Abstract
During the last decade, the development of biomarkers for the complications seen after allogeneic hematopoietic stem cell transplantation has expanded tremendously, with the most progress having been made for acute graft-versus-host disease (aGVHD), a common and often fatal complication. Although many factors are known to determine transplant outcome (including the age of the recipient, comorbidity, conditioning intensity, donor source, donor-recipient HLA compatibility, conditioning regimen, posttransplant GVHD prophylaxis), they are incomplete guides for predicting outcomes. Thanks to the advances in genomics, transcriptomics, proteomics, and cytomics technologies, blood biomarkers have been identified and validated for us in promising diagnostic tests, prognostic tests stratifying for future occurrence of aGVHD, and predictive tests for responsiveness to GVHD therapy and nonrelapse mortality. These biomarkers may facilitate timely and selective therapeutic intervention. However, such blood tests are not yet available for routine clinical care. This article provides an overview of the candidate biomarkers for clinical evaluation and outlines a path from biomarker discovery to first clinical correlation, to validation in independent cohorts, to a biomarker-based clinical trial, and finally to general clinical application. This article focuses on biomarkers discovered with a large-scale proteomics platform and validated with the same reproducible assay in at least 2 independent cohorts with sufficient sample size according to the 2014 National Institutes of Health consensus on biomarker criteria, as well as on biomarkers as tests for risk stratification of outcomes, but not on their pathophysiologic contributions, which have been reviewed recently.
Introduction
Allogeneic hematopoietic stem cell transplantation (allo-HSCT) is the most effective tumor immunotherapy available to date. Although allo-HSCT can induce beneficial graft-versus-leukemia effects, the adverse effect of graft-versus-host disease (GVHD), which is closely linked to graft-versus-leukemia, is a major source of morbidity and mortality following HSCT. Clinically significant acute GVHD (aGVHD) continues to affect up to 50% of allo-HSCT recipients.1 Also, the incidence of chronic GVHD (cGVHD) has been as high as 70% in HSCT recipients who survived 100 days.2 Severe cGVHD results in high nonrelapse mortality (NRM; reaching 12%), significant morbidity, organ dysfunction, impaired quality of life, and increased incidence of secondary malignancies.3 The conditioning regimen, underlying primary disease, alloreactivity induced by donor T cells, and prophylactic immunosuppressive drugs lead to other less common but still potentially fatal complications post-HSCT such as hepatic sinusoidal obstruction syndrome (SOS), previously known as veno-occlusive disease, thrombotic microangiopathy (TMA), idiopathic pneumonia syndrome (IPS), and posttransplant diabetes mellitus (PTDM). Until recently, available diagnostic and staging tools frequently failed to identify patients at elevated risk of disease progression or death, but the past decade has seen an explosive evolution of “-omics” technologies, largely due to important advances in chemistry, engineering, high-throughput technical devices, and bioinformatics. Building on these opportunities, blood biomarkers have been identified and validated in several cohorts for the main posttransplantation outcomes. This article summarizes current information on biomarkers for posttransplantation outcomes and proposes future directions for biomarker-based clinical trials and ultimately biomarker use in standard practice. According to the type of article, I focus on biomarkers discovered with large-scale proteomics platforms and validated with the same reproducible assay in at least 2 independent cohorts with sufficient sample size according to the 2014 National Institutes of Health (NIH) consensus on biomarker criteria,4 and on biomarkers as tests for risk stratification of outcomes and not their pathophysiologic contributions.1,2
Advances in technologies available to explore biomarkers
Ideal clinical tests involve noninvasive sample collection, which allows for repetitive collection from a patient in a short timeframe. Therefore, biofluids, such as plasma, sera, or urine, are preferable. Plasma and sera are the most frequently collected samples in repositories due to relatively easy processing and storage, and are a good source of information related to systemic diseases such as GVHD because the levels of individual blood proteins represent a summation of multiple, disparate events that occur in every organ system. Urine represents an alternative medium for noninvasive biomarker discovery, but has disadvantages such as the protein mixture being inherently biased by renal filtration. Advances in omics technologies and bioinformatics throughput have made possible analysis of the entire spectrum of molecular changes in an organism or even a single cell to provide insights into disease mechanisms with few a priori assumptions. Figure 1 summarizes the blood omics available for identifying posttransplantation candidate biomarkers.
How can we assess GVHD through blood omics? The number of high-throughput omics tools that can be used to profile the human immune system in the blood is increasing rapidly. Genomic approaches consist of measuring abundance of cellular RNA and also microRNAs (miRNAs) that are present in cells or in the serum/plasma. Other genomic approaches consist of determining gene sequence and function (eg, genome-wide association studies, RNA-interference screens, exome sequencing, next-generation whole-genome sequencing). Proteomic assays are used to determine antibody specificity or measure changes in serum/plasma levels of cytokines or chemokines using multiplex assays or global profiles through tandem-mass spectrometry. Cellular profiling assays or cytomics are used to phenotype immune cells based on intracellular or extracellular markers using polychromatic flow cytometry or mass cytometry. CircRNA, circular RNA; lnRNA, long noncoding RNA; mRNA, messenger RNA; RNAseq, RNA sequencing; ULS, Universal Linkage System.
How can we assess GVHD through blood omics? The number of high-throughput omics tools that can be used to profile the human immune system in the blood is increasing rapidly. Genomic approaches consist of measuring abundance of cellular RNA and also microRNAs (miRNAs) that are present in cells or in the serum/plasma. Other genomic approaches consist of determining gene sequence and function (eg, genome-wide association studies, RNA-interference screens, exome sequencing, next-generation whole-genome sequencing). Proteomic assays are used to determine antibody specificity or measure changes in serum/plasma levels of cytokines or chemokines using multiplex assays or global profiles through tandem-mass spectrometry. Cellular profiling assays or cytomics are used to phenotype immune cells based on intracellular or extracellular markers using polychromatic flow cytometry or mass cytometry. CircRNA, circular RNA; lnRNA, long noncoding RNA; mRNA, messenger RNA; RNAseq, RNA sequencing; ULS, Universal Linkage System.
Genomics
Strategies to improve outcomes after allo-HSCT can be divided into those to reduce pretransplantation risk and those to facilitate the diagnosis and prognosis of posttransplantation complications. Advances in pretransplantation risk stratification have been made through detailed evaluations based on HLA genetics5 as well as genome-wide association studies (GWASs) of polymorphisms that either increase transplantation risk or protect against complications. A recent GWAS showed that the number of minor histocompatibility antigen mismatches doubles in unrelated vs sibling HLA-matched transplants, but has less impact on aGVHD than mismatching at HLA-DP.6 Another recent GWAS including ∼3000 donor-recipient pairs (Discovery-BMT study) showed that functional single-nucleotide polymorphisms (SNPs) in the major histocompatibility complex class II region are associated with overall survival after HLA-matched unrelated donor HSCT.7 However, studies of candidate-genetic polymorphisms in large cohorts have been unable to replicate findings from previous smaller studies for both aGVHD and cGVHD, suggesting that most published SNP associations have not held up or have been reproducible either because they were nonfunctional or were in linkage with more important genetic elements.8,9 As an exception, donor SNPs in IL1RL1 showed strong correlations with pretransplantation serum/plasma concentrations of soluble Stimulation-2 (ST2), also called interleukin 33 (IL-33) receptor, as well as an association with the risk of aGVHD with potential implications for donor selection.10
Transcriptomics
As with genomic analysis, studies of gene expression signatures of GVHD can be categorized as candidate-gene studies and genome-wide studies, which may offer a less biased approach to identifying genes, pathways, and gene expression networks active in this disease. In the past decade, large transcriptomic initiatives have enabled major discoveries in the fields of infectious disease, vaccinology, and solid organ transplantation.11-15 Transcriptomic analysis is mainly performed on bulk peripheral blood mononuclear cells, avoiding contamination by granulocytes seen with whole-blood approaches. In allo-HSCT, a 20-gene-set classifier distinguishing tolerant and nontolerant subjects was discovered, although not validated independently.16 Later, the multicenter Chronic Disease Consortium published an additional gene expression study of cGVHD and found 3 RNA biomarkers (IRS2, PLEKHF1, and IL1R2) and 2 clinical variables (recipient cytomegalovirus serostatus and conditioning-regimen intensity) that accurately segregated cGVHD cases from controls.17 A whole peripheral blood mononuclear cell approach can scan all circulating cells, but the resulting transcriptome is often dominated by the largest cell population, which does not always represent pathogenic cell types. Thus, some groups have purified cell populations prior to RNA isolation. Studies related to aGVHD have used sorted T cells, given their prominent role in disease pathogenesis. In mice, isolated CD8+ and CD4+ T cells have been applied in gene array, RNA sequencing (RNAseq), and micro-RNA (miRNA) analyses that identified novel drivers of GVHD, including programmed death ligand 1 (PDL-1) on donor T cells, proinflammatory cytotoxic T cell 17 (Tc17), and several miRNAs.18-25 Using sorted CD3+ T cells in nonhuman primates (NHPs) and CD4+ and CD8+ T cells in humans in supervised as well as unsupervised gene expression analyses to identify pathways controlling GVHD, Leslie Kean’s group made several discoveries such as: in both NHPs and humans, aGVHD is characterized by distinct “hyperacute” and “breakthrough” mechanisms, with hyperacute aGVHD driven by helper T cell (Th)/Tc1-mediated dysfunction and breakthrough aGVHD driven by inflammatory IL17-dominated pathways.26 They further discovered that Aurora Kinase A and the OX40:OX40L pathway are novel mediators of aGVHD induced in both NHP and human alloreactive T cells that can be blocked in combination with mammalian target of rapamycin inhibition with sirolimus to induce long-term control of both hyperacute and breakthrough aGVHD.27,28 In cGVHD, gene expression in circulating cGVHD monocytes (vs monocytes from normal subjects and non-CGVHD control patients) identified 2 upregulated pathways: interferon (IFN)-inducible genes (MX1, CXCL9, CXCL10) and innate receptors for cellular damage (Toll-like receptor 7 and DDX58).29 The knowledge gained from the studies described in this section was almost exclusively derived from gene array experiments performed on bulk cell populations, and such approaches are still in their infancy. In the coming years, new techniques, particularly single-cell RNAseq, will provide insights for mechanistic questions only answerable by single-cell analysis, allowing studies on low-frequency cells, particularly in low-cell input samples such as gut biopsies.30 Recently, intestinal tract bacterial floral diversity, as represented by the inverse Simpson index, was suggested as a risk-stratification biomarker. Fecal specimens were collected from 80 allo-HSCT recipients at stem cell engraftment, and the low-diversity group (inverse Simpson <2) had the highest rate of transplant-related death.31 A similar approach was used to stratify patients at risk for relapse after HSCT.32 The presence of specific species such as Blautia that correlate with reduced death from GVHD has also been proposed as a potential biomarker.33 Microbiome-host interactions and their potential as biomarkers were recently and extensively reviewed by Andermann et al.34
Proteomics
DNA alone does not determine cell fate, as complex regulatory processes occur at both the transcriptional and translational levels. Indeed, for ∼20 000 genes, they are ∼100 000 RNA transcripts and ∼1 000 000 proteins, increasing the complexity of analysis at each step toward the proteome. Although genomics and transcriptomics techniques have become routine, proteomics is performed only in specialized laboratories due to the complexity of data acquisition and analysis, with even the best platform revealing only the tip of the iceberg (at best 10 000 out of nearly 1 million possible proteins). However, discovery of a protein disease marker is immensely valuable, as it represents the actual state of disease.35 Here, we focus on the use of proteomics for the molecular diagnosis of GVHD post-HSCT. Both non–mass spectrometry (MS)-based, such as antibody arrays, and MS-based proteomic approaches have been used to identify potential GVHD biomarkers. Here, I discuss only large-scale studies that investigated qualitative and quantitative differences in complete protein profiles among samples from patients with and without GVHD or other complications post-HSCT. Although antibody arrays are quantitative and highly sensitive for low-abundance proteins such as cytokines, their main disadvantage is the restricted number of antibodies on the array, which thus limits the candidates to “usual suspects.” In contrast, next-generation MS is a powerful tool for qualitative and quantitative characterization of proteins in complex protein mixtures.36 Such approaches use gel-free separation methods (typically liquid chromatography) in the first steps, with MS as the final step offering reliable identification of proteins and determination of their isoforms and posttranslational modifications. MS, particularly tandem MS, allows unambiguous quantification and has been used most recently for quantification with either label-free methods or isotopically labeled tags. Mass spectra are matched to a sequence database to identify proteins.37 At present, these approaches are too time-consuming for use in validation, but they remain the most efficient methods for biomarker discovery in clinical research.
Although next-generation MS holds great promise for biomarker discovery, gaps remain between biomarker discovery and validation. Notably, the paucity of affinity-capture reagents has led to bias in the prioritization of candidate markers, and the sample numbers required for validation increase through each test phase, augmenting the need for high-throughput assays. The most applicable approach for quantitation of individual proteins for validation remains the highly specific sandwich enzyme-linked immunosorbent assay (ELISA). ELISAs are relatively simple and highly reproducible, limiting both interassay and intra-assay variability. Ideally, multiplex customized antibody arrays could validate several candidate proteins at once within the same sample. However, they are limited by (1) high cross-talk between antibodies, (2) lower sensitivity, (3) lower throughput than ELISA, and (4) the need for a specific platform.
Cytomics
Profiles of immune cell populations are obtained by high-throughput flow cytometry or mass cytometry. CyTOF is a time-of-flight MS approach for measuring many markers on cells similar to flow cytometry except the antibodies are labeled with heavy metal ion tags instead of fluorochromes. Its main advantage over flow cytometry is the combination of more antibody specificities in a single sample (classically 30-40 antibodies), without significant spillover between channels. This technology and its software tools permit discovery studies of new populations, although it is limited by the markers used. Flow cytometry and more recently mass cytometry have enabled identification of several important immune cells: regulatory T cells (Tregs),38-41 B cells,42,43 T follicular helper (TFH) cells,44 T follicular regulatory (TFR) cells,45 and invariant natural killer T frequencies.46 Proteomics with flow cytometry or flow and mass cytometry has been used to discover new cell populations in GVHD such as CD146+CD4+ T cells in aGVHD and cGVHD or blood mucosal-associated T cells (CD161+ TCRVα7.2+ T cells) and CD38+ T cells in cGVHD.47-49 Although the frequencies and absolute numbers of such immune cells provide insight into the pathophysiology of GVHD and these cells are excellent therapeutic targets, the relatively low throughput of cytomics, lack of a standard curve for quantification, and need for large samples of fresh blood, make them less ideal biomarkers than soluble factors measurable by ELISA. Their best use is as markers of response to a specific treatment (eg, Tregs, TFH cells, and TFR cells after IL-2 therapy).41,45
Definition of biomarkers, applications, and major phases of biomarker development
Known risk factors pre-HSCT are related to genetic factors, including HLA disparities between donor and recipient, age, unrelated donor, conditioning-regimen intensity, malignant disease status, and donor graft content. Before the advent of biomarkers, aGVHD diagnosis relied entirely on clinical signs in 1 of 3 major target organs, skin, liver, and/or gastrointestinal (GI) tract as confirmed by biopsy.50 The types of biomarkers and their potential applications that were devised by the 2014 NIH Chronic GVHD Consensus Biomarker Working Group are summarized in Table 1.4 Surrogate markers are not ready in GVHD. Indeed, the US Food and Drug Administration (FDA) perspective on surrogate markers is that they will be used as primary measures of the effectiveness of investigational drugs in definitive drug trials (ie, PDL-1 in checkpoint inhibitor trials). The primary difference between a biomarker and a surrogate marker is that a biomarker is a “candidate” surrogate marker, whereas a surrogate marker is a test used, and taken, as a measure of the effects of a specific treatment. In the 2014 NIH consensus on biomarkers,4 experts in the HSCT field determined that there were not yet GVHD surrogate markers that can be used. Even since the FDA approval of ibrutinib as a second-line cGVHD drug, there are still no surrogate markers in GVHD.51
Definitions of biomarkers and potential applications
| Types of biomarkers . | Definition . | Applications . |
|---|---|---|
| Diagnostic | An assay that identifies patients at the onset of clinical disease | To help in rapid diagnosis and initiation of therapy |
| To distinguish patients with the disease from those without the disease but similar symptoms (ie, GI GVHD vs infectious colitis or bronchiolitis obliterans vs infectious pulmonary) | ||
| Prognostic | An assay that categorizes patients by degree of risk for disease occurrence | To help design a biomarker-based preemptive trial |
| To determine whether an intervention based on high- or low-risk biomarkers before the clinical signs reduce the anticipated incidence of the disease | ||
| Predictive | An assay that categorizes patients by their likelihood of response to or outcome of a particular treatment when measured prior to the treatment | To intensify treatment in high-risk patients |
| To decrease treatment in low-risk patients | ||
| Response to treatment | An assay performed after initiation of therapy | To monitor the response to treatment |
| Types of biomarkers . | Definition . | Applications . |
|---|---|---|
| Diagnostic | An assay that identifies patients at the onset of clinical disease | To help in rapid diagnosis and initiation of therapy |
| To distinguish patients with the disease from those without the disease but similar symptoms (ie, GI GVHD vs infectious colitis or bronchiolitis obliterans vs infectious pulmonary) | ||
| Prognostic | An assay that categorizes patients by degree of risk for disease occurrence | To help design a biomarker-based preemptive trial |
| To determine whether an intervention based on high- or low-risk biomarkers before the clinical signs reduce the anticipated incidence of the disease | ||
| Predictive | An assay that categorizes patients by their likelihood of response to or outcome of a particular treatment when measured prior to the treatment | To intensify treatment in high-risk patients |
| To decrease treatment in low-risk patients | ||
| Response to treatment | An assay performed after initiation of therapy | To monitor the response to treatment |
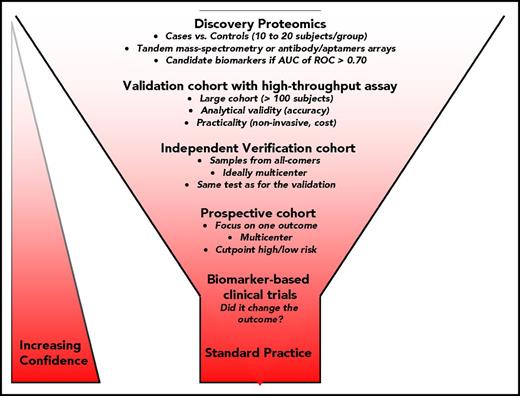
Biomarker development also entails multiple phases, from the identification of promising molecular targets to routine use in clinical practice.4,52 Validation with training and verification cohorts, followed by independent cohorts and multicenter cohorts is required before prospective studies.53-56 Figure 2 shows the workflow of biomarker development post-HSCT. For reporting on observational study design and diagnostic accuracy, 2 guidelines were established by the Strengthening the Reporting of Observational Studies in Epidemiology (STROBE) and Standards for Reporting of Diagnostic Accuracy (STARD) initiatives.57,58 Table 2 summarizes key characteristics for evaluating the analytical performance of an assay and key statistical analyses for evaluating test(s) accuracy. Although other statistical analyses are possible, receiver operating characteristic (ROC) curve analysis and the derived indexes of accuracy, particularly the area under the curve (AUC), have become a popular method for evaluating the accuracy of biomarkers.59 The most desirable property on ROC analysis is that the accuracy indices derived from this technique are not distorted by fluctuations caused by the use of arbitrarily chosen decision criteria or cutoffs. The AUC is the derived summary measure of accuracy and determines the inherent ability of the test to discriminate between cases and controls. Using this as a measure of diagnostic or prognostic performance, one can compare individual tests or judge whether various combinations of tests can improve diagnostic or prognostic accuracy.60 However, if a biomarker is not highly correlated with other biomarkers, 1 or 2 biomarkers could be sufficient. For example, soluble ST2 predicts responsiveness to GVHD treatment and subsequent 6-month NRM as well as a panel of 12 biomarkers.53
Major phases of biomarker development. Five major steps are required to develop a clinical test for standard practice. First, the discovery or pilot phase compares 20 to 40 cases and controls using antibody-based arrays or MS-based approaches or flow/mass cytometry. It is recommended that the term “candidate biomarker” be used to refer to findings of early phase studies when additional validation is needed. A protein will be considered a candidate marker worth being pursued if the area under the curve (AUC) of the receiver operating characteristic (ROC) is >0.70. Next, the validation phase is usually performed with a high-throughput immunoassay, and the cohort is created from retrospective longitudinal case-control repositories from multiple institutions. The fourth step focuses on few biomarkers and requires a prospective multicenter validation, typically on a large multicenter cohort with the goal to define cutoff for high and low risk for a specific outcome. The hope is that this step will lead to a clinical test that will be approved by the FDA. Finally, the biomarker can be used in a clinical trial. If the use of biomarker changes the patients’ outcome, it is likely that it will be used in standard practice and its impact on the reduction of disease burden in the population of interest is quantified.
Major phases of biomarker development. Five major steps are required to develop a clinical test for standard practice. First, the discovery or pilot phase compares 20 to 40 cases and controls using antibody-based arrays or MS-based approaches or flow/mass cytometry. It is recommended that the term “candidate biomarker” be used to refer to findings of early phase studies when additional validation is needed. A protein will be considered a candidate marker worth being pursued if the area under the curve (AUC) of the receiver operating characteristic (ROC) is >0.70. Next, the validation phase is usually performed with a high-throughput immunoassay, and the cohort is created from retrospective longitudinal case-control repositories from multiple institutions. The fourth step focuses on few biomarkers and requires a prospective multicenter validation, typically on a large multicenter cohort with the goal to define cutoff for high and low risk for a specific outcome. The hope is that this step will lead to a clinical test that will be approved by the FDA. Finally, the biomarker can be used in a clinical trial. If the use of biomarker changes the patients’ outcome, it is likely that it will be used in standard practice and its impact on the reduction of disease burden in the population of interest is quantified.
Characteristics of assay analytical performance and statistical analyses for evaluating test(s) accuracy
| Characteristics . |
|---|
| Specimen and analytical performance characteristics |
| • Specimen stability with freeze/thaw |
| • Specimen type and matrix complexity (undiluted plasma/serum may have circulating antibodies that can cross-react with the specific antibodies) |
| • Assay specificity (interference, cross-reactivity) |
| • Assay sensitivity (lower limit of detection) |
| • Assay range |
| • Precision within runs and between runs |
| • Accuracy: Recovery of spiked analyte is within 75%-125% at all ranges |
| Analysis for test(s) accuracy evaluation |
| • Sensitivity: Proportion of subjects in a sample of patients with the target condition in whom the test is positive |
| • Specificity: Proportion of subjects in a sample of patients without the target condition in whom the test is negative |
| • Receiver operator characteristic curve: A plot of the true-positive rate vs the false-positive rate for all possible cut points of a biomarker |
| • Positive predictive value: Proportion of patients in the overall population with a positive test who have the target condition |
| • Negative predictive value: Proportion of patients in the overall population with a negative test who do not have the target condition |
| Characteristics . |
|---|
| Specimen and analytical performance characteristics |
| • Specimen stability with freeze/thaw |
| • Specimen type and matrix complexity (undiluted plasma/serum may have circulating antibodies that can cross-react with the specific antibodies) |
| • Assay specificity (interference, cross-reactivity) |
| • Assay sensitivity (lower limit of detection) |
| • Assay range |
| • Precision within runs and between runs |
| • Accuracy: Recovery of spiked analyte is within 75%-125% at all ranges |
| Analysis for test(s) accuracy evaluation |
| • Sensitivity: Proportion of subjects in a sample of patients with the target condition in whom the test is positive |
| • Specificity: Proportion of subjects in a sample of patients without the target condition in whom the test is negative |
| • Receiver operator characteristic curve: A plot of the true-positive rate vs the false-positive rate for all possible cut points of a biomarker |
| • Positive predictive value: Proportion of patients in the overall population with a positive test who have the target condition |
| • Negative predictive value: Proportion of patients in the overall population with a negative test who do not have the target condition |
Most validated and recent biomarkers
Figure 3 shows the most validated biomarkers by chronological transplantation outcome. Table 3 summarizes proteins that have moved from candidate to biomarker for different posttransplantation outcomes according to the 2014 NIH consensus on biomarkers.4
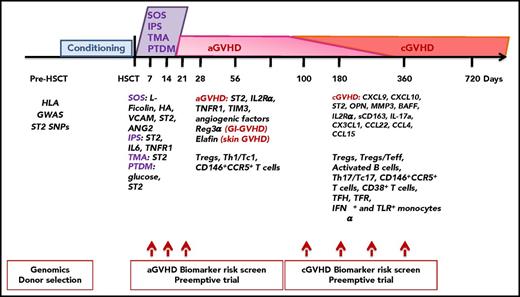
Most validated posttransplantation biomarkers by chronological outcomes. Chronological outcomes posttransplantation and biomarkers are all discussed in “Most validated and recent biomarkers.” BAFF, B-cell–activating factor; HA, hyaluronic acid; MMP3, matrix metalloproteinase 3; OPN, osteopontin; Reg3α, regenerating islet-derived 3-α; sCD163, soluble CD163; Teff, T effector; TIM3, T-cell immunoglobulin mucin-3; TNFR1, tumor necrosis factor receptor-1.
Most validated posttransplantation biomarkers by chronological outcomes. Chronological outcomes posttransplantation and biomarkers are all discussed in “Most validated and recent biomarkers.” BAFF, B-cell–activating factor; HA, hyaluronic acid; MMP3, matrix metalloproteinase 3; OPN, osteopontin; Reg3α, regenerating islet-derived 3-α; sCD163, soluble CD163; Teff, T effector; TIM3, T-cell immunoglobulin mucin-3; TNFR1, tumor necrosis factor receptor-1.
Plasma biomarkers for posttransplantation outcomes
| Protein . | Study year . | No. of patients in the study . | Association direction . | Diagnosis time point (median day post-HSCT) . | Prognostic time point (median day post-HSCT) . | References . |
|---|---|---|---|---|---|---|
| aGVHD | ||||||
| 4-protein panel (sIL-2Ra, TNFR1, HGF, IL-8) | 2009 | 42 + 282* + 142* | Increased | 28 | ND | 60 |
| ST2 | 2013 | 20 + 381* + 673† + 75† | Increased | 28 | 14 | 53 |
| 2015 | 328 + 164* + 300* | Increased | 28 | ND | 64 | |
| 2015 | 74* + 76* | Increased | 28 | Not significant | 63 | |
| 2016 | 211 (independent cohort following validation) | Increased | 28 | ND | 54 | |
| 2017 | 620 + 309† + 358† | Increased | ND | 7 | 56 | |
| TIM3 | 2013 | 20 + 127* + 22* | Increased | 28 | ND | 72 |
| 2015 | 74* + 76* + 167† | Increased | 28 | 14 | 63 | |
| 2016 | 211 (independent cohort following validation) | Increased | 28 | ND | 54 | |
| IL-6 | 2014 | 53 (1 cohort but subsequently validated) | Increased (3-14) then decreased | 30 | 7-14 | 66 |
| 2015 | 74* + 76* | Increased | 28 | Not significant | 63 | |
| GI specific | ||||||
| Reg3α | 2011 | 20 + 871* + 143* | Increased | 28 | ND | 69 |
| TIM3 | 2013 | 20 + 127* + 22* | Increased | 28 | ND | 72 |
| Liver specific | ||||||
| Reg3α > HGF and KRT18 | 2007 | 55 + 826* + 128* | Increased | 28 | ND | 71,73 |
| 2011 | ||||||
| Skin specific | ||||||
| (Elafin) | 2010 | 20 + 492* | Increased | 28 | ND | 67 |
| 2015 | 59 | Increased in skin | 28 | ND | 68 | |
| Late aGVHD | ||||||
| AREG-to-EGF ratio | 2016 | 105 + 50* | Increased | 160 | ND | 79 |
| cGVHD | ||||||
| (sBAFF) | 2007 | 104 | Increased | 480 | NA | 103 |
| 2008 | 80 (Pediatric) | Increased | 171 (early), 429 (late) | NA | 104 | |
| 2014 | 35 + 109* + 211* | Increased, and not validated in independent cohort | 154, 256 (early), 619 (late) | NA | 105 | |
| 2016 | 23 + 198* + 83* | Increased | 203, 174 | NA | 86 | |
| 2017 | 341 | Increased/decreased number | 189 | NA | 83 | |
| CXCL9 | 2014 | 35 + 109* + 211* | Increased | 154, 256 (early), 619 (late) | NA | 105 |
| 2016 | 53 + 211* + 180† | Increased | 210, 203 | 100 | 55 | |
| 2016 | 23 + 198* + 83* | Increased, and not validated in independent cohort | 203, 174 | NA | 86 | |
| 2016 | 26 + 83* | Increased | 132 | NA | 29 | |
| 2016 | 211† | Increased | NA | 100, 180, 365 (time-dependent analysis) | 54 | |
| CXCL10 | 2016 | 23 + 198* + 83* | Increased | 203, 174 | NA | 86 |
| 2016 | 26 + 83* | Increased | 132 | NA | 29 | |
| 4-protein panel (CXCL9, ST2, OPN, MMP3) | 2016 | 53 + 211* + 180† | Increased | 210, 203 | 100 | 55 |
| (MMP3) | 2016 | 76 (BOS) | Increased | 531 | NA | 85 |
| (CCL15) | 2018 | 211* + 792† | Increased at onset but not prognostic | 203 | 100 | 89 |
| SOS | ||||||
| ST2, ANG2, HA, VCAM1 L-Ficolin | 2015 | 40 + 45* + 35* | All increased but L-Ficolin that was decreased | 14 | NA | 90 |
| HA, VCAM1 L-Ficolin | 2015 | 26† + 24† | All increased but L-Ficolin that was decreased | NA | 0 | 90 |
| L-Ficolin | 2017 | 211 | Decreased | 28 | NA | 54 |
| TMA and aGVHD | ||||||
| ST2 | 2017 | 95† + 110† + 107† | Increased | NA | 14 | 91 |
| EASIX | 2017 | 239 + 141* + 173* + 89* | Increased (significant only in reduced-intensity conditioning) | 30-44 | ND | 93 |
| Protein . | Study year . | No. of patients in the study . | Association direction . | Diagnosis time point (median day post-HSCT) . | Prognostic time point (median day post-HSCT) . | References . |
|---|---|---|---|---|---|---|
| aGVHD | ||||||
| 4-protein panel (sIL-2Ra, TNFR1, HGF, IL-8) | 2009 | 42 + 282* + 142* | Increased | 28 | ND | 60 |
| ST2 | 2013 | 20 + 381* + 673† + 75† | Increased | 28 | 14 | 53 |
| 2015 | 328 + 164* + 300* | Increased | 28 | ND | 64 | |
| 2015 | 74* + 76* | Increased | 28 | Not significant | 63 | |
| 2016 | 211 (independent cohort following validation) | Increased | 28 | ND | 54 | |
| 2017 | 620 + 309† + 358† | Increased | ND | 7 | 56 | |
| TIM3 | 2013 | 20 + 127* + 22* | Increased | 28 | ND | 72 |
| 2015 | 74* + 76* + 167† | Increased | 28 | 14 | 63 | |
| 2016 | 211 (independent cohort following validation) | Increased | 28 | ND | 54 | |
| IL-6 | 2014 | 53 (1 cohort but subsequently validated) | Increased (3-14) then decreased | 30 | 7-14 | 66 |
| 2015 | 74* + 76* | Increased | 28 | Not significant | 63 | |
| GI specific | ||||||
| Reg3α | 2011 | 20 + 871* + 143* | Increased | 28 | ND | 69 |
| TIM3 | 2013 | 20 + 127* + 22* | Increased | 28 | ND | 72 |
| Liver specific | ||||||
| Reg3α > HGF and KRT18 | 2007 | 55 + 826* + 128* | Increased | 28 | ND | 71,73 |
| 2011 | ||||||
| Skin specific | ||||||
| (Elafin) | 2010 | 20 + 492* | Increased | 28 | ND | 67 |
| 2015 | 59 | Increased in skin | 28 | ND | 68 | |
| Late aGVHD | ||||||
| AREG-to-EGF ratio | 2016 | 105 + 50* | Increased | 160 | ND | 79 |
| cGVHD | ||||||
| (sBAFF) | 2007 | 104 | Increased | 480 | NA | 103 |
| 2008 | 80 (Pediatric) | Increased | 171 (early), 429 (late) | NA | 104 | |
| 2014 | 35 + 109* + 211* | Increased, and not validated in independent cohort | 154, 256 (early), 619 (late) | NA | 105 | |
| 2016 | 23 + 198* + 83* | Increased | 203, 174 | NA | 86 | |
| 2017 | 341 | Increased/decreased number | 189 | NA | 83 | |
| CXCL9 | 2014 | 35 + 109* + 211* | Increased | 154, 256 (early), 619 (late) | NA | 105 |
| 2016 | 53 + 211* + 180† | Increased | 210, 203 | 100 | 55 | |
| 2016 | 23 + 198* + 83* | Increased, and not validated in independent cohort | 203, 174 | NA | 86 | |
| 2016 | 26 + 83* | Increased | 132 | NA | 29 | |
| 2016 | 211† | Increased | NA | 100, 180, 365 (time-dependent analysis) | 54 | |
| CXCL10 | 2016 | 23 + 198* + 83* | Increased | 203, 174 | NA | 86 |
| 2016 | 26 + 83* | Increased | 132 | NA | 29 | |
| 4-protein panel (CXCL9, ST2, OPN, MMP3) | 2016 | 53 + 211* + 180† | Increased | 210, 203 | 100 | 55 |
| (MMP3) | 2016 | 76 (BOS) | Increased | 531 | NA | 85 |
| (CCL15) | 2018 | 211* + 792† | Increased at onset but not prognostic | 203 | 100 | 89 |
| SOS | ||||||
| ST2, ANG2, HA, VCAM1 L-Ficolin | 2015 | 40 + 45* + 35* | All increased but L-Ficolin that was decreased | 14 | NA | 90 |
| HA, VCAM1 L-Ficolin | 2015 | 26† + 24† | All increased but L-Ficolin that was decreased | NA | 0 | 90 |
| L-Ficolin | 2017 | 211 | Decreased | 28 | NA | 54 |
| TMA and aGVHD | ||||||
| ST2 | 2017 | 95† + 110† + 107† | Increased | NA | 14 | 91 |
| EASIX | 2017 | 239 + 141* + 173* + 89* | Increased (significant only in reduced-intensity conditioning) | 30-44 | ND | 93 |
This table includes only proteins that have been discovered with a large-scale proteomics platform, are identifiable, and that have reached the point of validation as biomarkers with the same reproducible assay in at least 2 independent cohorts of sufficient sample sizes from different institutions according to the 2014 NIH consensus on biomarkers criteria. Candidate biomarkers of interest that have not met these criteria are indicated in parentheses.
ANG2, angiopoietin-2; AREG, amphiregulin; BOS, bronchiolitis obliterans syndrome; EASIX, Endothelial Activation and Stress Index; EGF, epidermal growth factor; HA, hyaluronic acid; HGF, hepatocyte growth factor; MMP3, matrix metalloproteinase 3; NA, not applicable; ND, not done; OPN, osteopontin; sBAFF, soluble B-cell–activating factor; TIM3, T-cell immunoglobulin mucin-3; TNFR1, tumor necrosis factor receptor-1.
Patient number in validation cohort 1 and cohort 2.
Prognostic cohort.
Acute GVHD biomarkers
The first biomarker panel identified and validated for aGVHD diagnosis is a 4-protein biomarker panel (IL-2 receptor α chain [sIL-2Rα/sCD25], tumor necrosis factor receptor-1 [TNFR1], IL-8, and hepatocyte growth factor [HGF]) discovered by screening aGVHD patient plasma samples by competitive hybridization to arrays of antibodies for 130 proteins.60
ST2 is the most validated biomarker for aGVHD and NRM measured alone53,61,62 or with other markers.54,63,64 ST2 was discovered by analyzing therapy-resistant aGVHD samples with a state-of-the-art proteomic technology that is gel-free and based on high-resolution MS.53 ST2 was also tested and validated on several platforms such as with nonmyeloablative conditioning,65 in cord blood transplantation,61 and in HLA-haploidentical or HLA-matched transplantation with subsequent cyclophosphamide use.62 ST2 as early as day 7 or 14 post-HSCT was also validated as a prognostic marker for aGVHD and NRM in large cohorts.53,56,63
Regenerating islet-derived 3-α (Reg3α) and T-cell immunoglobulin mucin-3 (TIM3) that were discovered as GI GVHD biomarkers were also validated as prognostic biomarkers of aGVHD when measured at day 7 and 14, respectively.56,63
IL-6 levels were measured from days 3 to 60 posttransplant in HCT 53 patients and found elevated early during the transplant course.66 This was subsequently validated at onset of GVHD.63
Target-specific biomarkers that can differentiate skin GVHD from other rashes and GI GVHD from other forms of enteritis were discovered and could replace invasive biopsies in this fragile population.
Elafin was discovered using next-generation proteomics and samples from skin aGVHD and supports the specific diagnosis of aGVHD in plasma and skin biopsies.67,68 However, the ELISA for elafin lacked reproducibility, and elafin as a prognostic biomarker has been difficult to reproduce in large cohorts, although it remains a good marker for individual patients at time of diagnosis of skin GVHD and for monitoring of response (Stephanie J. Lee, International applied biology in chronic GVHD working group meeting, oral communication, 24 March 2017).
Regenerating islet-derived 3-α (Reg3α) and T-cell immunoglobulin mucin-3 (TIM3) were discovered using next-generation proteomics and samples from lower GI aGVHD and validated in several cohorts alone or combined with other markers.63,69-72
HGF and cytokeratin-18 fragments (KRT18) were correlated with liver GVHD, although Reg3α had a much better AUC for diagnosis of liver GVHD than these 2 biomarkers.71,73
Urine proteomics identified patterns of peptides that correlated with aGVHD and cGVHD in European cohorts74,75 but were not validated in current US cohorts. In addition, a multicenter double-blinded, placebo-controlled trial of preemptive treatment of aGVHD using the aGVHD pattern showed no differences between groups.76 One possible explanation is that classifiers using a machine learning-based algorithm can be overfitted. Thus, these classifiers have not yet met criteria for FDA approval as biomarkers. Low urinary levels of indoxyl-sulfate, a metabolite of indole that reflects GI microbiome diversity, have been correlated with poor outcome in a single-center cohort of 131 patients.77
Fecal proteins such as calprotectin and α-1-antitrypsin have been suggested as candidate biomarkers, as they correlated with response to corticosteroids in GI-GVHD in a single-center cohort of 72 patients.78 Although increases in fecal proteins were reported by multiple studies, these were small sample-size, single-center cohort studies using different tests. Thus, these proteins have yet to be confirmed as biomarkers.
Circulating angiogenic factors were correlated with late aGVHD with some inconsistencies in findings between cohorts, GI biopsies, and experimental models.79-81 Importantly, the authors compared the AREG-to-EGF ratio in classic aGVHD and cGVHD, and found that the AREG-to-EGF ratio was also elevated in classic aGVHD, but not in cGVHD.79
Chronic GVHD biomarkers
The clinical manifestations of cGVHD often resemble those of autoimmune diseases, such as scleroderma and Sjogren syndrome. Its diagnosis is based on clinical symptoms (ie, inflammatory and fibrotic components) involving almost any target organ (eg, skin, nails, mouth, eyes, genitalia, skeletal muscle, GI tract, liver, and lung). The pathophysiology of cGVHD is complex, as recently reviewed.2 Blood biomarkers (cellular and protein) have been evaluated. Some noteworthy and novel biomarkers reported since the 2014 NIH consensus biomarker and biology papers are listed.
High levels of soluble B-cell–activating factor and the balance of B-cell subsets during B-cell reconstitution were the first biomarkers correlated with cGVHD.42,82,83
Prolonged imbalance of CD4+CD25+FOXP3+ Tregs vs conventional CD4+ T cells post-HSCT was associated with a loss of tolerance and significant cGVHD manifestations.40,84
Using a quantitative proteomics approach, a biomarker panel of 4 proteins (ST2, CXCL9, matrix metalloproteinase 3 [MMP-3], and osteopontin) showed significant correlation with cGVHD diagnosis. Moreover, at day +100 post-HSCT, this panel allowed patient stratification according to cGVHD risk.55 MMP-3 was also correlated with bronchiolitis obliterans diagnosis.85 Recently, both CXCL9 and CXCL10 were significantly correlated with cGVHD diagnosis in the first replication cohort, but only CXCL10 was in the second.86 In another study, gene expression profiling of circulating monocytes from cGVHD patients revealed significant upregulation of IFN-inducible (including CXCL9 and CXCL10) and damage-response genes in cGVHD patients compared with controls. These pathways were confirmed in plasma ELISAs showing elevated CXCL9 and CXCL10 levels.29 Together, the IFN-inducible chemokines CXCL9 and CXCL10, which are responsible for CXCR3-expressing Th1/natural killer lymphocyte recruitment,87 are upregulated at diagnosis and warrant further testing in prospective studies.
An activated Th17-prone T-cell subset expressing both CD146 and CCR5 was found to be involved in cGVHD and sensitive to pharmacological inhibition.48
Circulating TFH cells were shown to correlate with cGVHD and exhibit a Th17 profile.44
Plasma CD163 concentration was associated with de novo–onset cGVHD.88
Among 42 patients who received ibrutinib after failure of prior therapy, responders had decreased levels of sIL-2Rα, CX3CL1, CXCL9, CXCL10, CCL22, and CCL4.51
CCL15 was recently discovered as a novel biomarker in patients via murine cGVHD proteome profiling.89
Hepatic SOS
Hepatic SOS is a major complication during the early post-HSCT period, caused by both toxic injury of conditioning therapy to sinusoidal endothelial cells and inflammation, with clinical symptoms of hyperbilirubinemia, tender hepatomegaly, ascites, and weight gain. The incidence and severity of SOS have decreased significantly in recent years, but SOS-related deaths are still observed in clinical practice. Biomarkers for SOS diagnosis (ST2, angiopoietin-2 [ANG2], L-Ficolin, hyaluronic acid [HA], and VCAM1) and prognosis (L-Ficolin, HA, and VCAM1) were identified by a proteomics study and validated in several cohorts.54,90
TMA
TMA is associated with endothelial injury in vivo and was recently linked to complement activation in vitro. ST2 was shown to be a reliable early biomarker of TMA independent of aGVHD in several cohorts.91,92 Routine laboratory measurements (lactate dehydrogenase, creatinine, and thrombocytes) can be used to create a formula called the Endothelial Activation and Stress Index (EASIX), which was found as a predictor of survival in patients with reduced-intensity conditioning.93
IPS
IPS is a noninfectious pulmonary post-HSCT complication that is difficult to diagnose. A recent study showed that ST2 and IL-6 are diagnostic and prognostic biomarkers of IPS, and TNFR1 is a marker for differential diagnosis from viral pneumonia. ST2 at onset and at day 7 post-HSCT had the highest positive predictive value for IPS occurrence.94
New-onset PTDM
New-onset PTDM occurs commonly post-HSCT and is associated with reduced survival. In a recent study, high ST2 at engraftment predicted increased PTDM and NRM risk, independent of conditioning and grades 3-4 aGVHD.95
Incorporating posttransplantation biomarkers in clinical trials
Different possible clinical trial applications for aGVHD biomarkers have been reviewed.96,97 Given the progress being made in GVHD biomarker identification and validation, it is not surprising that clinical trial designs have already begun incorporating aGVHD biomarkers in trials design at time of onset. Two approaches are possible: (1) use only biomarkers that provide information not available from the clinical status of the patient (ie, ST2, which classifies patients’ risk of NRM independent of clinical grade).53 However, the majority of the other biomarkers discovered so far do correlate with clinical data; or (2) in the approach currently used by the Blood and Marrow Transplant Clinical Trials Network (BMT CTN), clinical data at onset of aGVHD are combined with biomarkers to randomize between sirolimus and prednisone patients with a Minnesota standard risk (https://web.emmes.com/study/bmt2/protocol/1501_protocol/1501_protocol.html). Trials are also under development with high-risk biomarkers using intensified treatment of newly diagnosed GVHD. Biomarkers have been used at onset of GVHD not only to predict the future response to treatment,53,67,69-71 but can also be used as potential response measures to adapt the treatment.98
Currently, there are no “preemptive intervention” clinical trials using biomarkers measured prior to clinical manifestation of GVHD. Although a schema for a preemptive trial to decrease the aGVHD incidence using biomarkers has been reported,96,97 cGVHD biomarkers still lack the sensitivity and specificity required for clinical trial use. Considering recent major strides in the discovery and validation of cGVHD biomarkers, I expect some will be ready for clinical testing in the next 3 to 5 years. A possible schema for newly diagnosed cGVHD could involve measuring biomarkers at cGVHD onset for risk stratification. Randomization of low-risk patients to compare standard treatment to a reduced toxicity or steroid-free treatment will show whether steroid-free treatment is as efficient as steroid treatment and whether it can reduce relapse and infection rates. The toxicity of the intervention should be considered in trial design, particularly for cGVHD, as excess toxicity from preemption will reduce acceptance of the strategy. Randomization of high-risk patients to compare a standard to an intensified treatment will show whether treatment intensification increases effectiveness against cGVHD signs without increasing relapse and infection rates. In a preemptive strategy, biomarkers will be measured starting at day 100 post-HSCT before the occurrence of any clinical signs and repeated every 3 months, with cut points used to identify low- and high-risk patients. In low-risk patients, comparison of no intervention with rapid tapering of immunosuppressive drugs will indicate whether rapid tapering reduces toxicity and infection rates while achieving tolerance sooner. In high-risk patients, comparison of no intervention with preemptive treatment with a steroid-free agent, in 1 possible option, will show whether the incidence of cGVHD is reduced with preemptive intervention. In another option, randomization to compare no intervention vs slow taper of immunosuppressive drugs will show whether the slower taper reduces the incidence of cGVHD to below the expected incidence. As with any preemptive test, improvements in sensitivity come at the expense of specificity and vice versa, and which aspect should be emphasized is a matter of clinical judgment. The potential toxicity of an intervention may influence the threshold used for inclusion by prioritizing high specificity over sensitivity to avoid treatment with corticosteroids or other immunosuppressants in false-positive cases that will not need it. Notably, specificity and sensitivity are used because PPV and NPV are dependent on incidence of an outcome.
Future research on posttransplantation biomarkers
Future directions include a blinded evaluation of new biomarkers in parallel with “validated biomarkers” using samples collected in a current multicenter prospective study such as the BMT CTN protocol 1202: a prospective multicenter cohort for the evaluation of biomarkers predicting risk of complications and mortality following allogeneic HSCT. Trials for newly diagnosed aGVHD have been initiated, but a randomized trial to assess the effectiveness of aGVHD preemption is also needed, specifically in biomarker-identified high-risk patients. Efforts to discover better cGVHD biomarkers and target-specific cGVHD biomarkers are under way through American and European initiatives.4,99
Therapeutic approaches for aGVHD and cGVHD have been largely limited to nonspecific targeting of effector cells. Thus, corticosteroids remain the first-line treatment of patients presenting GVHD symptoms. Biomarkers can provide insight into disease pathophysiology and represent promising targets for new therapeutics. In addition, we propose that aGVHD-specific drugs discovered based on biomarkers will target the appropriate effector T cells to increase efficacy and lower toxicity. For example, ST2 is a potential therapeutic target for aGVHD, based on findings that intestinal stromal cells and donor T cells producing IFN-γ and IL-17 are major sources of soluble ST2 during aGVHD.100 Furthermore, blockade of soluble ST2 in the peritransplantation period with a neutralizing monoclonal antibody reduced GVHD severity and mortality by increasing the availability of IL-33 to T cells expressing membrane-bound ST2 (T helper 2 cells and ST2+FoxP3+ Tregs) and decreasing production of soluble ST2 by type 1 CD4+ and CD8+ T cells.100 Adoptive transfer of cells expressing membrane-bound ST2 (Tregs, IL-9–expressing T cells, innate lymphoid cells type 2) leads to similar results in murine models and is currently in trials.80,101,102
Conclusions
Omics is a revolutionary field with technologies for detecting RNA and proteins, the molecules most proximal to the real-time pathophysiology of alloreactivity compared with genes. In a short time, the use of transcriptomics and proteomics has led to major strides in posttransplantation biomarker discovery and validation. It is unlikely that these biomarkers would have been discovered by traditional hypothesis-driven research. These novel biomarkers have also enabled deeper understanding of mechanisms involved in the pathophysiology following allo-HSCT. In view of the scarcity of treatment options, aside from glucocorticoids, proposed for these patients in the last 30 years, the development of biomarker-based clinical trials is needed. The biomarker findings presented in this Perspective offer the potential for exploring targeted therapeutics, and the discovery of additional drug-targetable biomarkers for GVHD-specific immunosuppression is a main focus of current studies. Another promising proteomics approach is to use protein biomarkers in risk stratification to better use current disease treatment modalities. In summary, major advances in the posttransplantation field have made risk stratification to prevent or treat patients a reality, and the future looks increasingly bright for finally promoting tumor immunity without fatal immunity following allo-HSCT.
Acknowledgments
This work was supported by the National Institutes of Health, National Cancer Institute (R01CA168814), National Institutes of Health, National Heart, Lung, and Blood Institute (R21HL139934), National Institutes of Health, Eunice Kennedy Shriver National Institute of Child Health and Human Development (R01HD074587), the Leukemia & Lymphoma Society Scholar Award (1293-15), and the Lilly Physician Scientist Initiative Award.
Authorship
Contribution: S.P. conceived and wrote the paper.
Conflict-of-interest disclosure: S.P. is an inventor on a patent on “Methods of detection of graft-versus-host disease” (13/573766).
Correspondence: Sophie Paczesny, School of Medicine, Indiana University, 1044 W. Walnut St, Room 425, Indianapolis, IN 46202; e-mail: sophpacz@iu.edu.


This feature is available to Subscribers Only
Sign In or Create an Account Close Modal