in aplastic anemia; and TET2, DNMT3A, and ASXL1 in CHIP. HSC, hematopoietic stem cell.
Secondary MDS/AML after chemotherapy
MDS and AML are dreaded late effects of chemotherapy or radiation treatments for cancer. These are clinically obvious, readily dated, and powerful perturbations of the stem-cell compartment with well-defined outcomes in recovery from inevitable cytotoxicity (pancytopenia and marrow aplasia are common) and less frequent but unfortunately not rare late malignancies (predominantly secondary MDS and AML).93 Chemotherapy is a powerful environmental stress, skewing the stem-cell population toward cells that are more resistant to apoptosis and tolerant of DNA damage.94 Genotoxicity from cytotoxic therapies would also be expected, in the form of point mutations, double-stranded DNA breaks,95 and telomere attrition,96 to increase genetic diversity of surviving stem cells in a highly selective environment. NGS has been employed to track somatic clones in patients with lymphomas and solid tumors after chemotherapy.91,92,97-99 Small somatic clones were common before the initiation of cancer therapy and were associated with an increased risk of therapy-related MDS/AML years later. In particular, small clones with loss-of-function mutations in the tumor suppressor gene TP53 expanded and eventually evolved into leukemia.91,92 However, in other patients, somatically mutated clones (TP53, TET2) decreased or disappeared after chemotherapy.91 These results are the clearest examples of external environmental pressures shaping clonal hematopoiesis and promoting leukemic transformation.
Immune marrow failure
In AA, the immune system mediates the destruction of hematopoietic stem cells. Clonal evolution in AA is the development of MDS and AML in survivors, many months to years after initial, often successful immunosuppressive therapy.67 Cytotoxic T cells, their cytokines, and Fas-mediated apoptosis have been implicated in immune eradication of marrow targets.100 (Broadly similar mechanisms likely operate in some patients in the early stages of MDS,89,101 in large granular lymphocytosis, and in single-lineage marrow failure.)
The pathophysiology of immune destruction and its dependence on presentation of an (unknown) autoantigen by class I human leukocyte antigens is reflected in somatic mutation or copy-number–neutral loss of heterozygosity on chromosome 6p, the most common marker of clonal hematopoiesis in AA.102,103 Immune escape has been hypothesized to occur in PNH, uniquely associated with AA.104-106 Small populations of clonal stem cells of PNH phenotype are readily detectable in healthy individuals; PNH as a disease is exceedingly rare.61 PNH cells do not have an intrinsic growth advantage; external selective pressure must drive clonal expansion of glycosylphosphatidylinositol-negative hematopoietic stem cells.107 The glycolipid anchor itself might serve as the target antigen.62
Immune destruction creates hematopoietic stress. Thrombopoietin levels remain high for years in recovered patients with AA, providing a persistent drive on replication of stem cells.108 A relatively short course of eltrombopag, a thrombopoietin mimetic, seems to rapidly elicit proliferation of (presumably) dormant monosomy 7 cells in some patients with refractory or treatment-naïve disease, a dramatic example of clonal responsiveness to a discrete stimulus.109
Aging
Aging also selects for hematopoietic clones carrying specific mutations. CD34 cells accumulate mutations with chronologic aging, with random accumulation of ∼10 passenger exonic mutations per cell by late middle life.25 Aging of the marrow compartment has been difficult to study in humans, and mice may be particularly inapt as models.110 Diversity within the stem-cell compartment of the marrow and in all other tissues is the result of a lifetime of poorly defined but indisputable low-level toxic environmental exposures and harmful endogenous metabolites, as well as acute and chronic infections.111-114 Clones may also be blamed on a senescent immune system increasingly taxed with surveillance and elimination of genetically aberrant cells.115 Nevertheless, some of the same mechanisms implicated above likely operate to favor persistence of clones, combined with selection in an environment of continued regenerative demand from a shrinking stem-cell pool. C to T transitions, arising from deamination of cytosines, are the prevalent mutations in both normal aging and in patients with immune marrow failure.17,18,69 In both circumstances, cells unable to differentiate normally would passively accumulate over time, self-renewing rather than entering pathways of terminal differentiation; cells able to evade an aging immune system that has narrowed its repertory, especially among cytotoxic effector cells, will be favored. Increased genetic diversity and an altered local environment would be produced by the same genotoxic and selective factors but with more subtlety and over many more years.
Future directions
The culmination of the genome project justifiably created excitement and expectations. “Omics” technologies have produced precision medicine, with readily available molecular diagnostics, highly directed therapeutic strategies, and patient-specific treatments.116-118 CHIP follows this paradigm. For example, treatment regimens in future might be modified to minimize the risk of therapy-related MDS when somatic clones with TP53 mutations are detected at diagnosis. Small somatic clones in the hematopoietic compartment may be susceptible to targeted molecular therapies, preemptive introduction of hypomethylating agents, and even early use of myeloablative chemotherapy. However, as the original authors who defined CHIP prudently advised, the clinical significance and biological reasons for clonal hematopoiesis remain unclear, and currently widespread screening for CHIP would be of little benefit.32
We offer some additional cautions. The proposed definition of CHIP sets a lower VAF boundary for clonal hematopoiesis, but advances in sequencing capabilities reveal that small clones at a VAF <2.5% are present in most middle-aged individuals.42 It does not seem plausible that the risk of malignant transformation of a somatic clone is simply a function of its size. Rather, the clinical context in which clones appear likely strongly influences their risk of malignant transformation. For example, unexplained is the extraordinary discordance between the risk of MDS/AML conveyed by CHIP in a normal aging person compared with a similar clone in an older person with a cytopenia of unknown significance, in which CHIP is reported to be almost absolutely predictive of short-term development of MDS.78 There are other questions. Why are TP53-mutated clones prevalent in the marrow of patients with cancer before chemotherapy? What maintains stable clonality in a patient with AA, and why are clones not more prone to further evolution in so many cases? How do somatic mutations relate to chromosome aneuploidy, so common among malignancies in general and after chemotherapy and with immune marrow failure, as well as in aging?119 How does the immune system contribute to the generation of clonal diversity? Might T-cell responses in marrow failure occur in response to clonal expansion, with immune surveillance the normal constraint to clonal disease of the marrow, and failed immune surveillance part of the pathophysiology of early MDS and aplastic anemia? Which environmental factors favor clonal hematopoiesis? Are they always leukemogenic? Are they susceptible to manipulation?
Answering these questions requires an approach to studying clonal hematopoiesis beyond sequencing. Additional work is required to understand changes to the intrinsic properties of somatically mutated clones. Recent technical innovations have enabled the measurement of epigenetic (histone modifications,120 DNA accessibility,121,122 DNA methylation123 ) and gene expression patterns124 with limited cellular inputs or even single cells, allowing the study of rare cell populations from human sources. These methods may better differentiate how clonal hematopoiesis varies in various contexts, such as in aging, in marrow failure, and after cytotoxic therapy. The role of telomere attrition in chromosomal changes that are frequent in cancer in general and with clonal evolution requires further exploration.45,47,125 Understanding how cell-extrinsic factors shape clonal hematopoiesis will require longitudinal tracking of individuals with CHIP to identify the modulators of clonal stability, expansion, and extinction and their role in the early stages of oncogenesis. For example, tracking clones during immunomodulatory cancer therapy is an opportunity to better understand the still hypothetical relationship between somatic clones and the immune environment. Patients with chronic and episodic inflammatory disorders represent another opportunity to study CHIP in the context of inflammation and in the setting of therapy that affect the immune system.126
In this article, we have stressed the limitations of thinking of oncogenesis as a linear process that commences with a single mutation and progresses stepwise and inexorably toward leukemia. For this reason, we urge caution as clinicians incorporate sequencing into clinical practice. NGS is increasingly routine, and explaining the significance of a small clone to a patient is immensely difficult. What is certain is the anxiety among patients and physicians, as well as the risk of overtreatment in the absence of adequate evidence. Sophisticated testing of the genome (inherited and acquired mutations, telomere length, epigenetic changes, and others) may have the potential for broad predictive value. At worst, clinical testing of clonal hematopoiesis in the blood will only frighten the preoccupied patient with a dire prognosis and lead to reactive overtreatment by physicians. At best, clinical research will uncover those environmental causes of global DNA and cell damage that are reflected in genome testing and amenable to effective prevention.
Acknowledgments
The authors thank readers of draft versions of this manuscript, Cynthia Dunbar and Thomas Winkler.
Authorship
Contribution: J.N.C. and N.S.Y. conceived this review article, analyzed the literature, and wrote the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: James N. Cooper, Hatfield Clinical Center, Room 3E-5140, 10 Center Dr, NIH, Bethesda, MD 20892-1202; e-mail: cooperjam@nih.gov; Neal S. Young, Hatfield Clinical Center, Room 3E-5140, 10 Center Dr, NIH, Bethesda, MD 20892-1202; e-mail: youngns@mail.nih.gov.