TO THE EDITOR:
Clonal hematopoiesis of indeterminate potential (CHIP) describes an expansion of hematopoietic stem cells that harbor somatic mutations1-5 without an underlying hematologic malignancy or definitive morphologic evidence of dysplasia.6 CHIP can evolve to overt neoplasia, with a progression rate of 0.5% to 1.0% per year.2,3 CHIP was first identified through genomic profiling of peripheral blood from healthy individuals.1-5 Its incidence increases with age and has been detected in peripheral blood of patients with solid tumors.7,8 In elderly cancer patients, the presence of CHIP prior to chemotherapeutic exposure is associated with a higher incidence of subsequent therapy-related myeloid neoplasms.9,10
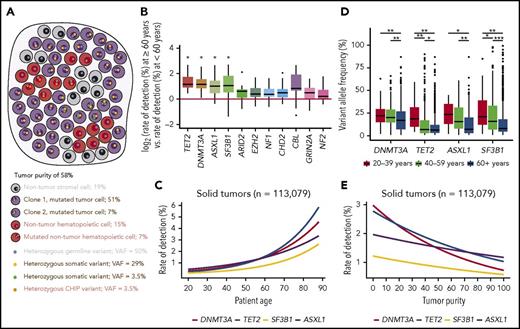
Hematopoietic cells permeate all tissues and are present in solid tumor specimens. The application of comprehensive genomic profiling (CGP) to tumor samples provides an unbiased view of heterogeneous cancer cells and admixed nontumor populations (Figure 1A). Here, we tested the hypothesis that CHIP-associated alterations could be detected in admixed hematopoietic elements within the solid tumor microenvironment. We analyzed CGP of 113 079 solid tumors, divided into 21 disease groups (median age, 61 years; range, 20-89 years) (supplemental Table 1 and supplemental Methods, available on the Blood Web site). While CHIP occurs at all ages, the prevalence begins to rise at age 50 years and increases significantly by age 60 years3,5,11,12 ; therefore, the rate of detection for CHIP-associated alterations should be correlated with age, independent of tumor type. After we corrected for multiple hypothesis testing across 257 genes, only TET2, DNMT3A, ASXL1, and SF3B1 had a significantly higher rate of pathogenic alterations detected in tumor specimens from patients >60 years across all disease groups (Figure 1B; supplemental Table 2). These 4 genes are known to be CHIP-associated,1-5 and remained significant when we stratified patients at age 50 or 70 years. However, at age 70 years, additional alterations in hematologic disease-associated CBL, U2AF1, IDH2,2,3 and MYD88,13 as well as EP300, CDKN2C, and HNF1A were significant (supplemental Figures 1). Furthermore, logistic regression analyses performed for each gene revealed significant correlations with increasing patient age (Figure 1B; supplemental Figure 2). The rate of detection for these alterations varied between tumor groups, likely secondary to the typical proportion of hematopoietic elements in each tumor type (supplemental Figure 3).
Detection of CHIP-associated alterations in comprehensive genomic profiling of solid tumor specimens. (A) In hybrid-capture, high-depth DNA sequencing, variants may arise from unfiltered germ line mutations present in all cells (blue dots), somatic mutations present in cancer cells (gold and green dots), and potentially somatic mutations present in nontumor cells (orange dots). If a somatic mutation is present in all cancer cells, then its expected VAF depends on tumor purity (ie, the percentage of tumor cells relative to all cells) and the chromosomal ploidy at its genomic locus. When the observed VAF of a mutation is significantly different from the frequency expected from tumor purity, it may only be present in a subpopulation of cancer cells or exist in nontumor elements. The VAFs in this sketch are calculated assuming a diploid genome in both tumor and nontumor cells. (B) The log odds ratio for rate of detecting genomic alterations in patients >60 years relative to those <60 years for 257 genes in each tumor type was calculated. Each data point on the boxplot represents the log odds for a single tumor type in a single gene. Genes presented have an adjusted P value of <1 by t test after Bonferroni correction for multiple hypothesis testing (*P < .05). (C) Logistic regression of rate of detection, by percentage of patients, for genomic alterations in DNMT3A, TET2, SF3B1, and ASXL1 as a function of patient age for all solid tumor samples. (D) VAF for genomic alterations in DNMT3A, TET2, SF3B1, and ASXL1 by age group (*P < .05, **P < 1e-05, and ***P < 1e-10, respectively as calculated by the Kruskal-Wallis test corrected with the Dunn test for multiple comparisons). Logistic regression of VAFs by age corroborates these results (supplemental Figure 5). (E) Logistic regression of the rate of detection versus tumor purity (DNMT3A, P < 2.0e-16; TET2, P < 2.0e-16; ASXL, P < 1.0e-06; and SF3B1, P < 1.9e-07).
Detection of CHIP-associated alterations in comprehensive genomic profiling of solid tumor specimens. (A) In hybrid-capture, high-depth DNA sequencing, variants may arise from unfiltered germ line mutations present in all cells (blue dots), somatic mutations present in cancer cells (gold and green dots), and potentially somatic mutations present in nontumor cells (orange dots). If a somatic mutation is present in all cancer cells, then its expected VAF depends on tumor purity (ie, the percentage of tumor cells relative to all cells) and the chromosomal ploidy at its genomic locus. When the observed VAF of a mutation is significantly different from the frequency expected from tumor purity, it may only be present in a subpopulation of cancer cells or exist in nontumor elements. The VAFs in this sketch are calculated assuming a diploid genome in both tumor and nontumor cells. (B) The log odds ratio for rate of detecting genomic alterations in patients >60 years relative to those <60 years for 257 genes in each tumor type was calculated. Each data point on the boxplot represents the log odds for a single tumor type in a single gene. Genes presented have an adjusted P value of <1 by t test after Bonferroni correction for multiple hypothesis testing (*P < .05). (C) Logistic regression of rate of detection, by percentage of patients, for genomic alterations in DNMT3A, TET2, SF3B1, and ASXL1 as a function of patient age for all solid tumor samples. (D) VAF for genomic alterations in DNMT3A, TET2, SF3B1, and ASXL1 by age group (*P < .05, **P < 1e-05, and ***P < 1e-10, respectively as calculated by the Kruskal-Wallis test corrected with the Dunn test for multiple comparisons). Logistic regression of VAFs by age corroborates these results (supplemental Figure 5). (E) Logistic regression of the rate of detection versus tumor purity (DNMT3A, P < 2.0e-16; TET2, P < 2.0e-16; ASXL, P < 1.0e-06; and SF3B1, P < 1.9e-07).
Because the incidence of CHIP increases with age, we hypothesized that CHIP-associated alterations were more likely to be present in hematopoietic elements as opposed to tumor cells in older individuals. Alterations found in tumor cells would be expected to be present at high variant allele frequency (VAF), whereas those associated with CHIP, which typically would be present in a subpopulation of hematopoietic cells, would be present at a lower VAF. Indeed, when we stratified the cohort into three age groups of 20-39, 40-59, and ≥60 years, the VAFs of CHIP-associated alterations were significantly decreased in the oldest group (Figure 1C). We also expected that detection of CHIP-associated alterations would inversely correlate with tumor purity, as an increased proportion of tumor cells requires decreased hematopoietic populations, and found this to be true (Figure 1D; supplemental Figure 4).
To test the hypothesis that certain CHIP-associated alterations identified in tumor-only sequencing may originate from hematopoietic cells admixed in the tumor microenvironment, we examined the CGP of 1636 solid tumor specimens analyzed at Rutgers University.14 TET2 and DNMT3A were the most commonly mutated CHIP-associated genes, consistent with prior results.15 We identified 49 patients with pathogenic alterations in TET2 and/or DNMT3A detected in their solid tumor specimens and labeled as tumor mutations in the sequencing reports. One patient was synchronously diagnosed with 2 lung carcinomas of different histology (Figure 2A-B) and distinct genomic alterations, except for 2 identical pathogenic variants in TET2 and DNMT3A detected at low VAFs (supplemental Figure 5). Sequencing of a patient-matched lymph node without evidence of tumor detected only the shared TET2 and DNMT3A alterations, confirming their hematopoietic origin (Figure 2C-D).
Validation of CHIP-associated mutations in hematopoietic elements. (A-B) A lung adenocarcinoma (A) and a lung squamous cell carcinoma (SCC) (B) were synchronously diagnosed in patient 11. Both specimens had extensive lymphocyte infiltration. CGP revealed distinct genomic alterations in each tumor except for identical genomic alterations in TET2 and DNMT3A. (C) A lymph node biopsy without histologic evidence of tumor was macrodissected to confirm the presence of CHIP mutations. (A-C) hematoxylin and eosin stain; scale bars represent 200 μm. (D) The same TET2 and DNMT3A genomic alterations were detected at a VAF of 1.5% and 1.7%, respectively, confirming their presence in hematopoietic elements; 3D scatter plot shows genomic alterations in all 3 samples with their respective VAFs. (E) Peripheral blood samples were available for 13 patients with TET2 and DNMT3A genomic alterations at VAFs lower than expected from purity. In 10 patients, targeted deep sequencing detected the identical genomic alterations in blood that were present in the associated solid tumor. There was no statistical difference between the VAFs of alteration in peripheral blood and solid tumor specimen (rank-sum P = .38). For the other 3 patients, we detected the genomic alterations in the macrodissected tumor cells enriched from the original samples, while the genomic alterations were absent or at significantly lower level in enriched macrodissected lymphocytes (supplemental Figures 7-9). (F) In the peripheral blood of only 1 of 7 sequenced patients with clonal TET2 and/or DNMT3A mutations, a genomic alteration previously found in the original solid tumor was detected (supplemental Figure 10).
Validation of CHIP-associated mutations in hematopoietic elements. (A-B) A lung adenocarcinoma (A) and a lung squamous cell carcinoma (SCC) (B) were synchronously diagnosed in patient 11. Both specimens had extensive lymphocyte infiltration. CGP revealed distinct genomic alterations in each tumor except for identical genomic alterations in TET2 and DNMT3A. (C) A lymph node biopsy without histologic evidence of tumor was macrodissected to confirm the presence of CHIP mutations. (A-C) hematoxylin and eosin stain; scale bars represent 200 μm. (D) The same TET2 and DNMT3A genomic alterations were detected at a VAF of 1.5% and 1.7%, respectively, confirming their presence in hematopoietic elements; 3D scatter plot shows genomic alterations in all 3 samples with their respective VAFs. (E) Peripheral blood samples were available for 13 patients with TET2 and DNMT3A genomic alterations at VAFs lower than expected from purity. In 10 patients, targeted deep sequencing detected the identical genomic alterations in blood that were present in the associated solid tumor. There was no statistical difference between the VAFs of alteration in peripheral blood and solid tumor specimen (rank-sum P = .38). For the other 3 patients, we detected the genomic alterations in the macrodissected tumor cells enriched from the original samples, while the genomic alterations were absent or at significantly lower level in enriched macrodissected lymphocytes (supplemental Figures 7-9). (F) In the peripheral blood of only 1 of 7 sequenced patients with clonal TET2 and/or DNMT3A mutations, a genomic alteration previously found in the original solid tumor was detected (supplemental Figure 10).
In 36 of 49 patients, TET2 and DNMT3A alterations were present at significantly lower VAFs than expected from tumor purity (supplemental Methods). These patients had a median age of 68 years (range, 51-90 years) and each harbored 1 to 4 pathogenic TET2 and/or DNMT3A alterations (supplemental Table 3). Conversely, in 13 patients, these alterations were detected at VAFs not significantly different than expected from tumor purity. The sequenced specimens from the patients with low VAF TET2 and/or DNMT3A alterations scored significantly higher for tumor-infiltrating lymphocytes compared with the patient specimens with high VAF alterations (rank-sum P = .022), as well as compared with 19 control solid tumor specimens without CHIP-associated mutations (rank-sum P = .035) (supplemental Figure 6). Tumor purity and patient blood cell counts at specimen collection were not significantly different between groups. We lacked statistical power to correlate clinical outcome with CHIP in this cohort.
Matched peripheral blood samples were available for 13 patients harboring TET2 or DNMT3A alterations at VAFs lower than expected from tumor purity; in 10 of 13, we detected the same TET2 or DNMT3A alteration found by solid tumor CGP (Figure 2E). Including the index patient, we confirmed the detection of CHIP in 79% of cases. For the other 3 patients, the alterations were detected in the macrodissected tumor cells and were either absent in the macrodissected lymphocytes or detected at very low levels (supplemental Figures 7-9), indicating their tumor-derived origin. We also sequenced seven peripheral blood samples from patients with TET2 and DNMT3A alterations with VAFs not significantly different than expected from tumor purity (Figure 2F). In only 1 patient was an alteration found in the original specimen detected (supplemental Figure 10).
Although the presence of CHIP in solid tumor microenvironment has been suggested,7 its prevalence and clinical associations have not been fully investigated. Our analyses of solid tumors from a broad array of cancer subtypes strongly support the hypothesis that many genomic alterations in DNMT3A, TET2, ASXL1, and SF3B1 detected in solid tumor samples arise from their presence in admixed hematopoietic elements, especially when their VAFs in the CGP are significantly lower than expected from tumor purity. Our empiric data suggest that detection of CHIP-associated alterations may be correlated with increased tumor-infiltrating lymphocytes. It is unclear whether this is simply a requisite to detect CHIP in solid tumors or whether patients with CHIP are more likely to have increased lymphocytic infiltrates, possibly due to CHIP’s proinflammatory effects.16 The differential rates of detecting mutations in some genes (eg, DNMT3A vs TET2) within the tumor microenvironment compared with those observed in peripheral blood samples suggests a distinct mutational spectrum for tumor-homing CHIP versus circulating clones. In contrast with paired tumor-and-blood sequencing, the hematopoietic origin of CHIP-associated alterations detected by tumor-only sequencing must be confirmed in patient-matched hematopoietic samples. Recognizing CHIP is critical for interpreting CGP of individual patients both to ensure that precision medicine strategies are not incorrectly focused on nontumor genomic alterations and to identify patients at risk for developing therapy-related myeloid neoplasms. CHIP-associated alterations detected in CGP of solid tumors are most likely to represent bona fide CHIP in patients who are older; have a history of smoking, prior chemotherapy, and/or radiotherapy exposure7 ; or have tumor samples with reduced tumor purity and increased immune infiltration. Therefore, as the clinical utilization of tumor-only sequencing assays increases, it is imperative to realize that CHIP may be detected in solid tumor microenvironment and, in the appropriate clinical context, perform a hematologic workup.
The online version of this article contains a data supplement.
Acknowledgments
This research was supported by the Comprehensive Genomics, Biomedical Informatics, and Biospecimen Repository Shared Resources at Rutgers Cancer Institute of New Jersey (National Institutes of Health, National Cancer Institute grant P30CA072720) as well as Rutgers Office of Advanced Research Computing (National Institutes of Health, Office of the Director grant 1S10OD012346-01A1). M.H. is a New Jersey Commission on Cancer Research postdoctoral fellow (DFHS17PPC007). H.K. acknowledges support from the American Cancer Society (IRG-15-168-01).
Authorship
Contribution: E.A.S., S.G., and H.K. designed the study, supervised research, and performed statistical analyses; G.M.R. evaluated histological data and performed macrodissection; E.A.S., G.M.R., S.G., and H.K. drafted the manuscript; C.F.C., J.-A.V., S.R., G.M.F., J.S.R., L.G., S.A., V.M., and J.E. integrated, analyzed, and interpreted the data from the FoundationOne and FoundationOneHeme assays; M.G., K.M.H., and L.R.-R. analyzed clinical data; A.F.-C. and M.H. analyzed high-throughput DNA sequencing data; and all authors wrote and approved the manuscript.
Conflict-of-interest disclosure: E.A.S., C.F.C., J.-A.V., S.R., G.M.F., J.S.R., L.G., S.A., V.M., and J.E. are employees of and have equity interest in Foundation Medicine, Inc. S.G. serves on a scientific advisory board and as consultant to Inspirata, Inc., has patents on digital imaging technology licensed to Inspirata, Inc., has equity in Inspirata, Inc., serves on advisory board for Novartis Pharmaceuticals, and consults for Roche. The remaining authors declare no competing financial interests.
Correspondence: Hossein Khiabanian, Rutgers Cancer Institute of New Jersey, Rutgers University, 195 Little Albany St, New Brunswick, NJ 08903-2681; e-mail: h.khiabanian@rutgers.edu; and Shridar Ganesan, Rutgers Cancer Institute of New Jersey, Rutgers University, 195 Little Albany St, New Brunswick, NJ 08903-2681; e-mail: ganesash@cinj.rutgers.edu.
References
Author notes
E.A.S., G.M.R., and C.F.C. contributed equally to this work.