Abstract
Background
Canonical JAK2 V617F and exon 12 mutations in myeloid neoplasms are well described. There are limited reports of other rare JAK2 variants of potential clinical relevance. We sought to survey JAK2 variants in a large cohort of MPN and AML patients and determine the likelihood of contribution to disease pathogenesis.
Methods
We retrospectively analyzed the JAK2 variant profile and mutational landscape of MPN and AML. Targeted, amplicon-based, next-generation sequencing of the coding region of JAK2 and 27 other leukemia-related genes was performed on DNA extracted from diagnostic bone marrow samples. The study population was classified into three cohorts: chronic myeloproliferative neoplasm-only (MPN); MPN transformed to AML (MPN>>AML); and AML-only, excluding MPN>>AML patients (AML).
Results
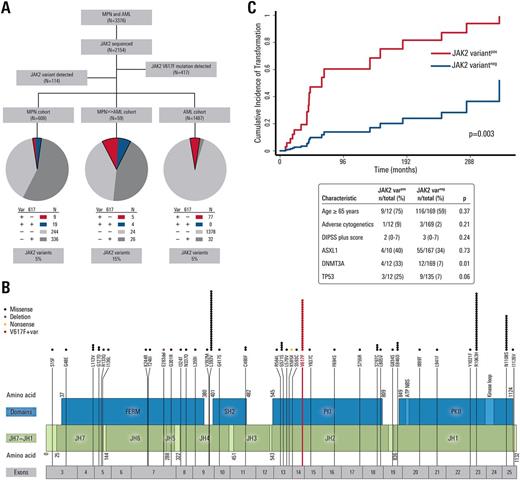
Of 3,376 MPN and AML patients, 2,154 patients underwent JAK2 sequencing. Non-V617F/non-exon 12 JAK2 sequence variants were identified in 114 (5.3%, Fig 1A). This included 35 unique JAK2 variants including several previously undescribed variants located within all functional domains (Fig 1B). Ten variants have been previously reported in cases of hematologic proliferation or shown to be activating alterations. Sixteen of 114 JAK2 variants occurred alone without somatic mutations in the remaining 27 somatic genes tested. JAK2 variants were detected at a significantly higher frequency in the MPN>>AML cohort (15.3%) compared with MPN (4.6%, p<0.001) and AML (5.2%, p<0.001) cohorts. Every detected variant occurred at higher-than-expected frequency in MPN and AML patients than in the general population, particularly L393V, R1063H, and N1108S (p<0.01 for each). Each missense variant was analyzed using 3 separate predictive computational platforms (FATHMM-MKL, MutationTaster, and DANN) to determine whether an amino acid substitution was likely to exert a functional effect on JAK2 . All variants were predicted to have functional consequence, and most were predicted to have a functional consequence (62.9%) by all 3 programs. The presence of a JAK2 variant in addition to JAK2 V617F (n=13) in myelofibrosis was associated with an increased cumulative risk of transformation from MPN to AML (p=0.003, Fig 1C). Some JAK2 variants, and variants in specific protein regions, were associated with increased frequency of chromosomal abnormalities. For example, L393V and N1108S variants were significantly associated with chromosome number alterations compared to other JAK2 variants (43.2% vs. 22.9%, p=0.002). Additionally, co-mutation analysis with the other 27 genes tested showed that 5/16 (31.3%) patients with JAK2 N1108S also had a TP53 mutation vs. 10/98 (10.2%) among other JAK2 variants (p=0.04).
Conclusions
Full exonic gene sequencing of JAK2 identified several JAK2 variants of potential clinical relevance. JAK2 variants may represent an important and potentially overlooked component of MPN/AML pathogenesis, evidenced by the prevalence of variants previously shown to have JAK2 activating effects, the frequencies of specific variants in MPN/AML compared to the general population, and the predicted functional effect of variant amino acid substitutions. Specific JAK2 variants detected in MPN patients may be meaningful predictors for transformation to AML.
DiNardo: Agios: Honoraria, Research Funding; Daiichi-Sankyo: Honoraria, Research Funding; AbbVie: Honoraria, Research Funding; Celgene: Honoraria, Research Funding; Novartis: Honoraria, Research Funding. Bose: Incyte Corporation: Honoraria. Pemmaraju: affymetrix: Research Funding; LFB: Consultancy, Honoraria; cellectis: Research Funding; stemline: Consultancy, Honoraria, Research Funding; Incyte Corporation: Consultancy, Honoraria; novartis: Consultancy, Honoraria, Research Funding; roche diagnostics: Consultancy, Honoraria; abbvie: Research Funding. Cortes: ARIAD: Consultancy, Research Funding; Novartis Pharmaceuticals Corporation: Consultancy, Research Funding; BMS: Consultancy, Research Funding; ImmunoGen: Consultancy, Research Funding; Teva: Research Funding; Pfizer: Consultancy, Research Funding; Sun Pharma: Research Funding. Kantarjian: Amgen: Research Funding; Bristol-Meyers Squibb: Research Funding; Delta-Fly Pharma: Research Funding; Pfizer: Research Funding; ARIAD: Research Funding; Novartis: Research Funding. Verstovsek: Galena BioPharma: Research Funding; Lilly Oncology: Research Funding; Seattle Genetics: Research Funding; NS Pharma: Research Funding; CTI BioPharma Corp: Research Funding; Bristol Myers Squibb: Research Funding; Astrazeneca: Research Funding; NS Pharma: Research Funding; Roche: Research Funding; Lilly Oncology: Research Funding; Gilead: Research Funding; Galena BioPharma: Research Funding; Seattle Genetics: Research Funding; Pfizer: Research Funding; Celgene: Research Funding; Incyte: Research Funding; Genentech: Research Funding; Genentech: Research Funding; Pfizer: Research Funding; Blueprint Medicines Corp: Research Funding; Roche: Research Funding; Astrazeneca: Research Funding; Bristol Myers Squibb: Research Funding; CTI BioPharma Corp: Research Funding; Incyte: Research Funding; Blueprint Medicines Corp: Research Funding; Gilead: Research Funding; Promedior: Research Funding; Celgene: Research Funding; Promedior: Research Funding.
Author notes
Asterisk with author names denotes non-ASH members.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal