Abstract
Introduction: Assays are needed to define the susceptibility of heterogeneous primary human tumor cells. We previously showed that single-cell mass accumulation rate (MAR) - conceptually similar to growth rate - can accurately define drug sensitivity ex vivo (Stevens et al. Nature Biotechnology, 2016). This live-cell assay is performed using a suspended microchannel resonator (SMR), which maintains cell viability and permits the downstream application of single-cell whole transcriptome sequencing (scRNA-seq) to identify molecular correlates of drug response. By comparing leukemias exposed to antineoplastic therapy or vehicle in vivo , we sought to identify unique transcriptional signatures that, when coupled with single-cell biophysical properties, provide a more nuanced view of therapeutic susceptibility.
Methods: We transplanted a patient-derived xenograft of BCR-ABL-rearranged, B-cell leukemia into NOD.Cg-PrkdcscidIl2rgtm1Wjl/SzJ recipients. This model was derived from a patient whose leukemia relapsed following combination chemotherapy and multiple tyrosine kinase inhibitors. Clinical sequencing revealed an ABL1 Y253H resistance mutation that was confirmed in the PDX. Upon engraftment, animals initiated therapy with ponatinib 40 mg/kg/day or vehicle via oral gavage. Splenocytes were harvested on treatment day 5, and immunomagnetically enriched leukemia cells were individually profiled for mass and MAR using the SMR, from which they were sequentially released directly into micro PCR tubes. Smart-Seq2 whole transcriptome amplification, library preparation, and 30-bp paired-end sequencing was then performed on these individual cells.
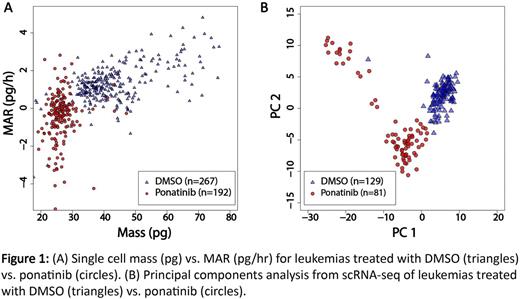
Results: Leukemia cells treated with ponatinib in vivo had significantly lower mass and normalized MAR (MAR/mass) than vehicle-treated controls (p<2e-16 by Welch two-sample t-test, Figure 1A). Single-cell RNA-seq libraries generated from both treatment conditions were of appropriate quality and complexity, with 5,540 ± 81 and 3,826 ± 92 genes mapped per cell for vehicle- and ponatinib-treated cells, respectively. These figures are in line with our published experience (Kimmerling et al. Nature Communications , 2016). Library quality was temporally stable, with consistent average single-cell complexities maintained over the full course of collection experiments.
Principal components analysis segregated cells by treatment status according to PC1 (positive enrichment for protein translation genes) and PC2 (negative enrichment for B-cell activation and B-cell receptor signaling genes) (Figure 1B). While single-cell measurements of biophysical properties or gene expression independently resolved treated and untreated cells at the population level (p<2e-16 by Welch two-sample t-test for both normalized MAR as well as PC1), layering single-cell mass and MAR on top of the transcriptomic data enabled us to identify and characterize subpopulations within treatment groups. For instance, biophysical measurements defined a subset of ponatinib-treated cells that maintained high mass and MAR despite continued therapeutic pressure. No subset of cells corresponding to this physically distinct subpopulation was identified by analysis of single-cell transcriptional profiles alone. Critically, linking biophysical and gene expression measurements from the same cells revealed gene sets with functional enrichment for cell cycle and translation - distinct from the gene sets identified from principal components analysis - that correlated with single-cell mass and mass-normalized MAR, respectively.
Conclusions: Integrating the biophysical and molecular profiles of human leukemias on a single-cell level using the SMR platform coupled with scRNA-seq permits assessment of in vivo therapeutic susceptibility within hours of tissue sampling, with the potential to resolve and characterize intratumoral heterogeneity with greater precision. We are now focused on defining the clinical significance of such physical-molecular tumor subpopulations and interrogating mechanisms of therapeutic resistance illuminated by these methods in the context of phase II-like in vivo treatment studies.
Stevens: Travera: Equity Ownership, Other: Co-founder. Kimmerling: Travera: Equity Ownership, Other: Co-founder. Manalis: Affinity Bio: Equity Ownership, Other: Co-founder; Travera: Consultancy, Equity Ownership, Other: Co-founder.
Author notes
Asterisk with author names denotes non-ASH members.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal