Key Points
Enasidenib inhibits mIDH2, leading to leukemic cell differentiation with emergence of functional mIDH2 neutrophils in rrAML patients.
RAS pathway mutations and increased mutational burden overall are associated with a decreased response rate to mIDH2 inhibition.
Abstract
Recurrent mutations at R140 and R172 in isocitrate dehydrogenase 2 (IDH2) occur in many cancers, including ∼12% of acute myeloid leukemia (AML). In preclinical models these mutations cause accumulation of the oncogenic metabolite R-2-hydroxyglutarate (2-HG) and induce hematopoietic differentiation block. Single-agent enasidenib (AG-221/CC-90007), a selective mutant IDH2 (mIDH2) inhibitor, produced an overall response rate of 40.3% in relapsed/refractory AML (rrAML) patients with mIDH2 in a phase 1 trial. However, its mechanism of action and biomarkers associated with response remain unclear. Here, we measured 2-HG, mIDH2 allele burden, and co-occurring somatic mutations in sequential patient samples from the clinical trial and correlated these with clinical response. Furthermore, we used flow cytometry to assess inhibition of mIDH2 on hematopoietic differentiation. We observed potent 2-HG suppression in both R140 and R172 mIDH2 AML subtypes, with different kinetics, which preceded clinical response. Suppression of 2-HG alone did not predict response, because most nonresponding patients also exhibited 2-HG suppression. Complete remission (CR) with persistence of mIDH2 and normalization of hematopoietic stem and progenitor compartments with emergence of functional mIDH2 neutrophils were observed. In a subset of CR patients, mIDH2 allele burden was reduced and remained undetectable with response. Co-occurring mutations in NRAS and other MAPK pathway effectors were enriched in nonresponding patients, consistent with RAS signaling contributing to primary therapeutic resistance. Together, these data support differentiation as the main mechanism of enasidenib efficacy in relapsed/refractory AML patients and provide insight into resistance mechanisms to inform future mechanism-based combination treatment studies.
Introduction
Somatic mutations in the isocitrate dehydrogenase 2 (IDH2) gene occur at conserved arginine residues (R140 and R172). These mutant proteins possess neomorphic enzymatic activity resulting in R-2-hydroxyglutarate (R-2-HG) accumulation.1-4 R-2-HG competitively inhibits a set of α-ketoglutarate-dependent enzymes, including the Ten-eleven translocation family of 5-methylcytosine hydroxylases and the Jumonji-C domain histone demethylases.5,6 This inhibition leads to DNA hypermethylation,7 increased repressive histone methylation,6 and impaired hematopoietic differentiation. Accordingly, inhibition of mutant IDH2 (mIDH2) reduces 2-HG levels and restores hematopoietic differentiation in vitro.6,8-10
Although both mutations are characterized by neomorphic enzymatic activity, myeloid malignancies with R140 and R172 IDH2 mutations are distinct with respect to clinical outcome, comutational profile, and molecular classification.11-13 In preclinical studies, enasidenib (AG-221/CC-90007), a small-molecule inhibitor of mIDH2, reduced serum 2-HG, DNA hypermethylation, and repressive histone marks and promoted hematopoietic differentiation in R140 and R172 mIDH2 models.14-17 In a phase 1/2 clinical trial, enasidenib demonstrated clinical activity in patients with both R140 and R172 mIDH2 relapsed/refractory AML (rrAML) with an overall response rate (ORR) of 40.3%.18 Here, we analyzed samples from this study to elucidate the mechanisms of action of enasidenib in R140 and R172 mIDH2 rrAML patients and to identify response biomarkers to targeted mIDH2 therapy.
Materials and methods
Study participants and treatment
Analyses were performed on rrAML patient samples collected from the AG-221-C-001 study with informed consent. Patients were included in the AG-221-C-001 trial and in the translational studies here based on IDH2 mutations detected by local testing. A retrospective central in vitro diagnostic test confirmed mutation status with a concordance rate above 95%. Enasidenib was administered to patients as described in the approved study protocol. Patient sample disposition indicating samples analyzed in each assay is shown in supplemental Figure 1 (available on the Blood Web site). Patient baseline characteristics and clinical responses of subpopulations analyzed in comparison with the rrAML cohort as a whole are provided in supplemental Table 1. Outcome data reflect a study cutoff date of 15 April 2016.
Measurement and analysis of 2-HG
Serum samples were collected from patients at screening within 28 days before the first dose of enasidenib or predose on day 1 of each treatment cycle. 2-HG concentration was determined with liquid chromatography tandem mass spectrometry by Covance, Inc. (formerly Tandem Labs), according to an analytically validated method. Baseline total 2-HG was determined to be either the average of the screening sample and the predose cycle 1 sample or either one or the other sample if both were not available. Maximum suppression of 2-HG levels was determined by comparing the lowest level of 2-HG observed on-treatment to baseline 2-HG level. Time to maximum suppression was the first time point at which the 2-HG level was within 5% of maximum suppression for that patient.
Sysmex OncoBeam digital polymerase chain reaction
Bone marrow, peripheral blood samples, or both were collected at screening 28 days before the first dose of enasidenib and during treatment and were processed to peripheral blood mononuclear cells and bone marrow mononuclear cells (BMMCs). Measurement of mutant and wild-type IDH2 was made with Sysmex BEAMing technology. Briefly, DNA was extracted from the samples, preamplified in a multiplex polymerase chain reaction (PCR) and amplified with nested primers in an emulsion PCR on the surface of magnetic beads in water-in-oil emulsions. Fluorescently labeled probes specific to the IDH2 mutation and to the wild-type sequence were hybridized to the uncovered DNA fragments on the bead surface. Fluorescently labeled beads were quantified with flow cytometry.
Blast percentage determination by flow and mIDH2 assessment
Multiparameter flow cytometry was performed on bone marrow aspirates at diagnosis and at day 1 of each treatment cycle. Abnormal populations were identified by antigen expression as described previously.19,20 Briefly, up to 1.5 million cells from freshly drawn bone marrow aspirate were stained with 3 10-“color” panels (supplemental Table 2), washed, and acquired on a fluorescence-activated cell sorter (FACS) Canto-10 cytometer (BD Biosciences, San Jose, CA). The results were analyzed with custom Woodlist software (generous gift of Wood BL, University of Washington). Following flow assessment, samples were lysed and assessed for mIDH2 as part of a 28-gene amplicon capture-based next-generation sequencing (NGS) assay at Memorial Sloan Kettering Cancer Center.
Hematopoietic immunophenotyping and mIDH2 assessment in flow-sorted cells
Viably frozen BMMCs from normal donors (N = 12) or rrAML patients (N = 9) were thawed, stained with antibodies listed in supplemental Table 3, and sorted on either a BD LSR Fortessa or a BD FACSAria Fusion (Becton Dickinson, Oxford, UK). Sorted fractions (>95% purity) of Lin-ve CD34−CD117− cells, which represent mature myelomonocytic cells, and unsorted mononuclear cells were processed for genomic DNA (gDNA). Whole-genome amplification (RepliG, Qiagen, UK) was carried out in samples with fewer than 104 cells or where extracted gDNA was inadequate. Analyses of mIDH2 at R140 and R172 codons were completed by PCR of exon 4 of the IDH2 gene, followed by NGS using a MiSeq (Illumina, UK).
Phagocytosis assay
Neutrophils were collected from fresh, citrated blood by centrifugation, red blood cell sedimentation, and Percoll gradient cell separation. Purified neutrophils (1 × 106) were incubated with 3 μL of fluorescent green latex beads (Sigma, France) for 15 minutes at 37°C in 1 mL of RPMI medium, supplemented with 10% heat-inactivated fetal bovine serum (Sigma). Cells were washed, fixed in 3.3% paraformaldehyde, stained with 4′,6-diamidino-2-phenylindole, and imaged with a laser-scanning TCS SP5 confocal microscope (Leica, France). The percentage of neutrophils containing latex beads was calculated by scoring 5 different fields of view for each sample. IDH2 mutations were confirmed in neutrophils with a TaqMan SNP Genotyping Assay (Life Technologies, Carlsbad, CA).
FoundationOne Heme panel
FoundationOne Heme analysis was conducted in a clinical laboratory improvement amendments–certified laboratory by Foundation Medicine, Inc. Briefly, fresh bone marrow, peripheral blood samples, or both were collected from patients, and DNA and RNA were extracted. Nucleic acid libraries were prepared, captured using custom bait sets, and sequenced to high depth by using Illumina HiSeq for 405 cancer-related genes by DNA sequencing and 265 frequently rearranged genes by RNA sequencing. Only known or likely gene mutations that are the targets of therapies—either approved or in clinical trials, or are otherwise known drivers of oncogenesis published in the literature—were included in this analysis.21 Comutational burden was calculated as the total sum of all unique known and likely somatic mutations, other than IDH2, identified in each patient.
Calculation of variant allele frequency (VAF)
In each assay, VAF was calculated as the measurement of a mutated allele (ie, mIDH2) over the measurement of a mutated allele + wild-type allele (ie, mIDH2 + wild-type IDH2) in each sample.
Statistical analysis
Statistical analyses were performed with GraphPad Prism software using methods noted in the figure and table legends. Mutational associations with either ORR or complete remission (CR) rate were assessed through a 2-tailed Fisher’s exact test on a 2 × 2 contingency table analyzing the sum of patients achieving a response or not (ORR ≥ partial remission [PR]; and CR ≥ morphologic leukemia-free state [MLFS]) versus the presence or absence of gene mutation identified by FoundationOne Heme. Associations between prognostic risk groups and response were assessed using a 2-tailed chi-square test for trend on a 3 × 2 contingency table analyzing the sum of patients achieving a response or not (ORR ≥ PR; CR = CR) versus risk classification as favorable, intermediate, or adverse, as stated in the figure legends.
Results
mIDH2 inhibition is associated with potent reduction of 2-HG in mIDH2 AML
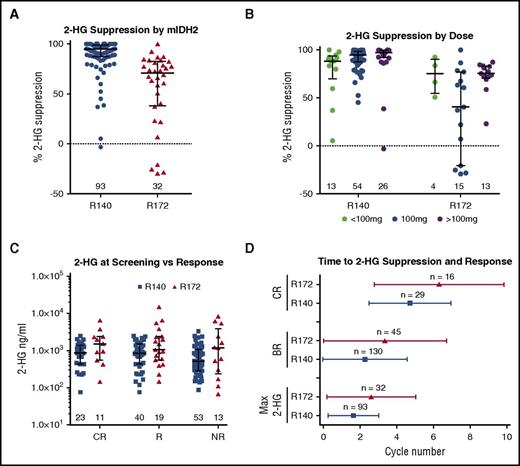
Total 2-HG measurements in blood correlate with R-2-HG levels, tumor mass, clinical response to cytotoxic therapy and are a proposed biomarker of IDH2 mutations.4 Therefore, we assessed total 2-HG levels in 125 rrAML patients with available samples prior to enasidenib treatment and every 28 days during therapy. Median 2-HG suppression (defined as the maximum extent of suppression in comparison with pretherapy) was 90.6%, consistent with potent target inhibition. 2-HG suppression in patients with R172 mIDH2 was less than in patients with R140 mIDH2 rrAML (median suppression of 70.9% and 94.9%, respectively [P < .001]), consistent with preclinical data and an interim analysis of samples from study AG-221-C-001 (Figure 1A).14-16,22-24 Of note, 5 patients during treatment had an increase in 2-HG. Two of these patients achieved a best response of PR despite never having 2-HG levels below baseline in multiple samples analyzed. None of these patients were observed to have co-occurring mutations in IDH1.
mIDH2 inhibition is associated with potent reduction of 2-HG in mIDH2 AML. (A) Dot plot with median and interquartile range showing maximum 2-HG suppression (percentage change from baseline) in blood observed in patients segregated by R140 and R172 mIDH2. Numbers indicate number of patients from each genotype graphed. (B) Dot plot with median and interquartile range showing maximum 2-HG suppression (percentage change from baseline) observed in patients segregated by total daily dose received (<100 mg in green, 100 mg in blue, >100 mg in purple) and stratified by R140 and R172 mIDH2. (C) Dot plot with median and interquartile range showing blood plasma 2-HG (ng/mL) at screening in patients segregated by best response achieved and stratified by R172 (red) and R140 (blue) mIDH2. Response (R) is defined as CR, CRi, CRp, MLFS, or PR. NR is defined as stable disease or progressive disease. (D) Whisker plot indicating mean and standard deviation of cycle to CR, BR, or maximum 2-HG suppression stratified by R172 (red) and R140 (blue) mIDH2. Max 2-HG, maximum 2-HG suppression.
mIDH2 inhibition is associated with potent reduction of 2-HG in mIDH2 AML. (A) Dot plot with median and interquartile range showing maximum 2-HG suppression (percentage change from baseline) in blood observed in patients segregated by R140 and R172 mIDH2. Numbers indicate number of patients from each genotype graphed. (B) Dot plot with median and interquartile range showing maximum 2-HG suppression (percentage change from baseline) observed in patients segregated by total daily dose received (<100 mg in green, 100 mg in blue, >100 mg in purple) and stratified by R140 and R172 mIDH2. (C) Dot plot with median and interquartile range showing blood plasma 2-HG (ng/mL) at screening in patients segregated by best response achieved and stratified by R172 (red) and R140 (blue) mIDH2. Response (R) is defined as CR, CRi, CRp, MLFS, or PR. NR is defined as stable disease or progressive disease. (D) Whisker plot indicating mean and standard deviation of cycle to CR, BR, or maximum 2-HG suppression stratified by R172 (red) and R140 (blue) mIDH2. Max 2-HG, maximum 2-HG suppression.
We next assessed 2-HG suppression in patients dosed with <100 mg, 100 mg, or >100 mg of enasidenib daily. Although we observed a trend toward greater 2-HG suppression at higher doses in R140 mIDH2 patients, there were no statistical differences in maximal 2-HG suppression among the 3 dosing groups (P = .054 for <100 mg vs 100 mg, and P = .094 for 100 mg vs >100 mg) (Figure 1B). In R172 mIDH2 patients, 2-HG suppression was more variable across dosing groups (95% confidence intervals: 43.9% to 102.9%, 10.7% to 61.0%, and 62.5% to 82.7% in the <100-mg, 100-mg, and >100-mg dosing groups, respectively). A statistical difference in 2-HG suppression was observed between the 100-mg and >100-mg dosing cohorts. However, the 100-mg group was confounded by 4 patients whose 2-HG levels increased, and no statistical difference was found in 2-HG suppression between <100-mg and >100-mg dosing groups (P = .152 for <100 mg vs 100 mg; P = .022 for 100 mg vs >100 mg; and P = .955 for <100 mg vs >100 mg). Furthermore, response rates for R172 mIDH2 patients were not statistically different between the 100-mg and >100-mg dosing groups analyzed (ORR, 44.0% and 58.8%, respectively; P = .530; Fisher’s exact test), suggesting no additional clinical benefit for R172 mIDH2 patients in dosing above 100 mg. Importantly, no difference in time to maximum 2-HG suppression between patients dosed with <100 mg, 100 mg, or >100 mg of enasidenib daily was observed either (supplemental Figure 2A). Together, 2-HG suppression and efficacy data indicate that the 100-mg enasidenib dose is biologically and therapeutically active in rrAML patients with mIDH2, regardless of the specific mutation.
We next assessed the relationship between pre-enasidenib–therapy 2-HG levels and clinical response. We observed no significant difference in baseline 2-HG levels between patients achieving a CR, patients obtaining any response (R = CR, CR with incomplete hematologic recovery [CRi], CR with incomplete platelet count recovery [CRp], MLFS, or PR) and patients who did not respond (no response [NR] = stable disease or progressive disease) (Figure 1C). We assessed whether timing of pharmacodynamic response, defined as maximal 2-HG suppression, correlated to the best clinical response. Although the mean cycle number to maximal 2-HG suppression, best response (BR), and CR was ∼1 treatment cycle later in R172 versus R140 patients (3 vs 2 cycles for maximal 2-HG suppression and BR, and 6 vs 5 cycles for CR) (Figure 1D), the ORR and CR rates for patients with R172 and R140 mutations were not statistically different.18 These data demonstrate that the kinetics of target 2-HG inhibition parallel the kinetics of clinical response without effect on response attainment.
Clinical responses to mIDH2 inhibition do not correlate with mIDH2 allele burden
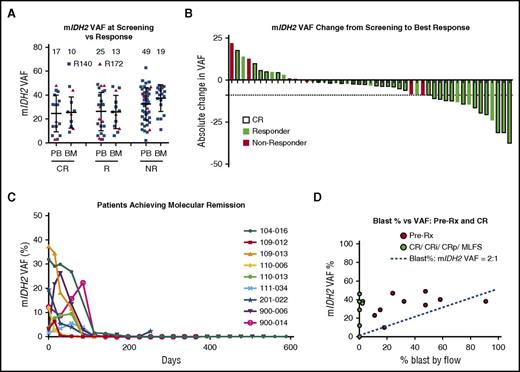
We evaluated whether mIDH2 allele burden at baseline or changes on-therapy correlated with response to enasidenib by quantification of mIDH2 VAF on unsorted samples by using digital PCR and NGS. We observed a significant correlation between digital PCR and NGS for 45 matched patient samples run on both assays (R2 = 0.59, P < .0001) (supplemental Figure 2B). A positive correlation trend was also observed between mIDH2 VAF and 2-HG levels at screening in 17 patients with available samples; however, it was not statistically significant (R2 = 0.21, P = .0636) (supplemental Figure 2C). Notably, mIDH2 allele burden was highly heterogeneous at screening among patients, ranging from low-level mutant positivity to fully clonal (∼50%) (Figure 2A; supplemental Figure 3A). No association between mIDH2 VAF at screening and clinical response was observed, and patients achieving CR had both low and high mIDH2 burden. To assess the possibility that ancestral (clonal) and nonancestral (subclonal) mIDH2 disease exhibit different responses, we analyzed the VAF of co-occurring mutations in the 30 patients with low mIDH2 VAF (<0.2). Eight of these patients, 5 responders and 3 nonresponders, had co-occurring mutations with VAF > 0.4, consistent with mIDH2 occurring as a nonancestral event (supplemental Figure 4). Taken together with data showing responses in both clonal and subclonal mIDH2, these data suggest that patients with ancestral or nonancestral mIDH2 clones can respond to enasidenib.
Clinical responses to mIDH2 inhibition do not correlate with mIDH2 allele burden. (A) Dot plot of mIDH2 VAF (R140 mIDH2 in blue and R172 mIDH2 in red) in patient samples measured at screening in either peripheral blood or bone marrow by FoundationOne Heme panel. Measurements are separated by the best response achieved by patients, as defined in Figure 1. Numbers indicate the number of patient samples in the graph. (B) Waterfall plot indicating absolute change in mIDH2 VAF from screening to achievement of best response measured by Sysmex OncoBeam digital PCR. Responders are plotted in green and nonresponders in red. Patients achieving CR are outlined in black. The dotted line indicates the largest VAF decrease observed in a nonresponder. (C) Line graph of mIDH2 VAF over time (days of treatment) in 9 patients achieving molecular remission (undetectable mIDH2) for at least 1 time point during treatment. (D) Scatter plot of bone marrow mIDH2 VAF versus blast percentage measured by flow cytometry in 9 responsive patients in samples taken pretreatment (red) and at response (CR, CRi, CRp, or MLFS; green). Blue line indicates expected ratio (2:1) between blast percentage:mIDH2 VAF in clonal mIDH2 disease. Three data points (in green) are superimposed with values close to or at zero. BM, bone marrow; PB, peripheral blood; pre-Rx, pretreatment.
Clinical responses to mIDH2 inhibition do not correlate with mIDH2 allele burden. (A) Dot plot of mIDH2 VAF (R140 mIDH2 in blue and R172 mIDH2 in red) in patient samples measured at screening in either peripheral blood or bone marrow by FoundationOne Heme panel. Measurements are separated by the best response achieved by patients, as defined in Figure 1. Numbers indicate the number of patient samples in the graph. (B) Waterfall plot indicating absolute change in mIDH2 VAF from screening to achievement of best response measured by Sysmex OncoBeam digital PCR. Responders are plotted in green and nonresponders in red. Patients achieving CR are outlined in black. The dotted line indicates the largest VAF decrease observed in a nonresponder. (C) Line graph of mIDH2 VAF over time (days of treatment) in 9 patients achieving molecular remission (undetectable mIDH2) for at least 1 time point during treatment. (D) Scatter plot of bone marrow mIDH2 VAF versus blast percentage measured by flow cytometry in 9 responsive patients in samples taken pretreatment (red) and at response (CR, CRi, CRp, or MLFS; green). Blue line indicates expected ratio (2:1) between blast percentage:mIDH2 VAF in clonal mIDH2 disease. Three data points (in green) are superimposed with values close to or at zero. BM, bone marrow; PB, peripheral blood; pre-Rx, pretreatment.
Next, we analyzed changes in absolute mIDH2 VAF from screening to best response. A decrease in mIDH2 VAF was more commonly observed in responding patients than was an increase; however, only one half of the patients showed a VAF change of more than 5 percentage points (Figure 2B; supplemental Figure 3B-C). Similar to recent results reported for a subset of mIDH1 AML patients treated with an mIDH1 inhibitor, longitudinal analysis by digital PCR in our cohort identified 9 of 29 CR patients for whom mIDH2 became undetectable with enasidenib treatment.25 Interestingly, all 9 patients had R140 mIDH2 rrAML (50% of the 18 R140 CR patients), and loss of both a minor mIDH2 subclone and a more substantive mIDH2 clone (∼40%) were observed. In 8 of these patients, mIDH2 remained undetectable with continued treatment, consistent with persistent molecular remission (Figure 2C). However, no significant difference was observed in an initial analysis of event-free survival between patients achieving molecular remission versus patients achieving CR without molecular remission (295.9 vs 259.9 days, respectively; P = .784; event-free survival was defined according to protocol as the interval from date of first dose to date of documented relapse, progression, or death due to any cause, whichever occurred first). Finally, we assessed the relationship between 2 measures of leukemic burden: bone marrow blast count (flow cytometry) and mIDH2 VAF (genetic) in 9 additional patients who responded to enasidenib (Figure 2D). In 6 of these patients, we observed marked blast count decreases to near 0% in aspirates with concomitant mIDH2 VAF above 10%. These data demonstrate that mIDH2 cells persist in most patients achieving CR and that a reduction in mIDH2 allele burden during treatment is neither necessary nor sufficient for clinical response to enasidenib.
Clinical response to mIDH2 inhibition is associated with induction of myeloid differentiation
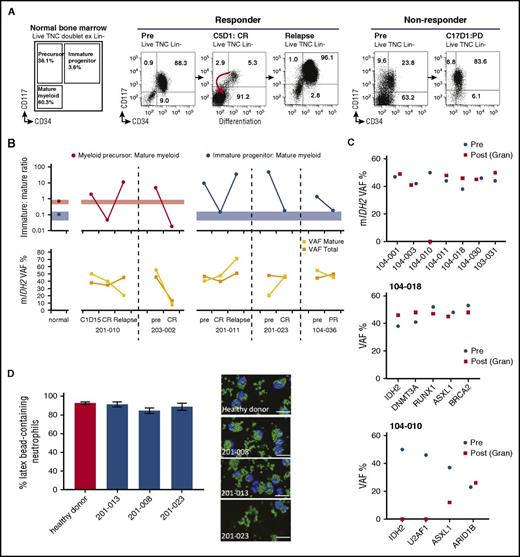
Given that a reduction in mIDH2 VAF was not required for CR, we hypothesized that enasidenib might induce a clinical response by promoting leukemic cell differentiation. We measured the magnitude of different immunophenotypic compartments in the hematopoietic hierarchy, before and during treatment, in 5 mIDH2 rrAML patients who achieved CR or PR (Figure 3A-B). Prior to treatment, all 5 patients had expanded myeloid leukemic progenitor or precursor populations. Enasidenib treatment resulted in near normalization of the immature-to-mature cell population ratio at CR and PR in patients with both R140 (201-023 and 203-002) and R172 (104-036, 201-010, and 201-011) mIDH2. In contrast, no improvement of immature-to-mature ratio was observed in 5 nonresponding patients (supplemental Figure 5). In the responding patients, we assayed mIDH2 VAF by NGS in bulk BMMC and flow-sorted mature myeloid cells (Figure 3B). In 4 of 5 patients achieving CR or PR, mIDH2 VAF remained stable (201-010, 201-011, 104-036) or increased (201-023) in both cell populations.
Clinical response to mIDH2 inhibition is associated with induction of myeloid differentiation. (A) Representative immunophenotypic analyses by flow cytometry on sequential bone marrow samples. Cell-surface markers studied are shown. Data from a responding patient (pretreatment to CR to relapse) (left). Data from a nonresponding patient (pretreatment to progressive disease) who remained in stable disease during treatment (right). Numbers in FACS plots refer to the size of the population as a percentage of lineage-negative bone marrow mononuclear cells. For normal bone marrow (n = 12), the standard deviation is ±2.7% for immature progenitor, ±9.6% for immature precursors, and ±9.7% for mature myeloid cells. (B) Graph showing ratio of immature to mature cell populations by flow cytometry from bone marrow over time (top): the average ratios of myeloid progenitor or myeloid precursors to mature myeloid cells in bone marrow from normal donors (n = 12) and 5 patients who had either a CR or a PR with enasidenib are shown. In patient 201-010, the changing size of myeloid precursor (red) cell populations in relation to mature myeloid cells is shown. In the remaining 3 patients, the changing size of myeloid progenitor (blue) populations to mature cells is shown. Colored bars represent the 95% confidence intervals in normal controls. The mIDH2 VAF in each patient at different time points in all bone marrow mononuclear cells (VAF total) and in FACS-sorted mature myeloid cells (CD34−CD117−) are shown (bottom). (C) mIDH2 VAF in bone marrow mononuclear cells prior to treatment (blue) and in sorted peripheral blood neutrophils at time of best response (red) in 7 patients achieving CR (top). VAF of indicated mutation in bone marrow mononuclear cells prior to treatment and in sorted peripheral blood neutrophils at time of best response in 2 patients achieving CR (middle and bottom). (D) Histogram of the percentages of functional neutrophils observed in ex vivo enasidenib-treated patient samples (left) and representative images (right) assessed by phagocytic assay quantifying neutrophils (blue) that contained latex beads (green). The percentage of neutrophils containing beads was measured by scoring 5 different fields of view per sample. BRCA2, breast cancer type 2; Gran, granulocyte; PD, progressive disease; Post, time of best response; Pre, prior to treatment; TNC, total nucleated cell count.
Clinical response to mIDH2 inhibition is associated with induction of myeloid differentiation. (A) Representative immunophenotypic analyses by flow cytometry on sequential bone marrow samples. Cell-surface markers studied are shown. Data from a responding patient (pretreatment to CR to relapse) (left). Data from a nonresponding patient (pretreatment to progressive disease) who remained in stable disease during treatment (right). Numbers in FACS plots refer to the size of the population as a percentage of lineage-negative bone marrow mononuclear cells. For normal bone marrow (n = 12), the standard deviation is ±2.7% for immature progenitor, ±9.6% for immature precursors, and ±9.7% for mature myeloid cells. (B) Graph showing ratio of immature to mature cell populations by flow cytometry from bone marrow over time (top): the average ratios of myeloid progenitor or myeloid precursors to mature myeloid cells in bone marrow from normal donors (n = 12) and 5 patients who had either a CR or a PR with enasidenib are shown. In patient 201-010, the changing size of myeloid precursor (red) cell populations in relation to mature myeloid cells is shown. In the remaining 3 patients, the changing size of myeloid progenitor (blue) populations to mature cells is shown. Colored bars represent the 95% confidence intervals in normal controls. The mIDH2 VAF in each patient at different time points in all bone marrow mononuclear cells (VAF total) and in FACS-sorted mature myeloid cells (CD34−CD117−) are shown (bottom). (C) mIDH2 VAF in bone marrow mononuclear cells prior to treatment (blue) and in sorted peripheral blood neutrophils at time of best response (red) in 7 patients achieving CR (top). VAF of indicated mutation in bone marrow mononuclear cells prior to treatment and in sorted peripheral blood neutrophils at time of best response in 2 patients achieving CR (middle and bottom). (D) Histogram of the percentages of functional neutrophils observed in ex vivo enasidenib-treated patient samples (left) and representative images (right) assessed by phagocytic assay quantifying neutrophils (blue) that contained latex beads (green). The percentage of neutrophils containing beads was measured by scoring 5 different fields of view per sample. BRCA2, breast cancer type 2; Gran, granulocyte; PD, progressive disease; Post, time of best response; Pre, prior to treatment; TNC, total nucleated cell count.
We extended these findings by measuring mIDH2 VAF in peripheral blood neutrophils prior to enasidenib treatment and at the time of CR in 7 additional patients who achieved CR (Figure 3C, top panel). In 6 of 7 cases, mIDH2 VAF remained constant between pretherapy leukemic cells and neutrophils at CR, consistent with differentiation of mIDH2 leukemia cells into mature neutrophils. Furthermore, in patient 104-018, the VAF of additional coassociated AML mutations remained unchanged in neutrophils at CR, consistent with enasidenib-mediated differentiation of a transformed leukemic clone (Figure 3C, middle panel). In contrast, in the 1 patient whose mIDH2 VAF dropped to 0% in mature neutrophils at CR (104-010), the VAF of other AML-associated mutations showed heterogeneous changes consistent with clonal selection rather than loss of all clonally derived leukemic cells (Figure 3C, top and bottom panels). Next, we investigated the functional status of differentiated leukemic neutrophils with mIDH2 in 3 patients who achieved CR (supplemental Table 4). In each case, mutant neutrophils demonstrated intact phagocytic activity consistent with restoration of normal granulocyte function (Figure 3D).
Genomic predictors of response to mIDH2 inhibition
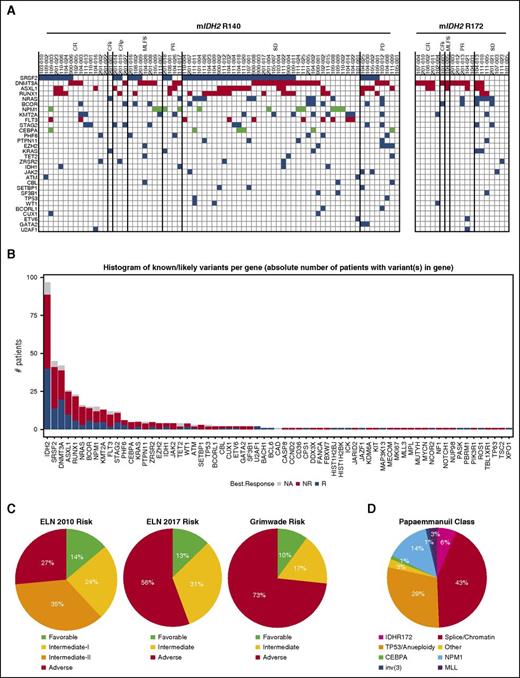
We assessed whether additional somatic mutations were associated with differential response by performing capture-based NGS with FoundationOne Heme mutational panel in 100 patients at screening. No overt biases in response, age, sex, prior treatments, bone marrow blast percentage, absolute neutrophil counts, or prior myelodysplastic syndrome diagnosis were observed between these patients and the 176 rrAML patients from the phase 1 portion of study AG-221-C-001 (supplemental Table 1). Ninety-eight percent of samples contained mutations other than mIDH2, and 17 co-occurring gene mutations were found in ≥5% of patients (Figure 4A-B). The most frequent co-occurring mutations were in serine/arginine-rich splicing factor 2 (SRSF2) (45%), DNA methyltransferase 3 alpha (DNMT3A) (42%), additional sex combs like 1 (ASXL1) (27%), runt-related transcription factor 1 (RUNX1) (24%), NRAS (17%), and BCOR (15%). Notably, the prevalence of mutations in this cohort differed from an analysis of 1376 de novo AML samples from Papaemmanuil et al (supplemental Figure 6A-B).13 Our cohort included a significant enrichment of adverse risk mutations (DNMT3A, ASXL1, RUNX1). Statistical significance of ASXL1 and RUNX1 enrichment was also upheld when restricted to the 130 mIDH2-positive patients in the Papaemmanuil dataset.13 A significantly lower level of favorable prognosis mutations (NPM1), as defined by Grimwade et al, were also observed.26 Differences in co-occurring mutations in patients with R140 mIDH2 AML, including increased comutational heterogeneity (60 different mutated genes vs 24 in R172 mIDH2 patients) and number of co-occurring mutations per patient (3.6 vs 2.6 mutations per patient in R172 mIDH2; P = .020) were observed (Figure 4A; supplemental Figure 6C-E). Additionally, some mutated genes were either exclusively observed (SRSF2, n = 45) or more prevalent in R140 mIDH2 rrAML (RUNX1, 27.3% vs 14.3% in R172 mIDH2 rrAML) or more prevalent in R172 mIDH2 rrAML (DNMT3A, 66.7% vs 36.4% in R140 mIDH2). These data extend work on de novo AML by showing that R140 and R172 mIDH2 rrAML are genetically distinct leukemia subtypes.13
Association of co-occurring mutations with clinical response and classification of patients from cytogenetic and molecular abnormalities. (A) Tile plot showing the number of co-occurring somatic mutations by gene identified in FoundationOne Heme panel from efficacy-evaluable patients separated by R140 and R172 mIDH2. Only mutated genes occurring in 2 or more patients are shown. Mutations associated with higher risk are in red and mutations associated with lower risk are in green, as defined by Grimwade et al.26 (B) Histogram of the number of mutations identified in each gene from all 100 patient samples analyzed. The number of mutations identified in responding patients are in blue, the number of mutations identified in nonresponding patients are in red, and the number of mutations in patients who were not efficacy evaluable are in gray. (C) Pie charts of proportions of patients in various risk categories according to European LeukemiaNet (ELN) 2010 AML risk stratification,29 ELN 2017 AML risk stratification,28 and Grimwade et al,26 on the basis of cytogenetic testing completed before the start of cycle 2 and mutations identified at screening. (D) Pie chart of proportions of patients in various genomic classifications according to Papaemmanuil et al,13 on the basis of cytogenetic testing completed before the start of cycle 2 and mutations identified at screening. CEBPA, CCAAT/enhancer-binding protein alpha; MLL, mixed-lineage leukemia.
Association of co-occurring mutations with clinical response and classification of patients from cytogenetic and molecular abnormalities. (A) Tile plot showing the number of co-occurring somatic mutations by gene identified in FoundationOne Heme panel from efficacy-evaluable patients separated by R140 and R172 mIDH2. Only mutated genes occurring in 2 or more patients are shown. Mutations associated with higher risk are in red and mutations associated with lower risk are in green, as defined by Grimwade et al.26 (B) Histogram of the number of mutations identified in each gene from all 100 patient samples analyzed. The number of mutations identified in responding patients are in blue, the number of mutations identified in nonresponding patients are in red, and the number of mutations in patients who were not efficacy evaluable are in gray. (C) Pie charts of proportions of patients in various risk categories according to European LeukemiaNet (ELN) 2010 AML risk stratification,29 ELN 2017 AML risk stratification,28 and Grimwade et al,26 on the basis of cytogenetic testing completed before the start of cycle 2 and mutations identified at screening. (D) Pie chart of proportions of patients in various genomic classifications according to Papaemmanuil et al,13 on the basis of cytogenetic testing completed before the start of cycle 2 and mutations identified at screening. CEBPA, CCAAT/enhancer-binding protein alpha; MLL, mixed-lineage leukemia.
Additionally, cytogenetics were overlaid with mutational data to characterize the cohort by 4 different risk classifications of de novo AML.13,26-28 Using the European LeukemiaNet (ELN) classification from 2010, 14% of patients were classified as favorable risk and 59% were classified as intermediate risk (Figure 4C).27,29 Using the recently revised ELN classification from 2017, which includes ASXL1 and RUNX1 mutations in the adverse risk group, a disproportionally large group of patients (56%) were classified as adverse risk.28 Furthermore, a classification scheme by Grimwade et al, which includes DNMT3A mutations in the adverse risk group, showed an even greater enrichment of adverse risk patients (73%).26 Finally, a classifier developed on a de novo AML cohort13 revealed an enrichment of patients with mutations in splicing/chromatin associated genes (43%) and with TP53 mutations or aneuploidy (29%). Only 14% of patients were categorized as NPM1 mutation-associated AML, which was reported as the largest AML subset in the study (Figure 4D). Importantly, clinical responses to enasidenib were observed across the risk spectrum in the 72 efficacy-evaluable patients with full genomic/cytogenetic data (Table 1).
Analysis of response in patients with favorable, intermediate, and adverse-risk rrAML
| Risk assessment . | ORR/CR, favorable risk profile, % (n) . | ORR/CR, intermediate risk (I + II) profile, % (n) . | ORR/CR, adverse risk profile, % (n) . | P value (ORR/CR)† . |
|---|---|---|---|---|
| Risk classification | ||||
| ELN 2010 | 22.2/0 (9) | 55.8/32.6 (43) | 30.0/10.0 (20) | .7322/.8493 |
| ELN 2017 | 37.5/0 (8) | 65.2/43.5 (23) | 34.1/14.6 (41) | .2049/.5816 |
| Grimwade | 33.3/0 (6) | 66.7/58.3 (12) | 40.7/16.7 (54) | .6121/.4487 |
| Molecular classification | ||||
| Papaemmanuil* | 80.0/60.0 (5) | 36.4/9.0 (11) | 42.8/21.4 (56) | .4631/.2605 |
| Risk assessment . | ORR/CR, favorable risk profile, % (n) . | ORR/CR, intermediate risk (I + II) profile, % (n) . | ORR/CR, adverse risk profile, % (n) . | P value (ORR/CR)† . |
|---|---|---|---|---|
| Risk classification | ||||
| ELN 2010 | 22.2/0 (9) | 55.8/32.6 (43) | 30.0/10.0 (20) | .7322/.8493 |
| ELN 2017 | 37.5/0 (8) | 65.2/43.5 (23) | 34.1/14.6 (41) | .2049/.5816 |
| Grimwade | 33.3/0 (6) | 66.7/58.3 (12) | 40.7/16.7 (54) | .6121/.4487 |
| Molecular classification | ||||
| Papaemmanuil* | 80.0/60.0 (5) | 36.4/9.0 (11) | 42.8/21.4 (56) | .4631/.2605 |
ORR (CR, CRi, CRp, MLFS, and PR) by risk assessment is based on cytogenetic testing, and mutations are identified by FoundationOne Heme panel according to ELN 2010,29 ELN 2017,28 Grimwade et al,26 and Papaemmanuil et al.13
Papaemmanuil risk was inferred from survival analysis of molecular classifications in which the mCEBPA and mIDH2-R172 groups were considered favorable; mNPM1 and others were considered intermediate; and chromosome 3 inversion, Chromatin-Spliceosome, MLL fusions, and mTP53-aneuploidy were considered adverse.
P values were for the contingency test for the logical trend from favorable to adverse risk.
Finally, we investigated whether the number of co-occurring mutations or specific mutant alleles correlated with enasidenib response. Patients who achieved a response (≥PR or CR) had significantly fewer co-occurring mutations than did nonresponders (P < .001; Figure 5A). Segregating patients in 3 tertiles (≤3, 3 to 6, and ≥6 comutations) revealed a significant difference in ORR for patients with the most versus least co-occurring mutations (ORR 21.9% vs 70.4%, respectively; P < .001) (Figure 5B). Analysis of comutated genes with response indicated a lower, but statistically nonsignificant, ORR with co-occurring SRSF2 (34%), ASXL1 (39%), RUNX1 (26%), and NRAS (19%) mutations (supplemental Table 5). However, significantly fewer patients with co-occurring NRAS mutations achieved CR (P = .0114; supplemental Table 5). When the most common mutations known to activate NRAS signaling (G12, G13, or Q61) were analyzed in comparison with ORR, the observed decrease in response rate was statistically significant (P = .002; Figure 5C). Notably, overall mutational burden was significantly higher (P < .001) in patients with mNRAS (G12, G13, or Q61), and mutations in NRAS were frequently subclonal (Figure 5D-E). Analysis of other gene mutations involved in activating MAPK signaling revealed that no patients with mPTPN11 responded and that mutant Kirsten rat sarcoma (mKRAS) was not associated with response (supplemental Table 5). Together, these data suggest that some RAS pathway mutations, either in the dominant or minor subclone, may directly or indirectly attenuate responses to mIDH2 inhibition.
Comutational burden and NRAS mutations are associated with lack of response. (A) Scatter plot showing mean and standard deviation of number of mutations found per patient, separated by response. P < .001, comparing the difference between nonresponders and either responders (R: CRi, CRp, MLFS, or PR) or patients achieving a CR. (B) Pie charts of response assessment and ORR (patients achieving CR, CRi, CRp, MLFS, or PR) in patients with the lowest third number of comutations (≤3 mutations) and the highest third (≥6 mutations). (C) Pie chart indicating proportion of responders and nonresponders in the 14 efficacy evaluable patients with NRAS comutations (mNRAS), specifically at G12, G13, and Q61. (D) Number of mutations found per patient separated by the presence of G12, G13, or Q61 mutant mNRAS, indicating that patients with mNRAS at G12, G13, or Q61 have an increased mutational burden in this cohort. The only mNRAS+ patient to achieve a CR is highlighted in green. (E) Dot plot of mIDH2 (blue) and mNRAS (red) VAF in the same patient in the 14 efficacy-evaluable patients with NRAS comutations specifically at G12, G13, and Q61. NRASwt, NRAS wild-type.
Comutational burden and NRAS mutations are associated with lack of response. (A) Scatter plot showing mean and standard deviation of number of mutations found per patient, separated by response. P < .001, comparing the difference between nonresponders and either responders (R: CRi, CRp, MLFS, or PR) or patients achieving a CR. (B) Pie charts of response assessment and ORR (patients achieving CR, CRi, CRp, MLFS, or PR) in patients with the lowest third number of comutations (≤3 mutations) and the highest third (≥6 mutations). (C) Pie chart indicating proportion of responders and nonresponders in the 14 efficacy evaluable patients with NRAS comutations (mNRAS), specifically at G12, G13, and Q61. (D) Number of mutations found per patient separated by the presence of G12, G13, or Q61 mutant mNRAS, indicating that patients with mNRAS at G12, G13, or Q61 have an increased mutational burden in this cohort. The only mNRAS+ patient to achieve a CR is highlighted in green. (E) Dot plot of mIDH2 (blue) and mNRAS (red) VAF in the same patient in the 14 efficacy-evaluable patients with NRAS comutations specifically at G12, G13, and Q61. NRASwt, NRAS wild-type.
Discussion
Our study of mIDH2 rrAML patient samples from the phase 1 trial of enasidenib indicates that this cohort is enriched for mutations more commonly seen in adverse-risk or secondary AML (eg, SRSF2, RUNX1, and ASXL1). Additionally, these studies extend previous observations in de novo R140 and R172 mIDH2 AML, confirming that the 2 mIDH2 subtypes are genetically distinct. Despite these observations, enasidenib exhibited potent target inhibition in both subtypes and an ORR of ∼40% with no statistical difference between R140 and R172 mIDH2 rrAML.18 Additionally, our data confirm the preclinical mechanism of action of mIDH2 inhibition by enasidenib and provide the first insight into the genetic basis of primary resistance.10,14-16
Evidence suggests that human AML comprises a hierarchy of both tumor-propagating leukemic stem cells (LSCs) arrested at a progenitor or precursor stage of hemopoiesis and more mature nontumor propagating leukemic cells.30,31 Our observations demonstrate that enasidenib promotes terminal differentiation of mIDH2 leukemic cells of granulocytic lineage in patients who achieve CR or PR. Furthermore, we observed ex vivo phagocytic function in differentiated mIDH2-containing neutrophils. These observations are clinically important and may explain the lower frequency of infections in patients achieving CR with enasidenib treatment.32 Our studies also demonstrate decreases in mIDH2 below a detectable limit in a subgroup of patients. Both observations support differentiation as the mechanism of action of enasidenib monotherapy. Where mIDH2 cells persist, it is most plausible that mIDH2 LSCs are not eradicated but differentiate to give rise to mIDH2-containing functional neutrophils. When molecular CR is achieved, mIDH2 inhibition may result in terminal or near-terminal exhaustion through differentiation of the mIDH2 clone. The differential effects may be due to the specific cellular and genetic contexts of the IDH2 mutation, and additional work will be required to dissect the mechanisms accounting for these observations. It is also intriguing and unclear how CR is achieved in the context of subclonal mIDH2; this requires further mechanistic studies of cell autonomous and cell nonautonomous effects of IDH2 mutations in AML.
In this study, we measured total 2-HG, which includes both L-2-HG and R-2-HG, whereas only R-2-HG is produced by neomorphic IDH mutations. It has been shown that total 2-HG levels correlate with R-2-HG, mIDH2 allelic burden, tumor mass, and clinical status (ie, CR vs absence of CR), consistent with the majority of 2-HG being derived from mIDH2 production of R-2-HG.4 When total 2-HG levels are low, measurement of R-2-HG, rather than total 2-HG, may improve sensitivity; however, this would not have changed our observations relating to the consistent, dose-dependent suppression of 2-HG seen in nearly all patients. Previously, serum 2-HG levels have been suggested as a biomarker for chemotherapy response in mIDH2-positive patients.1-4 Our data demonstrate that this is not the case in targeted therapy because enasidenib is able to inhibit the mIDH2 enzyme and suppress 2-HG regardless of clinical response. Consistent with this hypothesis the level of suppression of 2-HG is not prognostic of response. We also observed that in R172 mIDH2 patients, although the extent of 2-HG suppression is more variable and maximal suppression takes longer to achieve, clinical responses were equivalent to the R140 mIDH2 subtype. Paradoxically, in 4 R172 patients, 2-HG levels rose on enasidenib therapy. This anomaly could arise from differential production of 2-HG by different cell populations during enasidenib-induced differentiation, specific comutational patterns, or alterations in AML cell metabolism in these patients not seen in the larger cohort. Nevertheless, collectively, these data demonstrate that enasidenib potently suppresses 2-HG, the extent of 2-HG suppression does not predict clinical response, and primary resistance to enasidenib is not due to an inability to suppress mIDH2 enzyme activity.
These data also suggest that other factors determine clinical response to enasidenib. Patients with a higher mutational burden or co-occurring mutations in the RAS pathway were observed to be less likely to respond to mIDH2 inhibition. The observation that increasing number of driver mutations is associated with poorer outcome with mIDH2 inhibitor therapy mirrors similar observations in newly diagnosed AML patients treated with chemotherapy.13 From these data, it is also unclear whether constitutive activation of the RAS pathway imposes a 2-HG-independent differentiation block or whether mutations in RAS and other signaling pathways are a marker of overall higher mutational burden and other mechanisms of 2-HG-independent differentiation arrest. Notably, NRAS mutations were frequently present in a minor subclone, and this intriguing observation requires further investigation.
Although enasidenib responses are clinically durable, the genetic heterogeneity observed in our patients suggests that combination with other therapies may be required to achieve long-term disease remission in more patients. This is reminiscent of the impressive, but not durable, activity of all-trans-retinoic acid monotherapy in acute promyelocytic leukemia.33 Our data suggest that targeted therapies may optimally be delivered in combination with other therapies. Current clinical studies combining enasidenib with combination chemotherapy or azacitidine (NCT02677922 and NCT02632708) and future orthogonal targeted therapies will address this question. Although this is only a subgroup analysis of a large single-arm experience, taken together, the clinical response and translational data demonstrate that single-agent mIDH2 inhibition by enasidenib in rrAML represents a critical and novel differentiation therapy. It also provides the platform for future combination therapy regimens to optimize clinical response and further improve outcomes in mIDH2 AML.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors acknowledge helpful advice and review of drafts by Celgene and Agios colleagues, especially Krishnan Viswanadhan, Samantha Good, and Kyle Macbeth. The authors thank Sung Choe (Agios Pharmaceuticals) for helpful discussions, operational assistance, and data cleaning.
P.V. and L.Q. received funding from the Medical Research Council (MRC) (MRC Molecular Haematology Unit Award G1000729, MRC Disease Team Award 4050189188), Cancer Research U.K. (Program Grant C7893/A12796 [P.V.]), Bloodwise (Specialist Program 13001), and the Oxford Partnership Comprehensive Biomedical Research Centre (National Institute for Health Research Biomedical Research Centre Funding scheme). This work was supported by National Institutes of Health (NIH), National Cancer Institute (NCI) grant R01CA172636-01 (R.L.L.), by NIH Office of the Director grant U54OD020355-01 (R.L.L.), and by a supplement to NIH, NCI grant P30CA008748 (R.L.L.). The Memorial Sloan Kettering cores used to perform the studies included in this work are supported by grant P30CA008748. A.S. is supported by the Conquer Cancer Foundation and by grant NCIK08CA181507 from the NIH, NCI.
Authorship
Contribution: M.D.A., L.Q., A.S., M.R., B.M., and N.R.F. performed the experiments; E.M.S., S.d.B., and M.D.D. coordinated sample collection; P.V., R.L.L., and M.D.A. wrote the manuscript; A.T. led all translational studies and analysis efforts; and all authors contributed to experimental design, analyzed data, and edited the manuscript.
Conflict-of-interest disclosure: B.W. and K.E.Y. are employed by and own equity in Agios Pharmaceuticals, Inc. M.D.A. and A.T. are employed by and own equity in Celgene Corporation. L.Q. was supported by a fellowship from Celgene. R.L.L. is a scholar of the Leukemia and Lymphoma Society. The remaining authors declare no competing financial interests.
Correspondence: Anjan Thakurta, Translational Development, Celgene Corporation, 556 Morris Ave, Summit, NJ 07901; e-mail: athakurta@celgene.com.
References
Author notes
M.D.A., L.Q., and A.S. contributed equally to this study and are joint lead authors.
A.T., P.V., and R.L.L. contributed equally to this study and are joint senior authors.