Key Points
Genomic deletions of CDKN2A/2B are a new independent prognostic risk factor in adult Ph+ ALL.
Abstract
We investigated the role of copy number alterations to refine risk stratification in adult Philadelphia chromosome positive (Ph)+ acute lymphoblastic leukemia (ALL) treated with tyrosine kinase inhibitors (TKIs) and allogeneic stem cell transplantation (aSCT). Ninety-seven Ph+ ALL patients (median age 41 years; range 18-64 years) within the prospective multicenter German Multicenter ALL Study Group studies 06/99 (n = 8) and 07/2003 (n = 89) were analyzed. All patients received TKI and aSCT in first complete remission (CR1). Copy number analysis was performed with single nucleotide polymorphism arrays and validated by multiplex ligation-dependent probe amplification. The frequencies of recurrently deleted genes were: IKZF1, 76%; CDKN2A/2B, 45%; PAX5, 43%; BTG1, 18%; EBF1, 13%; ETV6, 5%; RB, 14%. In univariate analyses, the presence of CDKN2A/2B deletions had a negative impact on all endpoints: overall survival (P = .023), disease-free survival (P = .012), and remission duration (P = .036). The negative predictive value of CDKN2A/2B deletions was retained in multivariable analysis along with other factors such as timing of TKI therapy, intensity of conditioning, achieving remission after induction phase 1 and BTG1 deletions. We therefore conclude that acquired genomic CDKN2A/2B deletions identify a subgroup of Ph+ ALL patients, who have an inferior prognosis despite aSCT in CR1. Their poor outcome was attributable primarily to a high relapse rate after aSCT.
Introduction
Philadelphia chromosome positive (Ph+) acute lymphoblastic leukemia (ALL) is characterized by a reciprocal translocation that fuses the ABL1 oncogene on chromosome 9 to a breakpoint cluster region (BCR) on chromosome 22. Ph+ ALL is the largest genetically defined subtype in adult ALL and is traditionally considered the subtype with the worst prognosis. Substantial improvements in response and long-term survival have been achieved by combining tyrosine kinase inhibitors (TKIs) such as imatinib or dasatinib with an intensive chemotherapy regimen.1-6 However, allogeneic stem cell transplantation (aSCT) in first complete remission (CR1) is the only reliable curative post-remission therapy in adults.7 Following combined treatment strategies with TKIs, cytotoxic chemotherapy, and aSCT, long-term disease-free survival (DFS) ranges from 30% to 69%,4,5,8,9 with transplant-related mortality and relapse contributing approximately equally to treatment failure. This heterogeneity of prognosis suggests additional biologic pathomechanisms other than the BCR-ABL driver lesion that have not been systematically determined. This is further exemplified by the differences between adult and pediatric Ph+ ALL, with the majority of children appearing to be cured by combined TKI and chemotherapy, even without aSCT.10 To this end, microarray-based genome-wide profiling studies have revealed numerous submicroscopic genetic lesions in patients with BCR-ABL− as well as Ph+ ALL. The prognostic relevance of these alterations has been studied extensively in pediatric ALL and to a lesser degree in adult ALL.11-15 Most recurring genomic abnormalities involve single genes, which are implicated in B-cell differentiation, cell survival, or cell-cycle progression, such as CDKN2A/2B, IKZF1, PAX5, ETV6, RB1, BTG1, EBF1, and therefore, have been suggested to play a role in leukemogenesis. Deletions of IKZF1, which encodes the lymphoid transcription factor Ikaros, and of CDKN2A/2B and PAX5 genes have received particular attention because of their high frequency, particularly in Ph+ ALL. IKZF1 deletions have been linked to an inferior outcome in most pediatric ALL studies16-18 as well as in studies involving adult and adolescent Ph− BCP-ALL19 and adult Ph+ ALL,12 respectively. However, because these associations with prognosis were significant only in univariate but not in multivariate analyses in several initial studies, the additional prognostic value of IKZF1 deletions has been debated.20,21 Nevertheless, subsequent investigations taking into account leukemia subtype and therapeutic context have also retained IKZF1 deletions as an unfavorable prognostic marker.22,23
The prognostic relevance of IKZF1, CDKN2A/2B, PAX5, and other recurrent genomic aberrations in the setting of aSCT for adult patients with Ph+ ALL has hardly been examined. To this end, in adult Ph+ ALL, the presence of additional cytogenetic lesions in adult Ph+ ALL patients was shown to negatively influence prognosis in the therapeutic setting of TKI combined with chemotherapy24 or even beyond aSCT.25 Furthermore, TKI resistance in this patient group has been linked to ABL1 kinase mutations and/or the presence of genomic deletions such as IKZF1 or CDKN2A/2B.26,27 Kim et al recently investigated the prognostic relevance of IKZF1 deletions in adult Ph+ ALL patients in a uniform aSCT setting and combined treatment with imatinib. Therefore, the investigators could show a trend for higher incidence of relapse in patients with IKZF1 deletions, albeit not reaching significance nor independent statistical power in multivariate analyses.28 We therefore addressed the question whether in adult Ph+ ALL the putative adverse prognosis conferred by IKZF1 and/or other frequent microdeletions of genes associated with B-cell development can be overcome by aSCT, and whether these genomic alterations are predictive of outcome independently of, or in addition to, levels of minimal residual disease (MRD) during front-line therapy. By single nucleotide polymorphism (SNP) microarray analysis and validation with multiplex ligation-dependent probe amplification (MLPA), we examined adult Ph+ ALL samples obtained at initial diagnosis from n = 97 patients who received imatinib as part of their treatment and underwent aSCT in CR1, and correlated the genomic data with outcome after aSCT.
Patients and methods
Patients
The molecular and clinical outcome analyses in this study were carried out retrospectively on bone marrow (n = 61) or peripheral blood (n = 35), unknown (n = 1) samples from initial diagnosis of n = 97 adult patients with Ph+ ALL who were enrolled in the German Multicenter ALL Study Group (GMALL) studies 06/99 (NCT00199056) (n = 8) and 07/2003 (NCT00198991) (n = 89) from October 2001 to November 2009. Molecular analyses were carried out with written informed consent of all patients, approved by the Medical Ethics Committee of the University of Frankfurt, Germany in accordance with the Declaration of Helsinki. Characteristics of the 97 retrospectively analyzed patients are summarized in Table 1. All patients received imatinib and underwent aSCT within the study protocols.29 A detailed description of therapy is summarized by Wassmann et al.6 Briefly, all patients in the current study received an identical backbone of chemotherapy consisting of a prephase with dexamethasone and cyclophosphamide, followed by induction chemotherapies consisting of IP1 (weeks 1 to 3, containing dexamethasone, vincristine, daunorubicine and PEGylated-asparaginase) followed by induction phase 2 (IP2, weeks 4 to 6, containing cyclophosphamide, cytarabine, and 6-mercaptopurin). Consolidation consisted of dexamethasone, vindesine, high-dose methotrexate (MTX), etoposide, and cytarabine. Central nervous system prophylaxis consisted of intrathecal injection of MTX (15 mg) once during prephase and thrice during IP2 and a single intrathecal administration of MTX 15 mg, cytarabine 40 mg, and dexamethasone 4 mg after consolidation 1. Patients achieving CR in IP1 also received a prophylactic cranial irradiation of 24 Gy parallel to IP2. Patients in this current study received imatinib 600 mg once a day in 3 different schedules: in the cohort defined as “Imatinib late,” 5 patients received imatinib in an alternating schedule to chemotherapy either after IP2 or after the consolidation phase. The remaining 23 patients started imatinib treatment after IP1. The cohort defined as “Imatinib early” started imatinib directly after the prephase in parallel to IP1. All patients received aSCT in CR1. The recommendation within the GMALL protocol was to use a myeloablative conditioning (MAC) regimen (TBI-based) for patients undergoing aSCT. In the current study cohort, the following regimen was defined as MAC: TBI-based regimen with 12 Gy/cyclophosphamide (n = 40), TBI 12 Gy/etoposide (n = 27), TBI 12 Gy/etoposide/cyclophosphamide (n = 4), TBI 10 Gy/cyclophosphamide (n = 2), TBI 8 Gy/cyclophosphamide (n = 2), TBI 8 Gy/fludarabine (n = 2), TBI 8 Gy/cyclophosphamide/fludarabine (n = 1). The following regimen was considered as reduced intensity conditioning (RIC): fludarabine-based regimen such as fludarabine/busulfan/melphalan (n = 2), fludarabine/busulfan (n = 2), fludarabine/melphalan (n = 1), fludarabine/busulfan/thiotepa (n = 3), treosulfan-based regimen such as treosulfan/cyclophosphamide/etoposide (n = 7) or treosulfan/fludarabine (n = 1) or TBI-based regimen with 4 Gy TBI/cyclophosphamide (n = 1) and 2 Gy/fludarabine (n = 1), n.a. (n = 1).
Patient characteristics
| Parameter . | n . | % . |
|---|---|---|
| Sample size | ||
| All patients aSCT in CR1 | 97 | 100 |
| Age, y | ||
| Median | 41 | |
| Range | 18-64 | |
| Sex | ||
| Male | 58 | 60 |
| Female | 39 | 40 |
| Breakpoint BCR-ABL | ||
| M-Bcr-Abl (b2a2, b3a2) | 33 | 34 |
| m-Bcr-Abl (e1a2) | 52 | 54 |
| e1a3/b2a3 | 2/1 | 3 |
| n.a. | 9 | 9 |
| WBCs, ×109/L | ||
| <30 | 49 | 51 |
| >30 | 46 | 47 |
| n.a. | 2 | 2 |
| SCT donor type | ||
| Matched SIB/UD | 72 (27/45) | 74 |
| Mismatched SIB/UD | 21 (1/20) | 22 |
| Haplo | 1 | 1 |
| n.a. | 3 | 3 |
| Stem cell source | ||
| PBSCT | 92 | 95 |
| BM | 4 | 4 |
| n.a. | 1 | 1 |
| Conditioning regimen | ||
| TBI based (+Cy/+VP16/+other) | 80 | 82 |
| No TBI | 16 | 17 |
| n.a. | 1 | 1 |
| Imatinib therapy | ||
| From induction I | 69 | 71 |
| After induction I (day 24) | 23 | 24 |
| After induction II | 5 | 5 |
| Early clinical response | ||
| CR after IP1 (d26) | 78 | 81 |
| No CR after IP1 (d26) | 9 | 9 |
| n.a. | 10 | 10 |
| Follow-up (range), mo | ||
| Median follow-up OS | 35 (5.4-114.4) | |
| Median follow-up DFS and REM | 20.5 (0.6-108.3) |
| Parameter . | n . | % . |
|---|---|---|
| Sample size | ||
| All patients aSCT in CR1 | 97 | 100 |
| Age, y | ||
| Median | 41 | |
| Range | 18-64 | |
| Sex | ||
| Male | 58 | 60 |
| Female | 39 | 40 |
| Breakpoint BCR-ABL | ||
| M-Bcr-Abl (b2a2, b3a2) | 33 | 34 |
| m-Bcr-Abl (e1a2) | 52 | 54 |
| e1a3/b2a3 | 2/1 | 3 |
| n.a. | 9 | 9 |
| WBCs, ×109/L | ||
| <30 | 49 | 51 |
| >30 | 46 | 47 |
| n.a. | 2 | 2 |
| SCT donor type | ||
| Matched SIB/UD | 72 (27/45) | 74 |
| Mismatched SIB/UD | 21 (1/20) | 22 |
| Haplo | 1 | 1 |
| n.a. | 3 | 3 |
| Stem cell source | ||
| PBSCT | 92 | 95 |
| BM | 4 | 4 |
| n.a. | 1 | 1 |
| Conditioning regimen | ||
| TBI based (+Cy/+VP16/+other) | 80 | 82 |
| No TBI | 16 | 17 |
| n.a. | 1 | 1 |
| Imatinib therapy | ||
| From induction I | 69 | 71 |
| After induction I (day 24) | 23 | 24 |
| After induction II | 5 | 5 |
| Early clinical response | ||
| CR after IP1 (d26) | 78 | 81 |
| No CR after IP1 (d26) | 9 | 9 |
| n.a. | 10 | 10 |
| Follow-up (range), mo | ||
| Median follow-up OS | 35 (5.4-114.4) | |
| Median follow-up DFS and REM | 20.5 (0.6-108.3) |
BM, bone marrow derived stem cell transplantation; Cy, cyclophosphamide; DFS, disease-free survival; haplo, haploident; IP1, induction phase 1; n.a., not available; OS, overall survival; PBSCT, peripheral blood derived stem cell transplantation; REM, remission duration; SIB, sibling; TBI, total body irradiation; UD, unrelated donor; VP16, etoposide; WBCs, white blood cells.
Single nucleotide polymorphism microarray (SNP-A) analysis
Genomic DNA (500 ng per sample) from leukemic blasts was processed according to the Genome Wide Human SNP 6.0 Array protocol (Affymetrix, Santa Clara, CA). Genotyping files were imported into the CNAG 3.3 software to perform allele-specific copy number analysis with anonymous references as previously described.30,31 Known copy number polymorphisms were excluded from the data by comparison with known copy number polymorphisms registered in the University of California Santa Cruz genome browser (http://genome.ucsc.edu/, [hg-18]). Putatively acquired copy number alterations (CNAs) were validated in matched-pair SNP 6.0 array analysis of DNA from initial diagnosis and full remission samples in n = 11 cases.
MLPA analysis
To further validate the presence of deletions of commonly deleted genes in ALL detected by SNP array, the MLPA assay SALSA MLPA P335 ALL-IKZF1 probe mix kit (MRC Holland, Amsterdam, Netherlands) was used according to the manufacturer’s protocol. Thereby, deletions of the following genes were validated: IKZF1, CDKN2A/2B, PAX5, BTG1, EBF, ETV6, and RB. Electrophoresis and relative quantification of fluorescein-labeled amplicons were performed on an ABI-3130 genetic analyzer (Applied Biosystems, Carlsbad, CA). The resulting peak intensities were normalized to the manufacturer’s control probes and to normal DNA as a reference with the Coffalyser Software (MRC Holland). The resulting MLPA data were combined with SNP array data according to the following algorithm: homozygous deletions of the above listed commonly deleted genes were called if either the SNP array data called a homozygous deletion by Hidden Markov Model (HMM)31 and/or at least 2 adjacent MLPA probes measured an intensity ratio of <0.3. Heterozygous deletions were called if either SNP array data called a heterozygous deletion by HMM and/or at least 2 adjacent MLPA probes measured an intensity ratio of <0.7.
Analysis of MRD and BCR-ABL mutations
Analysis of MRD was performed using quantitative real-time polymerase chain reaction (PCR) analysis and confirmatory testing by nested PCR as previously described.32,33 Briefly, peripheral blood and bone marrow mononuclear cells were separated by Ficoll centrifugation. Total RNA was extracted with RNA Bee (Ambion; Invitrogen, Darmstadt, Germany) or RNeasy (Qiagen, Hilden, Germany). Complementary DNA was synthesized from 1 to 5 µg RNA using Moloney Murine Leukemia Virus reverse transcriptase (Promega, Mannheim, Germany) and random hexamers (Applied Biosystems, Darmstadt, Germany). Real-time PCR was performed with the Europe Against Cancer34 primer and probe set for BCR-ABL and ABL1. For samples earlier than 2004, in-house primers were used for GAPDH and BCR-ABL as described previously.35 ABL1 or glyceraldehyde-3-phosphate dehydrogenase (GAPDH) were used as housekeeping genes. Complete molecular response was defined as BCR-ABL/ABL ratio ≤10−4 or a BCR-ABL/GAPDH ratio ≤10−6. Historically, the difference of threshold for GAPDH was defined due to the higher intrinsic expression of this gene by ∼2 log.36
Major molecular response (MMR) for BCR-ABL/ABL was defined as >10−4 but <10−3 or BCR-ABL/GAPDH >10−6 but <10−5; “no MMR” was defined as >10−3 for BCR-ABL/ABL and >10−5 for BCR-ABL/GAPDH. For calling negative MRD, the ABL copy number had to be at least 104 and GAPDH had to be at least 106. MRD evaluation was performed at least once after consolidation for 69 patients. Another 9 patients were evaluated before consolidation. In 2 patients, only fluorescence in situ hybridization (FISH) results were available. These were added to the group “no MMR,” because they were FISH positive.
Statistical analyses
Relapse was defined by recurrence of >5% lymphoblasts in the bone marrow, in the peripheral blood, or by the appearance of extramedullary disease after achieving CR. OS was calculated from date of diagnosis to death of any cause. DFS was calculated from the time point of aSCT until relapse, death from any cause, or development of secondary malignancy. REM was calculated from the time point of aSCT until leukemia relapse. Patients were censored at last follow-up if no event of interest occurred. Univariate analyses of OS, DFS, and REM comparing 2 or more groups were made using the log-rank test. The median duration of follow-up for OS was 35 months (range 5.4-114.4) and 20.5 months (range 0.6-108.3) for REM and DFS. Univariate analyses were carried out using GraphPad Prism version 6.07 for Windows (GraphPad Software, San Diego, CA). For multivariable analyses, Cox proportional hazards models were calculated for OS, DFS, and REM. The following covariates were included in all models: IKZF1 deletion (presence vs absence), PAX5 deletion (presence vs absence), CDKN2A/2B deletion (presence vs absence), BTG1 deletion (presence vs absence), EBF1 deletions (presence vs absence), ETV6 deletions (presence vs absence), RB1 deletions (presence vs absence), total number of recurrent deletions (0 or 1 vs 2 or more), age (years), sex (male vs female), WBC count at diagnosis (<30 × 109/L vs >30 × 109/L), MRD pre-aSCT: complete molecular response or MMR vs no MMR, hematologic response after IP1 (yes vs no), transcript type (p210 BCR-ABL vs p190 BCR-ABL), type of donor (matched unrelated donor vs sibling/HLA-matched vs nonmatched). The imatinib schedule was as follows: imatinib early (starting with IP1) vs imatinib late (start either after IP1 or alternating after IP2 and after consolidation),6 MAC vs RIC, TBI-based conditioning vs non-TBI-based conditioning. Stepwise backward elimination was performed. The entry level for effects to enter the model was set at 0.10. The multivariable analyses were carried out at the Institute for Biomedical Statistics, Medical Faculty Mannheim, Heidelberg University, Germany with the SAS software, release 9.2 (SAS Institute, Cary, NC).
