Key Points
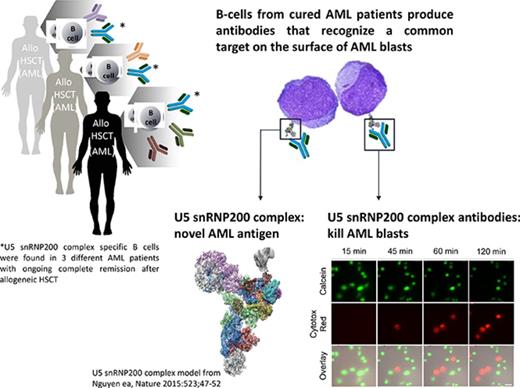
High-risk patients with AML with lasting GVL responses generate antibodies that kill leukemic blasts.
The target of these cytotoxic antibodies is the U5 snRNP200 complex, which is expressed on the membrane of AML blasts.
Most patients with acute myeloid leukemia (AML) can only be cured when allogeneic hematopoietic stem-cell transplantation induces a graft-versus-leukemia immune response (GVL). Although the role of T cells and natural killer cells in tumor immunology has been established, less is known about the contribution of B cells. From B cells of high-risk patients with AML with potent and lasting GVL responses, we isolated monoclonal antibodies directed against antigens expressed on the cell surface of AML cells but not on normal hematopoietic and nonhematopoietic cells. A number of these donor-derived antibodies recognized the U5 snRNP200 complex, a component of the spliceosome that in normal cells is found in the cell. In AML however, the U5 snRNP200 complex is exposed on the cell membrane of leukemic blasts. U5 snRNP200 complex–specific antibodies induced death of AML cells in an Fc receptor–dependent way in the absence of cytotoxic leukocytes or complement. In an AML mouse model, treatment with U5 snRNP200 complex–specific antibodies led to significant tumor growth inhibition. Thus, donor-derived U5 snRNP200 complex–recognizing AML-specific antibodies may contribute to antitumor responses.
Introduction
Allogeneic hematopoietic stem-cell transplantation (HSCT) is an established form of immunotherapy that is often applied in the treatment of patients with hematologic malignancies such as acute myeloid leukemia (AML).1 AML has a poor prognosis that can be significantly improved when the allogeneic immune system induces a potent graft-versus-leukemia (GVL) immune response.
Tumor immune responses are considered T cell mediated, and enhancing T-cell responses, for example with checkpoint inhibitors, has significantly improved clinical outcome in a number of solid tumors and lymphomas.2 In allogeneic HSCT, transplantation of T cell–depleted grafts was associated with high leukemia relapse rates,3 and this and other studies established the importance of T cells in GVL responses.1,4 Similarly, killer immunoglobulin receptor–mismatched natural killer (NK) cells that are alloreactive against AML blasts had a significant impact on allogeneic HSCT outcomes.5 Emerging evidence suggests that B cells and antibodies are also involved in antitumor immune responses. In colorectal tumors, B cells are 1 of the dominant infiltrating lymphocyte populations, and their interaction with follicular T cells was associated with survival.6 In ovarian carcinoma and other solid tumors, tumor B-cell infiltration was the single most predictive marker of response to neoadjuvant chemotherapy.7,8 In allogeneic HSCT recipients, serum analysis of patients with potent GVL responses demonstrated the appearance of antibodies against tumor-associated antigens.9,10 However, proof that donor-derived antibodies contribute to the antitumor immune response in allogeneic HSCT recipients is lacking.
Here we investigated the functionality of antibodies produced by donor-derived B cells in GVL responses. From allogeneic HSCT recipients who remained in complete remission despite high-risk AML, we established clonal B-cell lines producing antibodies that were highly specific for AML. A significant number of these antibodies, which were all donor derived, recognized the U5 snRNP200 complex, a component of the spliceosome that is expressed in the nucleus of normal cells but is exposed on the surface of AML cells. U5 snRNP200 complex–specific antibodies are directly cytotoxic for AML blasts in the absence of effector cells or complement, suggesting that these antibodies contribute to the antitumor response in HSCT recipients.
Materials and methods
Patients
Study protocols were approved by the Medical Ethical Committee of the Academic Medical Centre (Amsterdam, The Netherlands). All participants signed informed consent. Freshly isolated AML blasts were obtained from blood or bone marrow of patients with newly diagnosed AML. B-cell non-Hodgkin lymphoma cells were obtained as waste material from diagnostic punctures and biopsies. Endothelial cells were isolated from umbilical cords (human umbilical vein endothelial cells). Healthy bone marrow was acquired from the sternum of patients undergoing thoracotomy for cardiac surgery. Healthy peripheral blood mononuclear cells were isolated from buffy coats from blood donations (Sanquin, Amsterdam, The Netherlands).
Isolation of B cells and generation of AML-specific B-cell clones
B cells were sorted into CD27− CD19+ CD3− immunoglobulin M− (IgM−) IgA− B cells (naive IgG B cells), CD27+ CD19+ CD3− IgM− IgA− B cells (memory IgG B cells), CD27+ CD19+ CD3− IgM+ B cells, and CD27+ CD19+ CD3− IgA+ B cells on a FACSAria (Becton Dickinson), transduced with a B-cell lymphoma-6 (BCL-6) and BCL-xL construct, and expanded with interleukin-21 on CD40L-expressing fibroblasts as described previously.11
Recombinant antibody production and labeling
Total RNA of 2-5E5 cells was isolated with the RNeasy mini kit (Qiagen), and the heavy- and light-chain variable regions were cloned into the pCR2.1 TA cloning vector (Invitrogen). To rule out reverse transcriptase– or DNA polymerase–induced mutations, several independent cloning experiments were performed. Recombinant antibodies were produced with a C-terminal sortase (ST) tag.12 Triglycine probes with linked biotin or fluorescent dyes (JPT Peptides Technologies GmbH, Berlin, Germany) were linked to the ST-tagged antibodies by ST.
Flow cytometry
The following antibodies were used: CD45 (348815; clone2D1), CD33 (345798; cloneP67.6), and CD19 (348814; cloneSJ25C1; Becton Dickinson); CD34 (343516; clone581), CD14 (325618; cloneHCD14), and CD3 (317306; OKT3; Biolegend); IgM (2022-09) and IgG (2042-09; Southern Biotech); IgG (H+L; A-21445; Life Technologies); 7AAD (Beckman Coulter); 4′,6-diamidino-2-phenylindole, propidium iodide (PI), polyclonal rabbit snRNP200 antibody (HPA 029321; Sigma), and polyclonal rabbit snRNP200 antibody (A303-454A; Bethyl Laboratories); IgA (F0316) and hepatitis B virus core antigen antibody (B0586; Dako). Cells were analyzed on FACSAria (Becton Dickinson), FACSCanto (Becton Dickinson), FACS LSR Fortessa X20 (Becton Dickinson), and Guava (Millipore) flow cytometers, and flow cytometry data were processed with FlowJo software (Tree Star).
Target identification: immunoprecipitation and flow cytometry
THP-1 cells were lysed with 0.5% Triton X114 lysis buffer (10 mM Tris-HCl pH7.4, 150 mM sodium chloride, 1.5 mM MgCl2, 0.5% Triton X-114 [Sigma], and 1.5× complete protease and 1× PhosStop phosphatase [Roche] inhibitor cocktail) and cleared by centrifugation. To remove nonspecific binding proteins, the lysate was precleared with an irrelevant antibody (palivizumab; anti–RSV-F; Medimmune) bound to protein G dynabeads (Invitrogen) and streptavidin magnetic beads (Pierce). AT1331-ST-biotin, AT1223-ST-biotin, or AT1002-ST-biotin (control) antibodies were incubated with streptavidin beads and cell lysate (3 hours at 4°C). Proteins were eluted from the beads with glycine elution buffer (0.1 M glycine, 150 mM sodium chloride, 0.5 mM EDTA, and 0.5% Triton X100 pH2.7) and run on TGX gels (4% to 20%; Bio-Rad). Gels were stained for mass spectrometry with Coomassie (Bio-Rad) or immunoblotted with rabbit anti-snRNP200 (Sigma), rabbit anti-PRPF8 (Abcam), rabbit anti-snRNP200 (Bethyl), and rabbit anti-EFTUD2 (Abcam) and mouse anti-GAPDH (Sigma) antibodies. Intracellular staining for snRNP200 was performed after fixing cells with methanol (Sigma) and permeabilizing with Triton X-100 (Sigma), EDTA (Gibco)/bovine serum albumin (Roche) buffer, followed by incubation with rabbit anti-human snRNP200 antibody (Bethyl or Sigma) overnight at 4°C.
Trypsin digestion and mass spectrometry analysis
Proteins in sodium dodecyl sulfate–polyacrylamide gel electrophoresis gel bands were reduced and alkylated using dithiothreitol and iodoacetamide (Sigma), respectively, and digested with trypsin (Sequencing Grade Modified Trypsin; Promega). Peptides were analyzed by reversed-phase liquid chromatography–tandem mass spectrometry using an Ultimate 3000 RSLCnano system (Thermo Fisher Scientific) coupled to an amaZon ETD ion trap (Bruker Daltonics). Peak lists were generated using Data Analysis 4.0 (Bruker Daltonics) and exported as Mascot generic format files. Proteins were identified in the human Uniprot database using the Mascot search algorithm (Mascot 2.4.1; Matrix Science). Mass spectrometry analysis of the immunoprecipitated material yielded 17% coverage of snRNP200 with 21 peptides, and a second mass spectrometry experiment yielded 36% snRNP200 coverage with 77 peptides.
Target confirmation: snRNP200 ELISA
HEK 293T cells were transfected with an expression vector containing the full-length open reading frame of snRNP200 with an N-terminal FLAG tag or an H77 E1E2 FLAG-tagged construct (control). Cells were harvested and lysed with 0.5% Triton X114 lysis buffer. This lysate was cleared, and protein concentration was assessed. AML-specific antibodies, control antibodies, or commercially available snRNP200-specific antibodies (Bethyl) were coated on a goat anti-human IgG (Jackson) or anti-rabbit precoated enzyme-linked immunosorbent assay (ELISA) plate. The lysate was added at a concentration of 5 μg/mL for capturing. After extensive washing, captured snRNP200 flag was detected with a monoclonal mouse anti-flag horseradish peroxidase antibody (clone M2; Sigma).
Cell death assays
For imaging, calcein AM–labeled THP-1 cells (10 nM) were incubated with AML-specific antibodies (10 μg/mL) for 15 minutes and transferred to a 96-well flat-bottom SpectraPlate (PerkinElmer), where Cytotox Red (Essen Bioscience) was added (0.5 μM). Cells were imaged using a Leica DMi8 (Leica Microsystems) inverted wide-field microscope, with GFP and Texas Red filter cubes, 40× oil objective, at 37°C in an atmosphere containing 5% carbon dioxide. Time-lapse movies from 3 independent experiments were taken at 2-minute intervals for at least 3 hours using Adaptive Focus Control. Images were processed with LAS X software (Leica Microsystems). Cell death and adhesion quantification was performed with 1000 to 2000 (after 3 hours) and 150 to 300 (15 minutes and 1 hour) different cells per antibody, respectively. To measure apoptosis, THP-1 cells were stained with 40 nM DiOC6 (3,3-dihexyloxacabocyanine iodide; Molecular Probes/Invitrogen) for 20 minutes at 37°C, after which PI was added. To block apoptosis, THP-1 cells were preincubated with 100 μM Z-VAD-FMK or 10 μM Q-VD-OPh (R&D) and incubated with 1 to 10 μg/mL AML or control antibodies, dimethyl sulfoxide vehicle, or diclofenac for 24 hours.13,14 Cytocholasin D was used at 10 μg/mL, with 20 minutes incubation on ice (Sigma).
In vivo AML xenograft mouse model
SCID mice (8 per group) were inoculated with THP-1 subcutaneously. Antibody treatment (biweekly intraperitoneally at 10 mg/kg for 3 consecutive weeks) was started at an average tumor size of 106 mm3 3 weeks after inoculation. Treatment tolerability was assessed by body-weight measurements and clinical signs of treatment-related adverse effects. Tumors were measured twice weekly in 2 dimensions using calipers (tumor volume [cubic millimeters] = [width2 (millimeters) × length (millimeters)]/2). Tumor growth delay was determined by a time–to–end point analysis of individual mice. The end point was set as at a tumor size of 350 mm3 to prevent censored cases. Tumors of the 2 best and worst responders per group were harvested at day 25. Tissue sections were stained with hematoxylin and eosin and digitalized, and tumor and necrotic areas were measured using Philips Digital Pathology Solution (version 2.3.1.1).
Statistics
All statistical analyses were performed using GraphPad PRISM 6.0f. Significant differences in the snRNP200 ELISA between antibodies were determined with multiple Student t tests of a 2-way analysis of variance and between patients with multiple comparisons in a 1-way analysis of variance. Tumor growth delay in days to the defined end point of 350 mm3 was analyzed using survival curve statistics. Because a proportional hazard to reach the end point was assumed, a log-rank (Mantel-Cox) test was performed.
Results
Identification of AML-specific antibodies
We selected 3 patients with high-risk AML who achieved ongoing, durable (at the time of writing of this report, >7 years) remission after allogeneic HSCT (patients 58, 59, and 101; Table 1). To test whether these patients had generated AML-specific antibodies, we searched for AML-binding antibody-producing B cells. Peripheral blood B cells were transduced with BCL-xL and BCL-6 and cultured as described previously.11 We deposited 20 or 40 immortalized B cells per well of 96- or 384-well microtiter plates to generate minicultures and tested supernatants of these cultures for antibodies binding to AML cell lines using flow cytometry (Figure 1A). Because all 3 patients had been diagnosed with (myelo)monocytic AML, we selected the THP-1, Molm13, and MonoMac6 cell lines, which are morphologically and phenotypically similar to (myelo)monocytic AML, for these screening assays. Minicultures where the supernatant specifically bound the AML cell lines but not control cells were selected; single cells of these cultures were deposited in wells of microtiter plates, and after expansion, their supernatants were screened for binding against AML cell lines (Figure 1A). Using this approach, we identified 16 monoclonal B-cell lines, producing antibodies that bound to THP-1, Molm13, Monomac6, K562, and freshly isolated AML blasts of newly diagnosed patients (Figure 1B; Table 2) but not to non-AML cell lines or hematopoietic (including peripheral blood mononuclear cells, bone marrow, tonsil, and cord blood) or nonhematopoietic cells of healthy individuals (Figure 1C; data not shown). AML-binding clones were of the IgG1 or IgG3 isotype, and the variable heavy- and light-chain regions of most of the antibodies showed somatic hypermutations, indicative of in vivo antigen exposure of these B cells (Table 3). All AML-specific antibodies detected were of donor origin, as determined with microchimerism analysis of genomic polymorphisms by profiling of short tandem repeat DNA loci of B-cell clones (data not shown).
Patient characteristics
| . | Patient 58 . | Patient 59 . | Patient 101 . | Patient 102 . | Patient 184 . |
|---|---|---|---|---|---|
| AML | |||||
| FAB | M5 | M4 | M5 | M5 | M2 |
| WHO | AML-NOS | AML-NOS | AML-NOS | Mutated NPM1 | Mutated NPM1 |
| Age, y | 53 | 36 | 49 | 34 | 53 |
| Sex | F | F | M | F | M |
| Treatment | |||||
| Induction | Cytarabine, idarubicin, amsacrine | Cytarabine, idarubicin, amsacrine | Cytarabine, idarubicin, amsacrine | Cytarabine, idarubicin, amsacrine | Cytarabine, idarubicin, amsacrine |
| Consolidation | Mitoxantone, etoposide | MA-MUD | Busulfan/cyclophosphamide/autoSCT | FLAMSA-MUD* | RIST-sibling |
| Relapse | |||||
| Reinduction | HD cytarabine | HD cytarabine | HD cytarabine | — | HD cytarabine |
| Consolidation | RIST-MUD | None† | RIST-MUD | — | DLI |
| GVHD | |||||
| Organ | Liver, skin | Liver, skin | Liver, skin, intestine | Skin, intestine | Liver, lung |
| Time, mo | 5‡ | 2§ | 1.5‡ | 1‡ | 1|| |
| Remission¶ | 86 | 86 | 86 | 61 | 45 |
| B-cell cloning# | 28 | 24 | 38 | 12 | 26|| |
| . | Patient 58 . | Patient 59 . | Patient 101 . | Patient 102 . | Patient 184 . |
|---|---|---|---|---|---|
| AML | |||||
| FAB | M5 | M4 | M5 | M5 | M2 |
| WHO | AML-NOS | AML-NOS | AML-NOS | Mutated NPM1 | Mutated NPM1 |
| Age, y | 53 | 36 | 49 | 34 | 53 |
| Sex | F | F | M | F | M |
| Treatment | |||||
| Induction | Cytarabine, idarubicin, amsacrine | Cytarabine, idarubicin, amsacrine | Cytarabine, idarubicin, amsacrine | Cytarabine, idarubicin, amsacrine | Cytarabine, idarubicin, amsacrine |
| Consolidation | Mitoxantone, etoposide | MA-MUD | Busulfan/cyclophosphamide/autoSCT | FLAMSA-MUD* | RIST-sibling |
| Relapse | |||||
| Reinduction | HD cytarabine | HD cytarabine | HD cytarabine | — | HD cytarabine |
| Consolidation | RIST-MUD | None† | RIST-MUD | — | DLI |
| GVHD | |||||
| Organ | Liver, skin | Liver, skin | Liver, skin, intestine | Skin, intestine | Liver, lung |
| Time, mo | 5‡ | 2§ | 1.5‡ | 1‡ | 1|| |
| Remission¶ | 86 | 86 | 86 | 61 | 45 |
| B-cell cloning# | 28 | 24 | 38 | 12 | 26|| |
autoSCT, autologous stem-cell transplantation; DLI, donor lymphocyte infusion; FAB, French-American-British; FLAMSA, fludarabine, amsacrine, and cytarabine; HD, high dose; MA, myeloablative; MUD, matched unrelated donor; NOS, not otherwise specified; RIST, reduced-intensity stem-cell transplantation; WHO, World Health Organization.
Patient 102 received FLAMSA-MUD for primary refractory AML.
Patient 59 developed grade 3 GVHD of the liver before the planned DLI.
Number of months after allogeneic HSCT when GVHD was diagnosed.
Months after reinduction chemotherapy for AML relapse.
Months after DLI.
Duration of remission from date of allogeneic HSCT (patients 58, 101, and 102) or from start of remission induction (patients 59 and 184) until 1 June 2017.
Number of months between allogeneic HSCT (patients 58, 101, and 102) or reinduction chemotherapy/DLI (patients 59 and 184) and isolation of B cells from patient for current analysis.
Identification of AML-specific antibodies. (A) Antibodies in the supernatant of 1 of the minicultures (11C9; 40 cells per well) binding to the AML cell line THP-1 (top panels). B cells from this miniculture were plated into 1 cell per well solutions and supernatants again screened for antibodies binding to THP-1 (lower panels). Positive control: HLA antigen D related (HLA-DR); negative control: in-house generated, influenza-specific antibody AT1002.12 Recombinant-produced 11C9-22C11 was named AT1223. FSC, free secretory component. (B) Representative example of binding of 3 of the selected antibodies (AT1223, AT1331 [patient 59], and AT1337 [patient 58]) to the AML cell lines THP-1, Molm13, MonoMac 6, and K562, and primary AML blasts freshly isolated from a patient with AML (BL-061; NPM1+). (C) AML antibodies did not bind to bone marrow–derived CD34+ hematopoietic progenitor cells, peripheral blood mononuclear cells (PBMCs), monocytes, fibroblasts, or endothelial cells isolated from healthy individuals, nor to the colon cell line HT29 or the liver cell line HepG2. Negative control: AT1002 (gray-filled histograms). HUVEC, human umbilical vein endothelial cell.
Identification of AML-specific antibodies. (A) Antibodies in the supernatant of 1 of the minicultures (11C9; 40 cells per well) binding to the AML cell line THP-1 (top panels). B cells from this miniculture were plated into 1 cell per well solutions and supernatants again screened for antibodies binding to THP-1 (lower panels). Positive control: HLA antigen D related (HLA-DR); negative control: in-house generated, influenza-specific antibody AT1002.12 Recombinant-produced 11C9-22C11 was named AT1223. FSC, free secretory component. (B) Representative example of binding of 3 of the selected antibodies (AT1223, AT1331 [patient 59], and AT1337 [patient 58]) to the AML cell lines THP-1, Molm13, MonoMac 6, and K562, and primary AML blasts freshly isolated from a patient with AML (BL-061; NPM1+). (C) AML antibodies did not bind to bone marrow–derived CD34+ hematopoietic progenitor cells, peripheral blood mononuclear cells (PBMCs), monocytes, fibroblasts, or endothelial cells isolated from healthy individuals, nor to the colon cell line HT29 or the liver cell line HepG2. Negative control: AT1002 (gray-filled histograms). HUVEC, human umbilical vein endothelial cell.
Expression of U5 snRNP200 complex on AML blasts and AML cell lines
| . | WHO 2008 . | AT1223 . | AT1331 . | AT1337 . |
|---|---|---|---|---|
| AML with recurrent genetic abnormalities | ||||
| BL-p67 | t(8;21) | 405 | 120 | 231 |
| BL-065 | t(8;21) | 832 | 1016 | 841 |
| BL-059 | RUNX1+ FLT3/ITD+ | 688 | 20 | 247 |
| BL-038* | Inv16 | 292 | 344 | 937 |
| BL-025 | Inv16 | 126 | 449 | 471 |
| BL-037 | APL | 204 | 87 | 353 |
| BL-057 | EVI1 17p del | 20 | 333 | 426 |
| BL-061† | NPM1+ | 1079 | 766 | 775 |
| BL-051 | NPM1+ | 171 | 81 | 11 |
| BL-p78 | NPM1+ | 238 | 822 | 771 |
| AML with myelodysplasia-related changes | ||||
| BL-054 | Multilineage dysplasia | 786 | −22 | 289 |
| Therapy-related myeloid neoplasm | ||||
| BL-047 | t-AML (latency, 3 years) | 302 | 362 | 972 |
| AML-NOS | ||||
| BL-007 | NOS; M0 | 267 | 285 | 124 |
| BL-046 | NOS; M4 | 1104 | 569 | 296 |
| BL-034‡ | NOS; M5 | 785 | 502 | 694 |
| Myelodysplastic syndrome | ||||
| BL-032 | RCUD | 265 | 1512 | 261 |
| BL-011 | RCMD | 616 | 939 | 1225 |
| BL-062 | RAEB-1 | 628 | 766 | 367 |
| BL-022 | RAEB-2 | 205 | 464 | 565 |
| BL-033 | RAEB-2 | 1088 | 811 | 672 |
| BL-042 | RAEB-2 | 298 | 898 | 710 |
| AML cell lines, FAB classification | ||||
| THP-1 | FAB M5 | 1275 | 5323 | 1169 |
| Molm13 | FAB M5 | 162 | 1809 | 256 |
| MonoMac6 | FAB M5 | 594 | 627 | 1376 |
| K562 | CML | 2618 | 2497 | 973 |
| . | WHO 2008 . | AT1223 . | AT1331 . | AT1337 . |
|---|---|---|---|---|
| AML with recurrent genetic abnormalities | ||||
| BL-p67 | t(8;21) | 405 | 120 | 231 |
| BL-065 | t(8;21) | 832 | 1016 | 841 |
| BL-059 | RUNX1+ FLT3/ITD+ | 688 | 20 | 247 |
| BL-038* | Inv16 | 292 | 344 | 937 |
| BL-025 | Inv16 | 126 | 449 | 471 |
| BL-037 | APL | 204 | 87 | 353 |
| BL-057 | EVI1 17p del | 20 | 333 | 426 |
| BL-061† | NPM1+ | 1079 | 766 | 775 |
| BL-051 | NPM1+ | 171 | 81 | 11 |
| BL-p78 | NPM1+ | 238 | 822 | 771 |
| AML with myelodysplasia-related changes | ||||
| BL-054 | Multilineage dysplasia | 786 | −22 | 289 |
| Therapy-related myeloid neoplasm | ||||
| BL-047 | t-AML (latency, 3 years) | 302 | 362 | 972 |
| AML-NOS | ||||
| BL-007 | NOS; M0 | 267 | 285 | 124 |
| BL-046 | NOS; M4 | 1104 | 569 | 296 |
| BL-034‡ | NOS; M5 | 785 | 502 | 694 |
| Myelodysplastic syndrome | ||||
| BL-032 | RCUD | 265 | 1512 | 261 |
| BL-011 | RCMD | 616 | 939 | 1225 |
| BL-062 | RAEB-1 | 628 | 766 | 367 |
| BL-022 | RAEB-2 | 205 | 464 | 565 |
| BL-033 | RAEB-2 | 1088 | 811 | 672 |
| BL-042 | RAEB-2 | 298 | 898 | 710 |
| AML cell lines, FAB classification | ||||
| THP-1 | FAB M5 | 1275 | 5323 | 1169 |
| Molm13 | FAB M5 | 162 | 1809 | 256 |
| MonoMac6 | FAB M5 | 594 | 627 | 1376 |
| K562 | CML | 2618 | 2497 | 973 |
Mean fluorescence intensity (MFI) for antibody binding to blasts isolated from newly diagnosed patients in our clinic. Depicted is MFI for each antibody minus MFI of the negative control antibody (CD30). A difference in δ MFI of >100 was considered relevant.
CML, chronic myeloid leukemia; RAEB, refractory anemia with excess blasts; RCMD, refractory cytopenia with multilineage dysplasia; RCUD, refractory cytopenia with unilineage dysplasia; t-AML, therapy-related AML.
Example of antibody-induced cell death of these blasts is depicted in Figure 3D.
Example of antibody staining to blasts of this patient is depicted in Figure 1B.
Patient deferred further treatment; cytogenetic and molecular analyses were not performed. Morphologically, all patients (except BL-007) in this table had (myelo)monocytic AML (FAB classification M4 and M5). U5 snRNP200 complex–specific antibodies did not bind to 20 other patients with AML with morphologically undifferentiated or minimally differentiated types of AML (FAB classification M0-M2; data not shown).
Characteristics of AML-specific antibodies
| Antibody . | Isotype . | SHMs VH/VL . | Target . |
|---|---|---|---|
| Patient 59 | |||
| AT1219 | IgG1κ | 10/9 | TBD |
| AT1223 | IgG3λ | 0/6 | U5 snRNP200 complex |
| AT1225 | IgG3κ | 1/1 | U5 snRNP200 complex |
| AT1322 | IgG3λ | 0/0 | TBD |
| AT1324 | IgG3κ | 6/3 | TBD |
| AT1331 | IgG1κ | 1/1 | U5 snRNP200 complex |
| Patient 58 | |||
| AT1333 | IgG3λ | 18/5 | U5 snRNP200 complex |
| AT1334 | IgG3κ | 14/6 | TBD |
| AT1335 | IgG3λ | 0/0 | U5 snRNP200 complex |
| AT1336 | IgG3λ | 4/9 | TBD |
| AT1337 | IgG3κ | 4/8 | U5 snRNP200 complex |
| Patient 101 | |||
| AT1413 | IgG1κ | 26/11 | CD43s39 |
| AT1414 | IgG3κ | 9/6 | TBD |
| AT1415 | IgG3λ | 9/9 | TBD |
| AT1416 | IgG3λ | 9/5 | TBD |
| AT1508 | IgG1κ | 14/12 | U5 snRNP200 complex |
| Antibody . | Isotype . | SHMs VH/VL . | Target . |
|---|---|---|---|
| Patient 59 | |||
| AT1219 | IgG1κ | 10/9 | TBD |
| AT1223 | IgG3λ | 0/6 | U5 snRNP200 complex |
| AT1225 | IgG3κ | 1/1 | U5 snRNP200 complex |
| AT1322 | IgG3λ | 0/0 | TBD |
| AT1324 | IgG3κ | 6/3 | TBD |
| AT1331 | IgG1κ | 1/1 | U5 snRNP200 complex |
| Patient 58 | |||
| AT1333 | IgG3λ | 18/5 | U5 snRNP200 complex |
| AT1334 | IgG3κ | 14/6 | TBD |
| AT1335 | IgG3λ | 0/0 | U5 snRNP200 complex |
| AT1336 | IgG3λ | 4/9 | TBD |
| AT1337 | IgG3κ | 4/8 | U5 snRNP200 complex |
| Patient 101 | |||
| AT1413 | IgG1κ | 26/11 | CD43s39 |
| AT1414 | IgG3κ | 9/6 | TBD |
| AT1415 | IgG3λ | 9/9 | TBD |
| AT1416 | IgG3λ | 9/5 | TBD |
| AT1508 | IgG1κ | 14/12 | U5 snRNP200 complex |
SHM, somatic hypermutation; TBD, to be determined; VH, variable heavy domain; VL, variable light domain.
Target identification of AML-specific antibodies: U5 snRNP200 complex
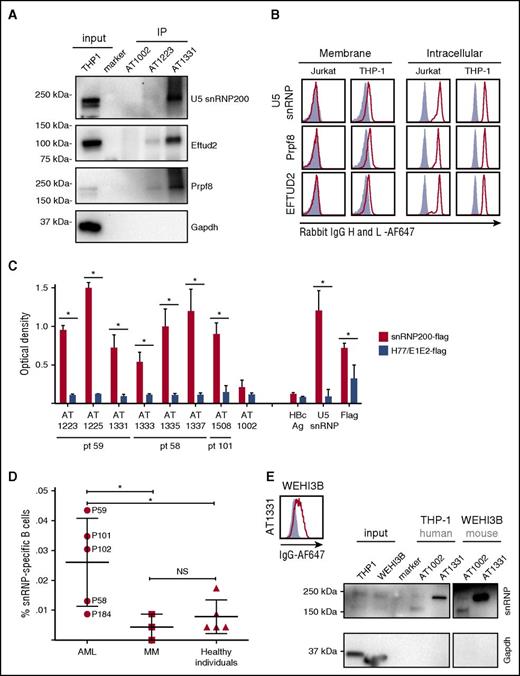
We selected 3 antibodies and generated recombinant forms for further analysis: AT1223 (IgG3), AT1337 (IgG3), and AT1331 (IgG1). Immunoprecipitation of cell membrane and total lysates of THP-1 cells by AT1223 and AT1331 yielded specific bands at ∼250 and ∼120 kDa. liquid chromatography–tandem mass spectrometry analysis of the ∼250-kDa AT1331 band identified the U5 small nuclear ribonucleoprotein helicase snRNP200 (Brr2; HELIC2) and its close relative helicase ASCC3 (HELIC1), complexed with the U5 spliceosome components Prpf8 (Prp8) and EFTUD2 (snRNP116) in the ∼120-kDa band as the major hits. Western blot analysis confirmed the U5 snRNP200 complex as the target of these antibodies (Figure 2A). The U5 snRNP200 complex is a large multiprotein component of the spliceosome indispensable for pre–messenger RNA splicing and is located in both nucleus and cytoplasm.15 We confirmed the intracellular localization of the U5 snRNP200 complex in the Jurkat T-cell leukemia cell line and in THP-1 cells with commercially available U5 snRNP200–, Prpf8-, and EFTUD2-specific antibodies. Contrary to Jurkat cells, THP-1 cells also expressed the U5 snRNP200 complex on the plasma membrane (Figure 2B).
Target identification of AML-specific antibodies: U5 snRNP200 complex. (A) Immunoblotting of THP-1 immunoprecipitates (IP) with AT1223, AT1331, and AT1002 with rabbit snRNP200–, PRPF8–, and EFTUD2– and mouse GAPDH–specific antibodies. (B) Jurkat and THP-1 cells were stained with a commercially available rabbit anti-PRPF8 (Abcam), -snRNP200 (Bethyl Labs), and -EFTUD2 (Abcam) antibodies on the membrane (left panels) and intracellularly (right panels). Polyclonal rabbit antihepatitis core antigen (HBcAg) was used as negative control (gray-filled histograms). (A-B) Depicted is 1 representative experiment of at least 3 independent experiments (Table 2). (C) Seven AML-specific antibodies recognize the U5 snRNP200 complex, as tested in ELISA. Depicted are means of 3 independent experiments and standard deviations. Flag-tagged hepatitis C–specific protein (H77/E1/E2-flag) was used as a negative control. SnRNP200, commercially available rabbit anti-snRNP200; snRNP200-flag, flag-tagged snRNP200 lysate; AT1002, influenza-specific antibody. *P < .003. (D) Screening of 20 cells per well cultures of 5 allogeneic HSCT recipients with lasting anti-AML responses (AML), 3 patients with multiple myeloma (MM) in long-time remission after allogeneic HSCT, and 5 healthy individuals for antibodies binding to snRNP200 (ELISA). In total, 23.040 B cells per patient were screened. NS, not significant. (E) Immunoblotting of THP-1 (human AML) and WEHI-3B (mouse AML) immunoprecipitates with AT1331 and AT1002 with rabbit snRNP200– and mouse GAPDH–specific antibodies. Specific U5 snRNP200 complex stain is seen for both human and mouse AML cells. (Right panel) Binding of AT1331 to WEHI-3B cells.
Target identification of AML-specific antibodies: U5 snRNP200 complex. (A) Immunoblotting of THP-1 immunoprecipitates (IP) with AT1223, AT1331, and AT1002 with rabbit snRNP200–, PRPF8–, and EFTUD2– and mouse GAPDH–specific antibodies. (B) Jurkat and THP-1 cells were stained with a commercially available rabbit anti-PRPF8 (Abcam), -snRNP200 (Bethyl Labs), and -EFTUD2 (Abcam) antibodies on the membrane (left panels) and intracellularly (right panels). Polyclonal rabbit antihepatitis core antigen (HBcAg) was used as negative control (gray-filled histograms). (A-B) Depicted is 1 representative experiment of at least 3 independent experiments (Table 2). (C) Seven AML-specific antibodies recognize the U5 snRNP200 complex, as tested in ELISA. Depicted are means of 3 independent experiments and standard deviations. Flag-tagged hepatitis C–specific protein (H77/E1/E2-flag) was used as a negative control. SnRNP200, commercially available rabbit anti-snRNP200; snRNP200-flag, flag-tagged snRNP200 lysate; AT1002, influenza-specific antibody. *P < .003. (D) Screening of 20 cells per well cultures of 5 allogeneic HSCT recipients with lasting anti-AML responses (AML), 3 patients with multiple myeloma (MM) in long-time remission after allogeneic HSCT, and 5 healthy individuals for antibodies binding to snRNP200 (ELISA). In total, 23.040 B cells per patient were screened. NS, not significant. (E) Immunoblotting of THP-1 (human AML) and WEHI-3B (mouse AML) immunoprecipitates with AT1331 and AT1002 with rabbit snRNP200– and mouse GAPDH–specific antibodies. Specific U5 snRNP200 complex stain is seen for both human and mouse AML cells. (Right panel) Binding of AT1331 to WEHI-3B cells.
In addition to AT1223 and AT1331, the antibodies AT1337, AT1225, AT1333, AT1335, and AT1508 specifically recognized the U5 snRNP200 complex, as determined by ELISA (Figure 2C). These antibodies were derived from all 3 patients screened, suggesting it is a commonly recognized antigen in patients with AML. Screening B cells of 2 other patients with AML with robust, lasting antitumor immunity after allogeneic HSCT (patients 102 and 184; Table 1) by ELISA identified U5 snRNP200 complex–specific B-cell clones in 1 of them (patient 102; Figure 2D), which was confirmed by fluorescence-activated cell sorting (FACS; data not shown). Thus, U5 snRNP200 complex–specific B cells were found in 4 of 5 patients with AML undergoing transplantation. In contrast, screening of 3 patients with multiple myeloma who remained in remission after allogeneic HSCT did not yield U5 snRNP200 complex–binding clones (Figure 2D). In the same ELISA screen, 1 of 5 healthy individuals showed U5 snRNP200 complex+ minibulk cultures (Figure 2D), but binding of these clones could not be confirmed with FACS (data not shown). Thus, none of the healthy individuals screened mounted significant B-cell responses against U5 snRNP200 complexes (Figure 2D). Because, in contrast to AML blasts, multiple myeloma cells do not express cell-surface U5 snRNP200 complex proteins, our data indicate the U5 snRNP200 complex to be immunogenic upon exposure on cell surfaces of tumor cells, explaining why antibodies specific for this complex are found only in patients with AML (Figure 2D). Immunoprecipitation of the mouse AML cell line WEHI-3B with AT1331 confirmed the U5 snRNP200 complex as the target antigen. Mouse and human snRNP200 are 99.7% homologous, confirming that the antibodies recognize a highly conserved epitope (Figure 2E). FACS analysis of these cells showed that the U5 snRNP200 complex antibodies bind these cells, indicating that the mechanism of extracellular expression of these proteins is conserved across species.
U5 snRNP200 antibodies induce nonapoptotic cell death in vitro
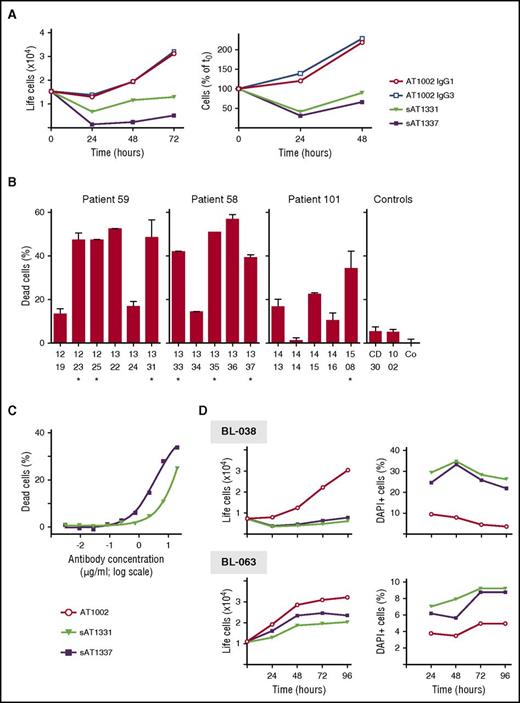
Incubation of THP-1 cells with the AML antibodies showed that some of the AML-specific antibodies prevented in vitro outgrowth of THP-1 cells, whereas other AML-specific antibodies or non–AML-specific antibodies did not (Figure 3A-B). Cells did not become resistant to killing; when THP-1 cells that survived the first incubation with AT1337 and AT1331 were again incubated with the U5 snRNP200 antibodies, outgrowth of cells was again prevented (Figure 3A, right panel). Screening of the killing capacity of all AML-specific antibodies revealed that all U5 snRNP200 complex–recognizing AML-specific antibodies killed THP-1 cells in vitro (Figure 3B). Two non–snRNP200-binding AML-specific antibodies, AT1322 and AT1336, also induced death of >40% of the THP-1 cells (Figure 3B); the specificities of these antibodies remain to be determined. U5 snRNP200 complex–specific antibodies killed target cells in a dose-dependent manner (Figure 3C). Importantly, expansion of primary AML cells (obtained from patients BL-038 and BL-063) was blocked when cultured in the presence of AT1331 or AT1337 (Figure 3D).
U5 snRNP200 antibodies induce death in vitro. (A) Addition of AML-specific U5 snRNP200 complex–binding antibodies sAT1331 (black triangles) and sAT1337 (black squares) to THP-1 cells at time point 0 blocked expansion of this cell line, whereas the flu antibodies AT1002 IgG1 (open circles) and AT1002 IgG3 (open squares) did not (left panel). (Right panel) Reculturing of THP-1 cells that survived antibody treatment (left panel), again in the presence of sAT1331, sAT1337, or control antibody. Antibodies were added at 24-hour intervals at a concentration of 7.5 μg/mL. Viable cells were 4′,6-diamidino-2-phenylindole− (DAPI−). (B) THP-1 cells were incubated with AML-specific antibodies for 4 hours at 37°C, after which cell death was measured using a standard bead controlled assay. Numbers on the x-axis denote each specific AML-specific antibody; asterisks indicate antibodies that are specific for the U5 snRNP200 complex. Co, heat-inactivated medium control. (C) THP-1 cells were incubated with increasing concentrations of cytotoxic AML antibodies sAT1331 and sAT1337, resulting in a concentration-dependent death of target cells. (D) Primary AML blasts isolated from newly diagnosed patients with AML (BL-038: upper panels; BL-063: lower panels) cultured in the presence of sAT1331, sAT1337, or control antibody. (Left panels) Numbers of life cells; (right panels) percentage of DAPI+ cells.
U5 snRNP200 antibodies induce death in vitro. (A) Addition of AML-specific U5 snRNP200 complex–binding antibodies sAT1331 (black triangles) and sAT1337 (black squares) to THP-1 cells at time point 0 blocked expansion of this cell line, whereas the flu antibodies AT1002 IgG1 (open circles) and AT1002 IgG3 (open squares) did not (left panel). (Right panel) Reculturing of THP-1 cells that survived antibody treatment (left panel), again in the presence of sAT1331, sAT1337, or control antibody. Antibodies were added at 24-hour intervals at a concentration of 7.5 μg/mL. Viable cells were 4′,6-diamidino-2-phenylindole− (DAPI−). (B) THP-1 cells were incubated with AML-specific antibodies for 4 hours at 37°C, after which cell death was measured using a standard bead controlled assay. Numbers on the x-axis denote each specific AML-specific antibody; asterisks indicate antibodies that are specific for the U5 snRNP200 complex. Co, heat-inactivated medium control. (C) THP-1 cells were incubated with increasing concentrations of cytotoxic AML antibodies sAT1331 and sAT1337, resulting in a concentration-dependent death of target cells. (D) Primary AML blasts isolated from newly diagnosed patients with AML (BL-038: upper panels; BL-063: lower panels) cultured in the presence of sAT1331, sAT1337, or control antibody. (Left panels) Numbers of life cells; (right panels) percentage of DAPI+ cells.
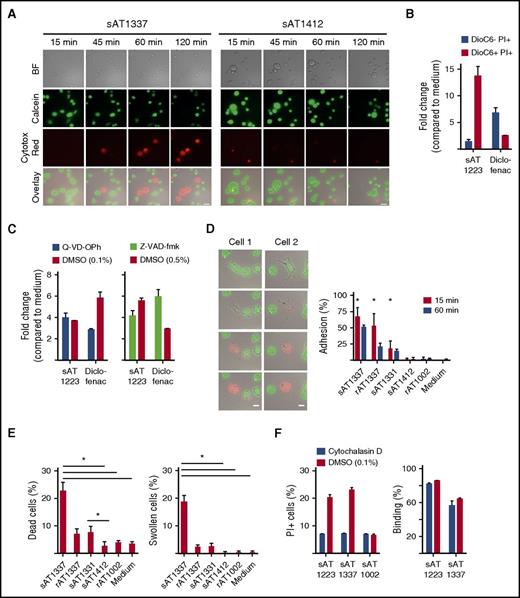
In investigating the mechanism of the inhibition of expansion or survival of THP-1 cells, we observed that U5 snRNP200 complex–recognizing AML-specific antibodies killed THP-1 cells in vitro independent of complement (serum was heat inactivated) and independent of effector cells such as NK cells. Cell death occurred within 1 to 2 hours as visualized by time-lapse microscopy. Upon incubation with the antibodies, cells started to swell, lost their membrane integrity, and died, as indicated by loss of green fluorescent calcein AM and gain of red fluorescent Cytotox Red (Figure 4A). The morphology of dying cells in the context of U5 snRNP200 complex antibodies was different than that observed with apoptotic cells, which is characterized by cell shrinkage and fragmentation into membrane-bound apoptotic bodies. This suggested that the antibodies did not induce apoptosis. To examine this further, we employed DiOC6, a cell-permeable, green fluorescent, lipophilic dye that in low concentrations binds mitochondria of live cells, and PI, which enters the cells after permeabilization of the membrane. Apoptotic cells first lose mitochondrial membrane potential (loss of DiOC6 binding) and next become PI+, but U5 snRNP200 complex antibodies induced PI influx in THP-1 cells without prior mitochondrial membrane potential changes (Figure 4B). Cell killing by snRNP200-specific antibodies was not inhibited by the pan-caspase inhibitors Q-VD-OPh or Z-VAD-FMK (Figure 4C), further confirming that these antibodies do not induce apoptosis.
U5 snRNP200 antibody–induced death is nonapoptotic. (A) Time-lapse phase contrast fluorescent imaging of THP-1 cells, labeled with calcein AM (green) and incubated with sAT1337 or the AML-binding, noncytotoxic IgG3 antibody sAT1412 in the presence of Cytotox Red to indicate dying cells (red). Stills were taken every 2 minutes. Dying cells lose calcein AM green and become Cytotox Red. Scale bars represent 25 μm. BF, bright field. (B) THP-1 cells incubated with sAT1223 led to increased membrane permeability (PI+) but not loss of mitochondrial membrane potential (DioC6+). Diclofenac, which induces apoptosis of THP-1 cells,14 was used as a positive control. Incubation with diclofenac led to loss of mitochondrial potential and increased membrane permeability (DioC6− PI+). (C) Cell death of THP-1 cells by U5 snRNP200 complex–specific antibodies (incubated overnight) could not be blocked with the pan-caspase inhibitors Z-VAD-fmk or Q-VD-OPh. DMSO, dimethyl sulfoxide. (D) Stills of time-lapse imaging of 2 dying THP-1 cells, with frames taken every 2 minutes. Dying cells became adherent, flattened out, and lost membrane integrity, as indicated by loss of green calcein AM dye and uptake of Cytotox Red dye. Scale bars represent 25 μm. Graph indicates percentage of cells adhering to the plate at 15 and 60 minutes (n = 3). (E) Cell death and induction of typical morphologic changes by B cell–derived (sAT1337, sAT1331) and recombinant (rAT1337) antibodies (n = 3). (F) Preincubation of the target cells with the membrane-stabilizing agent cytocholasin D (in 0.1% DMSO) protected the THP-1 cells against cell death by AML-specific cytotoxic antibodies (left panel); however, this did not affect binding of AML-specific antibodies (right panel). Cells were incubated with antibodies for 4 hours.
U5 snRNP200 antibody–induced death is nonapoptotic. (A) Time-lapse phase contrast fluorescent imaging of THP-1 cells, labeled with calcein AM (green) and incubated with sAT1337 or the AML-binding, noncytotoxic IgG3 antibody sAT1412 in the presence of Cytotox Red to indicate dying cells (red). Stills were taken every 2 minutes. Dying cells lose calcein AM green and become Cytotox Red. Scale bars represent 25 μm. BF, bright field. (B) THP-1 cells incubated with sAT1223 led to increased membrane permeability (PI+) but not loss of mitochondrial membrane potential (DioC6+). Diclofenac, which induces apoptosis of THP-1 cells,14 was used as a positive control. Incubation with diclofenac led to loss of mitochondrial potential and increased membrane permeability (DioC6− PI+). (C) Cell death of THP-1 cells by U5 snRNP200 complex–specific antibodies (incubated overnight) could not be blocked with the pan-caspase inhibitors Z-VAD-fmk or Q-VD-OPh. DMSO, dimethyl sulfoxide. (D) Stills of time-lapse imaging of 2 dying THP-1 cells, with frames taken every 2 minutes. Dying cells became adherent, flattened out, and lost membrane integrity, as indicated by loss of green calcein AM dye and uptake of Cytotox Red dye. Scale bars represent 25 μm. Graph indicates percentage of cells adhering to the plate at 15 and 60 minutes (n = 3). (E) Cell death and induction of typical morphologic changes by B cell–derived (sAT1337, sAT1331) and recombinant (rAT1337) antibodies (n = 3). (F) Preincubation of the target cells with the membrane-stabilizing agent cytocholasin D (in 0.1% DMSO) protected the THP-1 cells against cell death by AML-specific cytotoxic antibodies (left panel); however, this did not affect binding of AML-specific antibodies (right panel). Cells were incubated with antibodies for 4 hours.
Many pathways of nonapoptotic cell death have been described,16 but none of these pathways has been as clearly defined as apoptotic cell death. A distinctive feature that we observed was the rapid adherence and flattening out of target cells in the presence of U5 snRNP200 complex antibodies. Thus, upon incubation with AT1337, cells become adherent and then lost membrane integrity and died (Figure 4D-E; supplemental Video 1, available on the Blood Web site). AT1331 induced less adherence, but with this antibody as well, most adherent cells died (Figure 4D-E). Of note, AT1331 is an IgG1 antibody, whereas AT1337 is an IgG3 antibody. Because IgG3 antibodies bind to Fc receptors with higher affinity than IgG1 antibodies,17 the superior cytotoxicity of AT1337 could have been related to its IgG3 format. To test this, we produced AT1331 recombinant in an IgG3 format, but this did not improve cytotoxicity (data not shown). In fact, when AT1337 was produced as a recombinant IgG3 antibody (rAT1337), the antibody was significantly less cytotoxic compared with the natural (B cell derived) sAT1337 antibody (Figure 4E). Recombinant rAT1337 did only briefly reduce target-cell motility (Figure 4D), and the extent of adherence induced by the U5 snRNP200 complex antibodies seemed to relate to the amount of target-cell death induced. For these experiments, the IgG3 antibody sAT1412 was used as a control antibody. This antibody binds THP-1 cells but despite its IgG3 format does not induce target-cell adhesion or death. When target cells were incubated with the actin polymerization inhibitor cytochalasin D, cell death was prevented, while antibody binding to the cells was unaffected (Figure 4F). Together these data suggest that patient-derived, U5 snRNP200 complex–specific antibodies induced death of target cells via a nonapoptotic, Fc-dependent process that brought forth tracking forces leading to loss of target-cell membrane integrity.
U5 snRNP200 antibodies delay tumor growth and can induce tumor necrosis in vivo
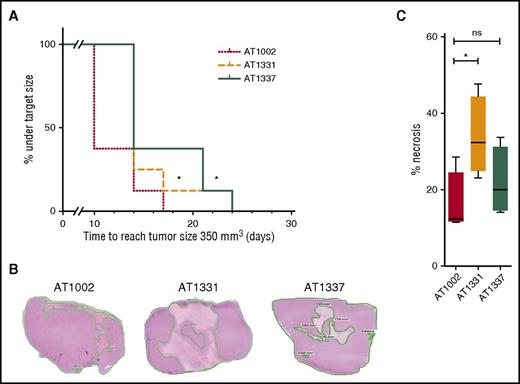
Given the specificity and the functional activities of the snRNP200-specific antibodies, we hypothesized that these antibodies might have contributed to the tumor-free status of the patients from whom the antibodies were derived. To test this hypothesis, we examined the capacity of these U5 snRNP200 complex–binding antibodies to affect tumor growth in vivo. Three weeks after subcutaneous injection of THP-1 cells into 24 mice, we treated the animals biweekly with either of 2 U5 snRNP200 complex–specific antibodies or a control antibody at a dose of 10 mg/kg. For these experiments, large amounts of antibody could only be produced in a recombinant, and thus less cytotoxic, format. Nevertheless, we found that treatment of mice with rAT1331 and rAT1337 resulted in significant tumor growth delay compared with treatment with isotype control (Figure 5A). When mice were euthanized 25 days after the start of treatment and 8 days after the last antibody injection, tumor sizes were no longer different between groups, but the amount of viable tumor tissue was significantly lower in the rAT1331-treated group (Figure 5B-C). These data demonstrate that U5 snRNP200 complex–specific antibodies affect growth of THP-1 cells in vivo. Interestingly, no obvious adverse effects were noted in the mice treated with the U5 snRNP200 antibodies, despite the antibodies reacting with mouse U5 snRNP200 complex, indicating that these antibodies do not act on normal cells in this model and could therefore be safe for therapeutic application, although additional studies in a setting with human hematopoietic cells present are needed to confirm this.
U5 snRNP200 complex antibodies delay tumor growth and can induce tumor necrosis in vivo. (A) Severe combined immunodeficient mice with THP-1 growing as a solid tumor under the skin were treated with cytotoxic AML-specific antibodies rAT1331 or rAT1337 or with an isotype control (AT1002). Treatment started when the tumors had a size of 100 mm3. Further tumor growth to a size of 350 mm3 was significantly delayed in mice treated with cytotoxic AML antibodies. (B) Mice were euthanized after 25 days, 8 days after the last antibody injection. Three representative hematoxylin and eosin stains of paraffin-embedded THP-1 tumors, 1 of each group, are shown. (C) Of each tumor, the area of necrosis was calculated as percentage of the total surface. Although total tumor sizes no longer differed between groups after 25 days, tumors from rAT1331-treated mice had significantly more necrotic tissue.
U5 snRNP200 complex antibodies delay tumor growth and can induce tumor necrosis in vivo. (A) Severe combined immunodeficient mice with THP-1 growing as a solid tumor under the skin were treated with cytotoxic AML-specific antibodies rAT1331 or rAT1337 or with an isotype control (AT1002). Treatment started when the tumors had a size of 100 mm3. Further tumor growth to a size of 350 mm3 was significantly delayed in mice treated with cytotoxic AML antibodies. (B) Mice were euthanized after 25 days, 8 days after the last antibody injection. Three representative hematoxylin and eosin stains of paraffin-embedded THP-1 tumors, 1 of each group, are shown. (C) Of each tumor, the area of necrosis was calculated as percentage of the total surface. Although total tumor sizes no longer differed between groups after 25 days, tumors from rAT1331-treated mice had significantly more necrotic tissue.
Discussion
Our data indicate that cell-surface exposure of U5 snRNP200 complexes is tumor specific and that engagement of this complex by antibodies can induce nonapoptotic cell death. The cell surface–exposed U5 snRNP200 complex as a target for tumor antibodies was unexpected, because this complex is normally expressed within the cell. Although U5 snRNP200 cell-surface expression on tumor cells has not been described before, some examples are known of ribonucleoproteins exposed on the cell membrane.18 Moreover, components of spliceosomes have been identified as autoimmune targets in systemic lupus erythematosus and mixed connective tissue disease.19,-21 Mutations in spliceosome components have been described in many cancers,22 including AML and myelodysplastic syndrome.23,-25 High-turnover cancers actually depend on the spliceosome to expand.26 U5 snRNP200 complex membrane expression, specifically observed for human and mouse AML blasts and not healthy cells, may reflect spliceosomal stress in leukemic blasts. However, the mechanism by which the U5 snRNP200 complex is exposed on the membrane of the tumor cells remains unclear.
Cytotoxic or killer antibodies that induce nonapoptotic programmed cell death have been described before. Antibodies directed against Lewis(y) and Lewis(b) that are expressed on a range of tumor types kill tumor cells in a nonapoptotic way.27 Anti-NeuGcGM3 antibodies kill non–small-cell lung cancer cells expressing this antigen by inducing membrane pores.28 The CD20 antibody obinutuzumab that is now being tested in the clinic kills leukemia and lymphoma cells via a nonapoptotic mechanism that depends on actin reorganization.29,30 A recent report describes the antibody TAG-A1, which selectively kills undifferentiated human embryonic stem cells via excess reactive oxygen species production.31 Similar to the U5 snRNP200 complex antibodies that we identified, these antibodies induce cell death in an autonomous way via a nonapoptotic mechanism, characterized by cell swelling, that cannot be blocked by pan-caspase inhibitors. Killer antibodies induce actin reorganization, resulting in the formation of pores that lead to the rapid disintegration of target cells. This process has been named oncosis,31,32 a nonapoptotic programmed death pathway that (similar to pyroptosis, necroptosis, and others) represents an area of active research.33
Some of the U5 snRNP200 complex antibodies harbored a significant number of somatic hypermutations in the heavy and light chains, implying antigen exposure induced affinity maturation in vivo. U5 snRNP200 complex antibodies were generated in 4 of 5 high-risk patients with AML but not in patients receiving allogeneic HSCT for multiple myeloma or in healthy individuals. All 9 allogeneic HSCT recipients screened had graft-versus-host disease (GVHD), but U5 snRNP200 complex antibodies were generated only in patients with AML, not in those with multiple myeloma. GVHD per se is therefore not responsible for the generation of U5 snRNP200 complex antibodies. Moreover, a patient with U5 snRNP200 complex–expressing AML who experienced relapse after allogeneic HSCT had not developed U5 snRNP200 complex–specific antibodies (data not shown). Finally, in a previous study, we described a patient who simultaneously developed an AML-specific antibody response and a potent GVL response; after depletion of B cells with rituximab for steroid-refractory GVHD, this patient experienced relapse of AML, suggesting the AML antibodies were functional in this case.34 Taken together, we hypothesize that leukemia-specific cytotoxic antibodies generated in patients with AML undergoing transplantation contributed to the tumor-free status of these patients. Although our observations point in the direction of an immune reaction elicited by AML cells, they should not be considered as definitive proof that this indeed occurred.
Immunological checkpoint blockade studies have established that tumor-specific cytotoxic T lymphocytes specifically recognizing neoantigens are important effectors in antitumor immunity. Neoantigens arise because of mutations in malignant cells, and the immunogenicity of cancers is correlated with their mutational status, with highly mutated cancers such as melanoma and non–small-cell lung carcinoma more susceptible to immunotherapy, particularly with checkpoint inhibitors.2,35 AML is characterized by a low mutational status36 and may therefore be less immunogenic for T cells and less sensitive to checkpoint inhibition than highly mutated cancers. Our study demonstrates that tumors with relatively few mutations can also be immunogenic in the setting of allogeneic HSCT, resulting in a tumor-specific antibody response.
Our data raise the possibility that antibodies obtained from patients who achieved long-term remission of their cancer have therapeutic utility. U5 snRNP200 complex antibodies may potentiate the effect of conventional chemotherapeutic regimens, as has been demonstrated for the CD20 antibodies rituximab and obinutuzumab in B-cell non-Hodgkin lymphoma. In addition, such antibodies may have a vaccinal effect by inducing long-term antitumor T-cells response, as reported recently.37,38 Moreover, the U5 snRNP200 complex has potential as a target for chimeric antigen receptor T cells, using the antibody as the chimeric antigen receptor. Together, our findings demonstrate that screening the antibody repertoire of patients with cancer enables the discovery of novel tumor-associated antigens that can be targeted by therapeutic or diagnostic antibodies or tumor-specific vaccines.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank the patients who participated in this study; Ludo Evers for chimerism analysis, the Department of Pathology for technical support, Przemek M. Krawczyk for time-lapse imaging support, and the Department of Hematology Trial Office for the collection of AML samples (Academic Medical Center, Amsterdam, The Netherlands); and Nico Smit, Leiden University Medical Center, Leiden, The Netherlands, for providing fibroblasts.
This study was supported by an intramural grant from the Academic Medical Center (PhD Scholarship to M.A.G.), a ZonMW Clinical Fellowship (#90700314; M.D.H.), and the Dutch Cancer Foundation (#UVA2014-6557; M.D.H. and H.S.).
Authorship
Contribution: M.A.G., M.K., G.d.J., and G.M. designed and performed the experiments and analyzed the data; M.A.G. and M.D.H. wrote the first draft of the paper; E.Y., K.W., M.B., and S.E.L. set up and performed the experiments; Y.B.C. and A.Q.B. performed the sequence and the subsequent analyses; P.J.H., D.S., and M.K. performed and analyzed the mass spectrometry data; P.M.v.H., T.B., and H.S. contributed critical input and helped in writing the paper; and M.D.H. initiated the study, designed the research, managed the patients, and analyzed the data.
Conflict-of-interest disclosure: M.A.G., M.K., G.M., S.E.L., A.Q.B., Y.B.C., K.W., M.B., P.M.v.H., T.B., and H.S. are employees of AIMM Therapeutics, a company that develops monoclonal antibodies to prevent and treat infectious diseases and cancer. The remaining authors declare no competing financial interests.
Correspondence: Mette D. Hazenberg, Department of Hematology F4-224, Academic Medical Center, Meibergdreef 9, 1105 AZ Amsterdam, The Netherlands; e-mail: m.d.hazenberg@amc.nl.

![Figure 1. Identification of AML-specific antibodies. (A) Antibodies in the supernatant of 1 of the minicultures (11C9; 40 cells per well) binding to the AML cell line THP-1 (top panels). B cells from this miniculture were plated into 1 cell per well solutions and supernatants again screened for antibodies binding to THP-1 (lower panels). Positive control: HLA antigen D related (HLA-DR); negative control: in-house generated, influenza-specific antibody AT1002.12 Recombinant-produced 11C9-22C11 was named AT1223. FSC, free secretory component. (B) Representative example of binding of 3 of the selected antibodies (AT1223, AT1331 [patient 59], and AT1337 [patient 58]) to the AML cell lines THP-1, Molm13, MonoMac 6, and K562, and primary AML blasts freshly isolated from a patient with AML (BL-061; NPM1+). (C) AML antibodies did not bind to bone marrow–derived CD34+ hematopoietic progenitor cells, peripheral blood mononuclear cells (PBMCs), monocytes, fibroblasts, or endothelial cells isolated from healthy individuals, nor to the colon cell line HT29 or the liver cell line HepG2. Negative control: AT1002 (gray-filled histograms). HUVEC, human umbilical vein endothelial cell.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/131/1/10.1182_blood-2017-02-768762/4/m_blood768762f1.jpeg?Expires=1767724548&Signature=DlM6b6KWZpKRCqdUqN~Ya9DGJchSX70bzJMF2AlfTc~3-IhBKHVzv04jDgVpQj56pwjVsYyRng6ovASqevU4c3mAoj6jweGb6Q5-4MGTBzGLwRCs~6ak81m4pghGEyB89XmlHTuJwbA-M3vwnKDKAD4Bu814A~saPlYptOeCTclyqD~nrk18e5Pc3Mra71V5M800Ut5SfshMm8kWqFToEaLhsowUJua7dyI7GiLDZur1vh25XwZWIikhaX3zJocMRLr4JdV9MuVYcjH2hjOxzfh4g6ITz3iKb5QQy8JsmJosXlauOzSV8pTDQlohBdVUNBsq1w-w7yjYqTfFHEU8vg__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
This feature is available to Subscribers Only
Sign In or Create an Account Close Modal