Results
Genome-wide copy number analysis and MLPA analysis of diagnostic Ph+ adult ALL samples
To assess the profile of acquired CNAs of adult Ph+ ALL samples, we analyzed initial diagnostic samples from the entire cohort of n = 97 patients with high-density Affymetrix SNP 6.0 arrays. For n = 11 samples, matched paired analysis with samples from a time point of CR as germ line controls could be performed. However, because of the unavailability of remission or other germ line control samples, the remaining n = 86 samples were analyzed with nonmatched anonymous references according to previously established robust algorithms.30,31 This analysis identified a total of 830 putatively acquired CNAs corresponding to a mean number of 8.6 CNAs per patient. The identified lesions consisted of 578 homozygous or heterozygous deletions, 240 duplications or amplifications, and 12 regions of copy number neutral loss of heterozygosity. A detailed summary of all CNAs detected by SNP array is depicted in supplemental Tables 1-3, available on the Blood Web site. The most common recurrent CNAs detected by SNP array analysis alone consisted of the known recurrently deleted genes in ALL such as IKZF1 (n = 72/74%), CDKN2A/2B (n = 41/42%), PAX5 (n = 39/40%), BTG1 (n = 14/14%), RB1 (n = 13/13%), EBF1 (n = 11/11%), and ETV6 (n = 5/5%) (Table 2). Of note, a total of n = 40 (41%) unbalanced breakpoints of BCR and/or ABL could be identified. The remaining bulk of recurrent lesions was mainly composed of whole chromosome aberrations such as deletion of chromosome 7 or trisomies of chromosomes 21, 4, 6, 8, and others in hyperdiploid samples. Because of the circumstance that with the exception of n = 11 cases, all SNP array copy number inferrals were carried out with nonmatched anonymous references, we validated the presence of the most common, ALL typical CNAs (IKZF1, CDKN2A/2B, PAX5, BTG1, RB1, EBF1, and ETV6) with MLPA as a second molecular method. Both methods, SNP array and MLPA, are semiquantitative methods, so expectedly there were some deviations of results in a few cases. However, the use of 2 complementary methods allowed the application of a higher stringency in calling the recurrent deletions listed above. Consequently, the MLPA data were combined with SNP array data according to the following algorithm: homozygous deletions of the above listed, 7 commonly deleted genes, were called positive if either the SNP array data called a homozygous deletion by HMM31 and/or at least 2 adjacent MLPA probes measured an intensity ratio of <0.3. Heterozygous deletions were called positive if either SNP array data called a heterozygous deletion by HMM and/or at least 2 adjacent MLPA probes measured an intensity ratio of <0.7. Using this algorithm, addition of MLPA analysis largely confirmed the SNP array data with the exception of single cases, which led to the identification of a slightly higher number of deletions of the 7 analyzed genomic regions by supplementation with MLPA (Table 2 right column).
Recurrent genomic alterations detected by SNP-A and in combination with MLPA
| Chromosome . | Genomic position . | Copy number . | Candidate genes . | Frequency (SNP-A) n/(%) . | Final frequency (SNP-A + MLPA) n/(%) . |
|---|---|---|---|---|---|
| 7p12.2 | 50433798-50449506 | 0/1 | IKZF1 | 72/(74) | 74/(76) |
| 9p21.3 | 21976218-21993223 | 0/1 | CDKN2A/B | 41/(42) | 44/(45) |
| 9q34.12; 22q11.21 | t(9;22) | 1/3 | Unbalanced BCR-ABL breakpoints | 40/(41) | — |
| 9p13.2 | 36920536-37014122 | 1 | PAX5 | 39/(40) | 42/(43) |
| 12q21.33 | 90806299-91060957 | 1 | BTG1 | 14/(14) | 17/(18) |
| 13q14.2 | 47885033-47960991 | 0/1 | RB1 | 13/(13) | 14/(14) |
| 21p11.2-q22.3 | Trisomy 21 | 3 | — | 13/(13) | — |
| 7p22.2-7q36.3 | Monosomy 7 | 1 | — | 12/(12) | — |
| 5q33.3 | 158373754-158464289 | 1 | EBF1 | 11/(11) | 12/(12) |
| 15q12 | 23587298-23635904 | 0/1 | ATP10A | 11/(11) | — |
| 4p16.3-q35.2 | Trisomy 4 | 3 | — | 10/(10) | — |
| 14q11.2-q32.33 | Trisomy/quatrosomy 14 | 3/4 | — | 10/(10) | — |
| 1q31.3 | 197072462-197162608 | 1 | MIR181A1/B1 | 9/(9) | — |
| 6p25.3-q27 | Trisomy 6 | 3 | — | 9/(9) | — |
| 8p23.3-q22.3 | Trisomy 8 | 3 | — | 8/(8) | — |
| 3q13.2 | 113640344-113678765 | 0/1 | BTLA/CD200 | 7/(7) | — |
| 2q32.3 | 196692024-196804136 | 1 | HECW2, STK17B | 7/(7) | — |
| 5p15.33-q35.3 | Trisomy 5 | 3 | — | 7/(7) | — |
| 20p12.2 | 10365416-10404586 | 0/1 | C20orf94 | 6/(6) | — |
| 2p25.3-q37.3 | Trisomy 2 | 3 | — | 6/(6) | — |
| 18p11.23-q23 | Trisomy 18 | 3 | — | 6/(6) | — |
| Xp22.33-q28 | Trisomy/quatrosomy X | 3 | — | 6/(6) | — |
| 12p13.2 | 11767661-11830795 | 1 | ETV6 | 5/(5) | 5/(5) |
| 17p13.3-q25.3 | Trisomy 17 | 3 | — | 5/(5) | — |
| 3p14.2 | 60037676-60351480 | 1 | FHIT | 4/(4) | — |
| 5q14.3 | 88090048-88251350 | 1 | MEF2C | 4/(4) | — |
| 10p15.3-q26.3 | Trisomy 10 | 3 | — | 4/(4) | — |
| Chromosome . | Genomic position . | Copy number . | Candidate genes . | Frequency (SNP-A) n/(%) . | Final frequency (SNP-A + MLPA) n/(%) . |
|---|---|---|---|---|---|
| 7p12.2 | 50433798-50449506 | 0/1 | IKZF1 | 72/(74) | 74/(76) |
| 9p21.3 | 21976218-21993223 | 0/1 | CDKN2A/B | 41/(42) | 44/(45) |
| 9q34.12; 22q11.21 | t(9;22) | 1/3 | Unbalanced BCR-ABL breakpoints | 40/(41) | — |
| 9p13.2 | 36920536-37014122 | 1 | PAX5 | 39/(40) | 42/(43) |
| 12q21.33 | 90806299-91060957 | 1 | BTG1 | 14/(14) | 17/(18) |
| 13q14.2 | 47885033-47960991 | 0/1 | RB1 | 13/(13) | 14/(14) |
| 21p11.2-q22.3 | Trisomy 21 | 3 | — | 13/(13) | — |
| 7p22.2-7q36.3 | Monosomy 7 | 1 | — | 12/(12) | — |
| 5q33.3 | 158373754-158464289 | 1 | EBF1 | 11/(11) | 12/(12) |
| 15q12 | 23587298-23635904 | 0/1 | ATP10A | 11/(11) | — |
| 4p16.3-q35.2 | Trisomy 4 | 3 | — | 10/(10) | — |
| 14q11.2-q32.33 | Trisomy/quatrosomy 14 | 3/4 | — | 10/(10) | — |
| 1q31.3 | 197072462-197162608 | 1 | MIR181A1/B1 | 9/(9) | — |
| 6p25.3-q27 | Trisomy 6 | 3 | — | 9/(9) | — |
| 8p23.3-q22.3 | Trisomy 8 | 3 | — | 8/(8) | — |
| 3q13.2 | 113640344-113678765 | 0/1 | BTLA/CD200 | 7/(7) | — |
| 2q32.3 | 196692024-196804136 | 1 | HECW2, STK17B | 7/(7) | — |
| 5p15.33-q35.3 | Trisomy 5 | 3 | — | 7/(7) | — |
| 20p12.2 | 10365416-10404586 | 0/1 | C20orf94 | 6/(6) | — |
| 2p25.3-q37.3 | Trisomy 2 | 3 | — | 6/(6) | — |
| 18p11.23-q23 | Trisomy 18 | 3 | — | 6/(6) | — |
| Xp22.33-q28 | Trisomy/quatrosomy X | 3 | — | 6/(6) | — |
| 12p13.2 | 11767661-11830795 | 1 | ETV6 | 5/(5) | 5/(5) |
| 17p13.3-q25.3 | Trisomy 17 | 3 | — | 5/(5) | — |
| 3p14.2 | 60037676-60351480 | 1 | FHIT | 4/(4) | — |
| 5q14.3 | 88090048-88251350 | 1 | MEF2C | 4/(4) | — |
| 10p15.3-q26.3 | Trisomy 10 | 3 | — | 4/(4) | — |
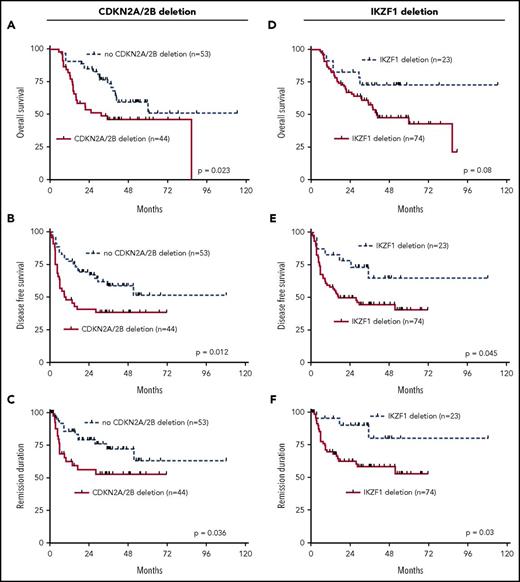
Correlation of recurrent genomic deletions with outcome in univariate analysis indicates that CDKN2A/2B, IKZF1, and BTG1 deletions are associated with inferior prognosis
The results of the combined genomic analysis of the 7 most commonly deleted genes, IKZF1, CDKN2A/2B, PAX5, BTG1, EBF1, ETV6, and RB1, were used to analyze their relationship to clinical outcome of the adult Ph+ ALL patients in the aSCT setting. Univariate analysis revealed that genomic deletions of CDKN2A/2B were significantly associated with an inferior performance in all 3 endpoints, OS, DFS, and REM (OS: P = .023; DFS: P = .012; REM: P = .036; Figure 1A-C). The presence of IKZF1 deletions was associated with a trend for higher risk in OS and with significantly inferior outcome for DFS (P = .045) and REM (P = .03) (Figure 1D-F). Similarly, the presence of deletions of all other above described commonly deleted genes mostly showed trends for worse performance in all 3 endpoints. Statistical significance was reached for BTG1 deletions for DFS (P = .037) and REM (P = .003) (supplemental Figures 1A-3C).
Univariate outcome data. Outcome in dependency of presence of CDKN2A/2B deletions, (A) OS, (B) DFS, and (C) REM. Outcome in dependency of presence of IKZF1 deletions, (D) OS, (E) DFS, and (F) REM.
Univariate outcome data. Outcome in dependency of presence of CDKN2A/2B deletions, (A) OS, (B) DFS, and (C) REM. Outcome in dependency of presence of IKZF1 deletions, (D) OS, (E) DFS, and (F) REM.
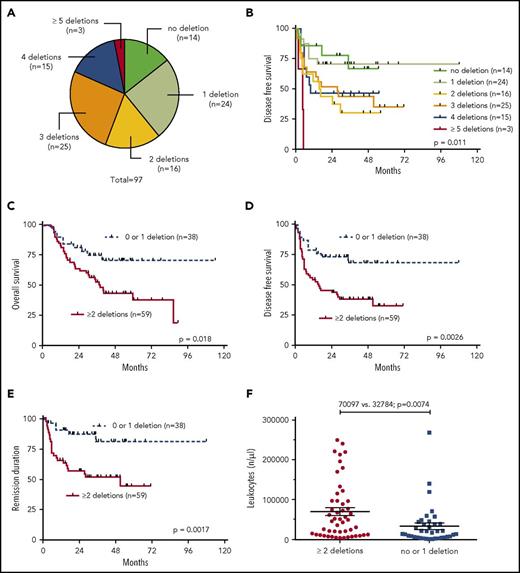
Cooccurrence of common deletions but not the presence of exclusive IKZF1 deletions alone is associated with inferior outcome
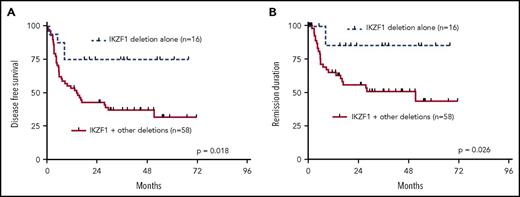
Our analysis showed that most patients (61%) had >1 of the recurrently deleted genes affected in the genomes of their leukemia cells. We therefore analyzed the clinical outcome data in dependency of the number of deletions of the 7 most recurrently deleted genes, IKZF1, CDKN2A/2B, PAX5, BTG1, EBF1, ETV6, and RB. As depicted in Figure 2A, n = 14 patients had none of these lesions; n = 24 had 1 of the above deletions; n = 16 had 2; n = 25 had 3; n = 15 had 4; and n = 3 had 5 or more cooccurring common deletions. Increasing numbers of cooccurring recurrent deletions per se were significantly associated with an increased risk of events after aSCT (P = .011) (Figure 2B). When dichotomized into 2 groups of patients with either 0 or 1 recurrent deletion vs 2 or more recurrent deletions, the latter group had a substantially worse outcome in all endpoints: OS (P = .018), DFS (P = .0026), and REM (P = .0017) (Figure 2C-E). Interestingly, this was also reflected in significantly higher numbers of leukocytes in the group with 2 or more recurrent deletions (P = .0074) (Figure 2F). In light of these results, we also asked whether a putative negative effect of IKZF1 deletions was influenced by the presence of accompanying lesions. This analysis showed that patients who had IKZF1 deletions as an exclusive lesion out of the 7 above described recurrent deletions had a significantly better prognosis in terms of DFS (P = .018) and REM (P = .026) as compared with patients who carried IKZF1 deletions in combination with any of the other recurrent deletions (Figure 3A-B).
Univariate outcome data in dependency of the numeric accumulation of the 7 most recurrent deletions (IKZF1, PAX5, CDKN2A/2B, BTG1, EBF1, ETV6, and RB1). (A) Proportionate distribution of the number concomitant recurrent deletions over all samples, (B) outcome of DFS in dependency of the number of the above described deletions, (C-E) outcome data in dependency of the presence of 0 or 1 deletion vs 2 or more, (C) OS, (D) DFS, (E) REM, and (F) leukocyte count in dependency of the number of deletions.
Univariate outcome data in dependency of the numeric accumulation of the 7 most recurrent deletions (IKZF1, PAX5, CDKN2A/2B, BTG1, EBF1, ETV6, and RB1). (A) Proportionate distribution of the number concomitant recurrent deletions over all samples, (B) outcome of DFS in dependency of the number of the above described deletions, (C-E) outcome data in dependency of the presence of 0 or 1 deletion vs 2 or more, (C) OS, (D) DFS, (E) REM, and (F) leukocyte count in dependency of the number of deletions.
Univariate outcome data in patients with IKZF1 deletions. (A) DFS and (B) REM in dependency of exclusive IKFZ1 deletions vs IKZF1 deletions in combination with other recurrent deletions.
Univariate outcome data in patients with IKZF1 deletions. (A) DFS and (B) REM in dependency of exclusive IKFZ1 deletions vs IKZF1 deletions in combination with other recurrent deletions.
Genomic CDKN2A/2B deletions, imatinib treatment schedule, and intensity of conditioning are strong independent negative predictors for adult Ph+ ALL treated with imatinib and aSCT
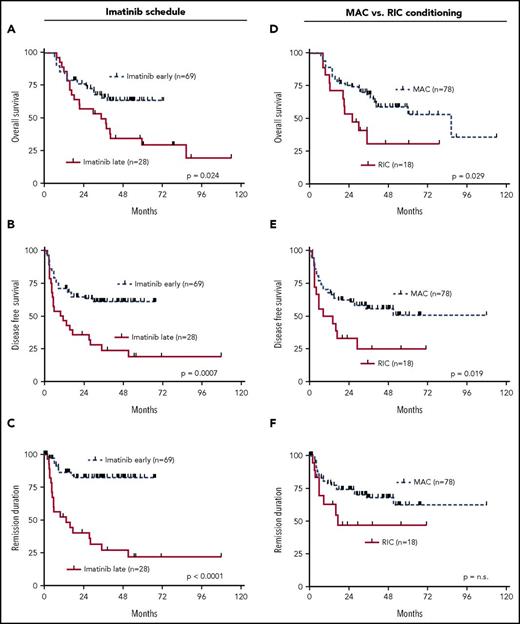
Risk stratification in adult ALL has so far been carried out with clinical parameters such as age, leukocyte counts at initial diagnosis, immunophenotypic ALL subtype, but also cytogenetic markers such as the t(9;22), t(4;11), other translocations or numeric chromosomal aberrations.37,38 Moreover, the time to reach CR after the initial induction chemotherapy has proven to be prognostically relevant.39 Expectedly, failure to reach CR after IP1 was highly predictive for relapse, DFS (P = .008), and REM (P < .0001) (supplemental Figure 3D-E). On the transcript level, highly sensitive and standardized detection of MRD by molecular measurement of BCR-ABL fusion products in Ph+ ALL has evolved as an important prognostic tool to determine how quickly patients reach complete molecular remissions (MRD < 10−4) after the first induction therapy (IP1).32,39,40 In our cohort, MRD data pre-SCT were available for n = 78 patients, and MRD response was not significantly associated with outcome regarding any of the tested endpoints (supplemental Figure 4A-C).
Previous data had shown that addition of imatinib to the treatment protocols of Ph+ ALL improved the outcome.6 In addition, an earlier start of imatinib within the protocol was associated with a higher benefit as compared with a later start of imatinib therapy.6 In the cohort of this current study, all patients achieved CR after IP2 following treatment with imatinib. In this context, starting imatinib early simultaneously with IP1 led to significantly improved outcome for all endpoints, OS (P = .024), DFS (P = .0007), and REM (P ≤ .0001) (Figure 4A-C). Because all patients in this study received aSCT, we also asked the question of whether the intensity of conditioning had an impact on outcome. This analysis revealed that RIC was associated with poorer OS (P = .029) and DFS (0.019) (Figure 4D-F).
Univariate outcome data in dependency of imatinib scheduling. (A) OS, (B) DFS, (C) REM and conditioning intensity, (D) OS, (E) DFS, and (F) REM.
Univariate outcome data in dependency of imatinib scheduling. (A) OS, (B) DFS, (C) REM and conditioning intensity, (D) OS, (E) DFS, and (F) REM.
To test whether the presence of any of the 7 recurrent genomic deletions, IKZF1, CDKN2A/2B, PAX5, BTG1, EBF1, ETV6, RB, or different cooccurrences of these such as in Figure 2C-E had independent prognostic value in comparison with the other clinical covariables described above, we performed a multivariable analysis using Cox proportional hazards models to assess their independent predictive power for OS, DFS, and REM. This analysis revealed that in addition to strong clinical and therapeutic covariables such as imatinib schedule, conditioning intensity, and failure to achieve CR after IP1, CDKN2A/2B deletions were retained as independent negative prognostic factors for all endpoints OS (hazard ratio 2.162, P = .014), DFS (hazard ratio 2.621, P = .0054), and REM (hazard ratio 4.066, P = .0061) (Table 3). To further exclude that this independent predictive effect was based on a biased distribution or confounding effects by connection other variables, we correlated the presence of CDKN2A/2B deletions with the most relevant other factors described above. Expectedly, in accordance with the multivariable analysis, this did not yield any significant associations (Table 4).
Results of multivariable analysis using Cox proportional hazards models
| Endpoint . | Covariable . | Hazard ratio . | P . |
|---|---|---|---|
| REM | Imatinib late schedule | 7.156 | <.0001 |
| No CR after IP1 | 4.122 | .01 | |
| CDKN2A/B deletion | 4.066 | .0061 | |
| DFS | Imatinib late schedule | 3.148 | .0004 |
| TBI-based conditioning | 2.915 | .0087 | |
| CDKN2A/B deletion | 2.621 | .0054 | |
| BTG1 deletion | 2.060 | .047 | |
| OS | CDKN2A/B deletion | 2.162 | .014 |
| RIC vs MAC | 1.934 | .069 | |
| Imatinib late schedule | 1.918 | .0429 |
| Endpoint . | Covariable . | Hazard ratio . | P . |
|---|---|---|---|
| REM | Imatinib late schedule | 7.156 | <.0001 |
| No CR after IP1 | 4.122 | .01 | |
| CDKN2A/B deletion | 4.066 | .0061 | |
| DFS | Imatinib late schedule | 3.148 | .0004 |
| TBI-based conditioning | 2.915 | .0087 | |
| CDKN2A/B deletion | 2.621 | .0054 | |
| BTG1 deletion | 2.060 | .047 | |
| OS | CDKN2A/B deletion | 2.162 | .014 |
| RIC vs MAC | 1.934 | .069 | |
| Imatinib late schedule | 1.918 | .0429 |
Comparison of distribution of clinical and treatment covariables with the presence or absence of CDKN2A/2B deletions
| Parameter . | CDKN2A/B deleted (n = 44) . | CDKN2A/B wild-type (n = 53) . | P . |
|---|---|---|---|
| Age | |||
| Median (range), y | 41 (19-64) | 41 (18-60) | .72 |
| Sex (female/male) (%) | 16 (39)/28 (64) | 23 (43)/36 (68) | .074 |
| WBC | .074 | ||
| Median (range), ×109/L | 41.0 (0.8-269) | 20.5 (1.1-250) | |
| n.a. (%) | 1 (2) | 1 (2) | |
| Breakpoint (%) | .539 | ||
| p190BCR-ABL1 | 21 (47) | 31 (59) | |
| p210BCR-ABL1 | 16 (36) | 17 (32) | |
| Other/n.a. | 3 (7)/2 (5) | 0/5 (9) | |
| Imatinib schedule (%) | .342 | ||
| Early | 30 (68) | 39 (74) | |
| Late | 14 (32) | 14 (26) | |
| Type of HSCT (%) | .598 | ||
| SIB | 12 (27) | 18 (34) | |
| MUD | 32 (73) | 34 (64) | |
| n.a. | 1 (2) | ||
| Conditioning regimen (%) | .134 | ||
| TBI-based | 36 (82) | 44 (83) | |
| No TBI | 8 (18) | 8 (15) | |
| n.a. | 1 (2) |
| Parameter . | CDKN2A/B deleted (n = 44) . | CDKN2A/B wild-type (n = 53) . | P . |
|---|---|---|---|
| Age | |||
| Median (range), y | 41 (19-64) | 41 (18-60) | .72 |
| Sex (female/male) (%) | 16 (39)/28 (64) | 23 (43)/36 (68) | .074 |
| WBC | .074 | ||
| Median (range), ×109/L | 41.0 (0.8-269) | 20.5 (1.1-250) | |
| n.a. (%) | 1 (2) | 1 (2) | |
| Breakpoint (%) | .539 | ||
| p190BCR-ABL1 | 21 (47) | 31 (59) | |
| p210BCR-ABL1 | 16 (36) | 17 (32) | |
| Other/n.a. | 3 (7)/2 (5) | 0/5 (9) | |
| Imatinib schedule (%) | .342 | ||
| Early | 30 (68) | 39 (74) | |
| Late | 14 (32) | 14 (26) | |
| Type of HSCT (%) | .598 | ||
| SIB | 12 (27) | 18 (34) | |
| MUD | 32 (73) | 34 (64) | |
| n.a. | 1 (2) | ||
| Conditioning regimen (%) | .134 | ||
| TBI-based | 36 (82) | 44 (83) | |
| No TBI | 8 (18) | 8 (15) | |
| n.a. | 1 (2) |
MUD, matched unrelated donor; SIB, HLA-matched sibling transplantation.
Discussion
In our study, we performed a comprehensive molecular analysis of a highly homogeneous and well-annotated patient cohort of adult Ph+ ALL patients. All patients received treatment according to the GMALL protocols with uniform induction and consolidation combined with different schedules of imatinib followed by aSCT in CR1. For a full and unbiased molecular analysis, we combined a global explorative SNP array copy number analysis with a concomitant validation of the 7 most common recurrent deletions in ALL by MLPA. Our discovery rate of a mean of 8.6 CNAs by SNP array per patient sample was consistent with previous studies assessing CNAs in ALL; Mullighan et al, 8.79 CNAs per sample,41 Okamoto et al 7.6 CNAs per sample,15 and yielded similar profiles of lesions as compared with these previous studies. Interestingly, our SNP array analysis revealed a relatively high percentage of unbalanced BCR-ABL breakpoints of >40%, which is considerably more frequent than in CML, where deletions of the reciprocal fusion product can be found in 10% to 15% of cases.42,43 A recent study investigating the role of additional chromosomal abnormalities by cytogenetics in adult Ph+ ALL found a lower frequency of +der(22)t(9;22) and −9/9p abnormalities and showed these patients as a poor risk group.24 In our hands, the presence of unbalanced BCR-ABL breakpoints did not have a negative effect (data not shown).
Because of limited access to germ line controls for validation for most of the SNP array data, we opted to validate the copy number data by the alternative molecular method MLPA, which enabled the confirmation of the 7 most common gene deletions of IKZF1, PAX5, CDKN2A/2B, BTG1, EBF1, ETV6, and RB1. This method had been employed as the sole detection method in several previous studies.22,23,28 As expected, this validation confirmed SNP array data in most cases. In a few exceptions, MLPA analysis led to a call of deletions, where SNP array analysis did not (ie, leading to slightly higher detection rates of deletions by addition of the supplementary MLPA analysis). These discrepancies can be explained by the circumstance that both methods are of semiquantitative nature and become ambiguous when the tumor samples are increasingly diluted with remaining normal DNA in the sample. In the discrepant cases, MLPA detected deletions of the respective regions by measuring intensity ratios <0.7, whereas the deletions were not distinct or large enough to enable a positive call by the HMM model of the more stringent SNP array analysis with anonymous references.31 Nevertheless, these deletions were also detectable upon visual inspection of the SNP array profiles even though they did not reach the threshold amplitude to result in a positive HMM signal. Therefore, our underlying data of the 7 most frequent and recurrent CNAs in ALL generated by the combination of SNP array and MLPA analysis in this study is of higher robustness as compared with previous studies, which only employed either one or the other method to determine profiles of CNAs.
Based on this high confidence data, we concentrated on integrating the 7 most common deletions (IKZF1, PAX5, CDKN2A/2B, BTG1, EBF1, ETV6, and RB1) with the clinical outcome results. Thereby, only the presence CDKN2A/2B deletions but not IKZF1 deletions were shown to be significantly adverse for all endpoints. On the one hand, this could possibly be attributed to the higher fraction of patients with IKZF deletions, 76% in our cohort vs 50% in the cohort of Kim et al,28 and as compared with the frequency of CDKN2A/2B deletions, which were more evenly distributed so that the prognostic value of IKZF1 deletions was weaker than that of CDKN2A/2B deletions. On the other hand, when considering that 65% of the samples carried >1 of the recurrent deletions simultaneously, IKZF1 deletions had to be interpreted in a cooperative context with other lesions. Samples that exclusively had IKZF1 deletions without any of the other deletions actually had a significantly lower risk for relapse in the group of patients with IKZF1 deletions (Figure 3). This was also corroborated by the observation that the mere numeric accumulation of any of the above described common deletions seemed to have a significant effect on survival and risk of relapse (Figure 2). Nevertheless, when subjected to a multivariable analysis in consideration of other prognostic markers, the numeration of genomic deletions alone did not retain statistical independency.
For prediction of relapse, the treatment parameter of late imatinib administration and failure to achieve CR after IP1 remained the strongest independent negative parameter in this cohort. In contrast, the level of MRD prior to aSCT had no significant impact on any of the outcome measures. Although the issue of MRD pretransplant is controversial, with several reports showing that MRD plays a predictive role in Ph+ ALL,44,45 our data are in agreement with other prospective studies8 and the largest retrospective analysis of Ph+ ALL to date,46 which clearly showed that the BCR-ABL1/ABL transcript ratio prior to aSCT had no impact on outcome. These data are also consistent with the results of a prospective study demonstrating that aSCT can override the otherwise adverse effect of elevated MRD.47
Interestingly, the presence of CDKN2A/2B deletions emerged as the third and only other independent marker for relapse. The result was similar for DFS. Therefore, timing of imatinib and TBI-based conditioning, which in principle was mostly myeloablative (MAC), were strong prognostically relevant covariables. However, CDKN2A/2B and BTG1 deletions also were retained as independent molecular markers. For OS, the presence of CDKN2A/2B deletions was even the strongest independent adverse risk marker. In comparison with previous studies, this is a novel and somewhat surprising result, because we show for the first time that a molecular marker (genomic deletion) independently supplements or even outperforms established risk stratifiers in Ph+ ALL, and this factor is not IKZF1. To this end, our results are largely in agreement with a recent study that revealed the presence of CDKN2A/2B deletions as determined by FISH analysis in a patient cohort of n = 135 adult Ph+ ALL patients as a negative predictor for survival and risk of relapse.27 Arguments possibly explaining an exceptionally strong role of CDKN2A/2B deletions in Ph+ ALL have been shown by functional biologic experiments indicating that absence of gene expression from the CDKN2A/2B locus cooperates with the BCR-ABL oncogene to enhance the aggressiveness of the disease by strongly increasing the self-renewal capacity of leukemia cells, increasing clonal turnover and thereby conferring resistance to TKI therapy.48-50 Furthermore, our results are also in agreement with the study by Kim et al,28 whereby a negative impact of IKZF1 deletions was shown for relapse, which was however not retained in multivariable analysis. Furthermore, in that study, no comparison regarding the prognostic value of IKZF1 deletions in combination with other deletions was made because other lesions were not examined.
In conclusion, in the adult Ph+ ALL cohort assessed in our study, CDKN2A/2B deletions for the first time emerged as a new strong and independent prognostic marker for predicting risk of relapse and OS on the background of TKI treatment and aSCT. In contrast, IKZF1 deletions did not prove to be a suitable marker because their effect was diluted by a dependency on the cooccurrence of other ALL typical genomic lesions. These results highly warrant the further prospective evaluation of ALL typical genomic deletions, especially CDKN2A/2B for risk stratification of adult Ph+ ALL to further improve therapeutic strategies for this high-risk patient group.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank Doreen Badowski, Brigitte Gehrke, and Sandra Markovic for excellent technical assistance.
This work was supported by grants from the Adolf Messer Stiftung, the Deutsche José Carreras Stiftung, the “Deutsche Krebshilfe,” the Gutermuth Foundation, the H.W. & J. Hector fund, Baden Wuerttemberg, and the Dr Rolf M. Schwiete Fund, Mannheim. O.G.O. was, and D.N. is, an endowed Professor of the Deutsche Jose Carreras Leukämie Stiftung (DJCLS H 03/01).
Authorship
Contribution: H.P., K.R., D.N., W.-K.H., and O.G.O. designed the study, analyzed data, and wrote the manuscript; S.M., V.N., S.F., J.O., J.P., V.B., and M.B. performed molecular SNP array and MLPA analyses; L.W., A.H., R.W., J.B., M.S., A.V., F.L., D.H., H.S., and N.G. performed clinical outcome analyses and patient follow-up; and C.W. performed multivariable statistical analysis.
Conflict-of-interest disclosure: O.G.O. received research support and honoraria for advisory board activities and scientific presentations from Novartis and Bristol-Myers-Squibb. The remaining authors declare no competing financial interests.
Correspondence: Daniel Nowak, Medizinische Klinik III, Haematologie, Onkologie, Universitaetsmedizin Mannheim, Medizinische Fakultaet Mannheim der Universitaet Heidelberg, Pettenkoferstr 22, 68169 Mannheim, Germany; e-mail: daniel.nowak@medma.uni-heidelberg.de; and Heike Pfeifer, Medizinische Klinik II, Universitaetsklinik Frankfurt, Theodor-Stern Kai 7, 60590 Frankfurt, Germany; e-mail: h.pfeifer@em.uni-frankfurt.de.
REFERENCES
Author notes
O.G.O. and D.N. contributed equally to this study.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal