Key Points
The mutational profile of patients with preclinical MDS is distinct from that reported in healthy individuals.
In the absence of morphologic disease, mutational analysis can predict those patients at high risk of disease progression.
Abstract
The diagnosis of myelodysplastic syndromes (MDS) remains problematic due to the subjective nature of morphologic assessment. The reported high frequency of somatic mutations and increased structural variants by array-based cytogenetics have provided potential objective markers of disease; however, this has been complicated by reports of similar abnormalities in the healthy population. We aimed to identify distinguishing features between those with early MDS and reported healthy individuals by characterizing 69 patients who, following a nondiagnostic marrow, developed progressive dysplasia or acute myeloid leukemia. Targeted sequencing and array-based cytogenetics identified a driver mutation and/or structural variant in 91% (63/69) of prediagnostic samples with the mutational spectrum mirroring that in the MDS population. When compared with the reported healthy population, the mutations detected had significantly greater median variant allele fraction (40% vs 9% to 10%), and occurred more commonly with additional mutations (≥2 mutations, 64% vs 8%). Furthermore, mutational analysis identified a high-risk group of patients with a shorter time to disease progression and poorer overall survival. The mutational features in our cohort are distinct from those seen in the healthy population and, even in the absence of definitive disease, can predict outcome. Early detection may allow consideration of intervention in poor-risk patients.
Introduction
The morphologic diagnosis of myelodysplastic syndromes (MDS) is problematic due to poor inter-observer concordance1 and the difficulty in distinguishing MDS from non-neoplastic conditions.2 Cytogenetics can provide objective evidence of disease, although reports of frequent driver mutations3,4 and/or structural variants detected by single nucleotide polymorphism (SNP) arrays5 have provided potential core criteria for the diagnosis of MDS. However, this approach has been complicated by reports of frequent somatic mutations6-8 and large chromosomal abnormalities9,10 in the aging healthy population. The term “clonal hematopoiesis of indeterminate potential” has been proposed for patients with somatic mutations but without evidence of hematologic malignancy, and “clonal cytopenias of undetermined significance” to encompass the subset with cytopenias.11 Although these individuals show an increased risk of developing hematologic malignancies,6-8 those with clinically significant mutations are currently indistinguishable from those who will not progress.
The aim of this study was to molecularly characterize those patients with the most clinically significant disease, who fail to meet diagnostic criteria using conventional techniques. We retrospectively identified patients who, despite having an initial bone marrow with nondiagnostic features, developed progressive dysplasia or acute myeloid leukemia (AML). We hypothesized that characterizing these patients would provide potential criteria to distinguish preclinical MDS from healthy individuals, and importantly, detect those patients at high risk of disease progression.
Study design
Patients and samples
A retrospective search was performed for new patients diagnosed with AML/MDS at the Haematological Malignancy Diagnostic Service between 2004 and 2012, with a previous nondiagnostic bone marrow performed for investigation of cytopenia (more recently termed “idiopathic cytopenia of undetermined significance”). The Haematological Malignancy Diagnostic Service provides a centralized hematopathology service for ∼6 million population with all cases dual-reported by experienced hematopathologists. Eighty-two patients were identified with both prediagnostic and diagnostic samples, representing 1.7% of patients with idiopathic cytopenia of undetermined significance during this time period (n = 4835). Sixty-nine patients had adequate molecular material at both time points for analysis (see supplemental Table 1 on the Blood Web site) and survival data were available for 59 patients. Samples were taken with informed consent, in accordance with the Declaration of Helsinki, for investigation of a suspected hematologic disorder. The study had local Institutional Review Board approval.
DNA extraction, targeted sequencing, and SNP-array analysis
DNA was either extracted at the point of referral or from stored, unstained smears. Targeted sequencing of 26 commonly mutated genes in myeloid malignancies was performed on MiSeq (Illumina, Chesterford, United Kingdom) using custom Fluidigm panels (Fluidigm, San Francisco, CA) to construct DNA libraries. Read alignment and variant calling were performed using MiSeq reporter (Illumina) and variants annotated using Ensembl Variant Effect Predictor software.12 Following exclusion of synonymous, noncoding variants and germline polymorphisms (unless recurrently reported in COSMIC database13 ), variants were validated by GS Junior (Roche, Burgess Hill, United Kingdom), conventional Sanger sequencing, or allele-specific oligonucleotide analysis (Janus kinase 2). All patients were screened for NPM1 and FLT3 mutations using conventional fragment analysis.
SNP-A analysis was performed on diagnostic samples using a HumanCytoSNP-12 BeadChip Kit (Illumina) with data visualized using KaryoStudio software (Illumina). Abnormalities were called using published guidelines.14 In those with a documented abnormality, SNP-A was performed on the corresponding prediagnostic sample.
See supplemental Methods for details.
Statistical analysis
Overall survival (OS) was estimated using the Kaplan–Meier method (censored on 01/06/2015). The impact of abnormalities on OS, and associations between genetic mutations and progression were investigated with Fisher’s exact test and Cox regression.
Results and discussion
Mutational profile in prediagnostic samples differs from healthy individuals
A somatic mutation and/or structural abnormality were identified in 91% (n = 63/69) of prediagnostic samples. This included 133 mutations across 62 patients, most commonly involving epigenetic regulators or spliceosome genes with TET2, SRSF2, and ASXL1 mutated in 39%, 26%, and 20% of patients, respectively (Figure 1A). The spectrum of mutations at this time point mirrored that reported in large MDS populations,3,4 with the exception of SF3B1 (n = 3), though these mutations are strongly associated with ring sideroblasts, which are easily identified morphologically.15 Although DNMT3A was the most frequently mutated gene in the healthy population,6-8 this was seen in only 10% (n = 7) of our patient cohort. A median of 2 mutations was detected per patient (range, 1-5; Figure 1C) and the frequency of multiple mutations was significantly greater here than reported in healthy individuals (≥2 mutations; 64% vs 8%6 ). The median variant allele fraction (VAF) and inferred clone size in our series was also notably greater at 40% (range, 2.31% to 100%; Figure 1B) than the reported median of 9% to 10% in healthy individuals.6 Importantly, only 1 patient harbored an isolated mutation with a VAF <20%. These differences suggest that the clone must expand to an appropriate level and/or acquire cooperating mutations to cause cytopenias and subsequent disease. This is supported by the greater mean VAF (25.2%) observed in healthy individuals who subsequently developed a hematologic malignancy.6 Therefore, although driver gene mutations with a high AF (>20%) and/or co-occurring mutations may not be disease-defining, it at least identifies clinically significant clonality requiring close follow-up.
Characteristics of mutations detected in a prediagnostic sample. (A) Frequency of driver mutations across all 69 patients in the prediagnostic sample. (B) VAF (%) of somatic variants in the prediagnostic sample, including median and mean. (C) Distribution of the number of mutations according to final disease subgroups, and across both prediagnostic and diagnostic samples. AF, allele fraction.
Characteristics of mutations detected in a prediagnostic sample. (A) Frequency of driver mutations across all 69 patients in the prediagnostic sample. (B) VAF (%) of somatic variants in the prediagnostic sample, including median and mean. (C) Distribution of the number of mutations according to final disease subgroups, and across both prediagnostic and diagnostic samples. AF, allele fraction.
In contrast, structural variants were identified in only 23% (16/69) of prediagnostic samples with all but one co-occurring with a somatic mutation (supplemental Table 5).
Mutations predict progression to high-risk disease and OS
Thirty-nine patients progressed to refractory anemia with excess blasts (RAEB) or AML, in a significantly shorter time than those with refractory cytopenia with multilineage dysplasia (median, 403 vs 606 days; hazard ratio [HR], 3.7; 95% confidence interval [CI], 2.1-6.6; P < .001). By analyzing the most frequently mutated genes (supplemental Methods), IDH2 was weakly associated with disease progression (P = .052). NPM1, CBL, and NRAS were mutated at low frequency, however, all progressed to AML or RAEB. Furthermore, IDH2 (HR, 4.2; 95% CI, 1.3-13.8; P = .017) and TP53 mutations (HR, 5.5; 95% CI, 1.1-27.7; P = .038) were associated with a more rapid time to progression. These observations require confirmation in larger cohorts.
Thirty patients (43%) acquired a mutation between samples, most commonly involving transcription factors and cell-signaling genes, and correlated strongly with progression to RAEB and AML (supplemental Results and supplemental Figure 2). Mutations across individual samples (supplemental Figure 1) and changes in VAF between samples (supplemental Figure 3) are presented in supplemental Results.
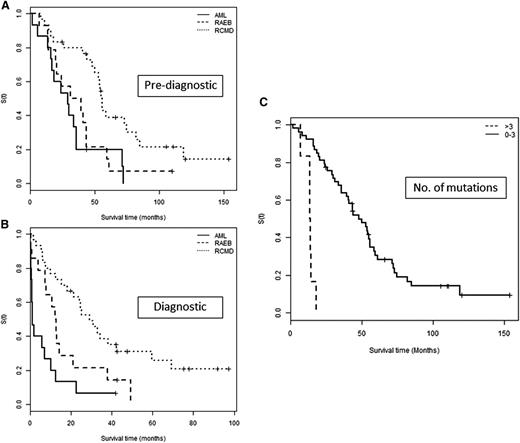
Of those with survival data available (n = 59), only 10 were alive at the point of analysis. Median OS from the prediagnostic sample was 43.6 months (95% CI, 33.8-55.8) and, as expected, much shorter from diagnosis (13 months; 95% CI, 9.9-24.6). This was most significant in those diagnosed with AML (1.28 months; 95% CI, 0.789-12.625; Figure 2) and importantly, all but 1 of these patients harbored a mutation in the prediagnostic sample.
OS according to sample time and mutation number. OS in patients grouped by final disease category from time of (A) prediagnostic sample and (B) diagnostic sample. The survival of the AML group is comparable to the RAEB group if determined from the prediagnostic sample (P = .442) and highlights a potential period for earlier intervention. (C) OS according to the number of mutations detected in a prediagnostic sample grouped into those with ≤3 and >3 mutations.
OS according to sample time and mutation number. OS in patients grouped by final disease category from time of (A) prediagnostic sample and (B) diagnostic sample. The survival of the AML group is comparable to the RAEB group if determined from the prediagnostic sample (P = .442) and highlights a potential period for earlier intervention. (C) OS according to the number of mutations detected in a prediagnostic sample grouped into those with ≤3 and >3 mutations.
Prediagnostic mutations were also associated with significantly worse OS, namely TP53 (HR, 21.68; 95% CI, 4.72-99.64; P < .001), U2AF1 (HR, 2.63; 95% CI, 1.0-6.4; P = .049), and the number of mutations (HR, 1.447; 95% CI, 1.12-1.88; P = .006). The latter was most significant in those with >3 mutations (Figure 2C).
The mutational profile in our cohort differs significantly from that of the healthy population, and has the potential to identify patients with clonal hematopoiesis of indeterminate potential/clonal cytopenias of undetermined significance who are at greater risk of progression, even in the absence of morphologic disease. Early detection would provide an increased window for therapeutic intervention in those with very poor prognosis. The diagnostic utility of these findings is however limited by the lack of a control group, including patients who did not progress to AML/MDS. Our study design included only those patients re-investigated for cytopenia, and both the mutation and disease frequency is likely to be greater. The optimal way to explore this and refine molecular criteria for diagnosis is to prospectively study an unselected cytopenic patient cohort, and this is currently in progress.
Presented as an oral presentation at the 56th annual meeting of the American Society of Hematology, San Francisco, CA, December 4-9, 2014.
The array data reported in this article have been deposited in the Gene Expression Omnibus database (accession number GSE73074).
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank Dr Jan Taylor for providing additional bioinformatic support.
Authorship
Contribution: C.A.C. and A.S.J. designed the study; C.A.C., N.R., P.A.E., and S.L.B. performed the research; C.A.C., S.L.B., and S.C. analyzed the data; and C.A.C., D.T.B., and A.S.J. wrote the paper.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Catherine A. Cargo, Haematological Malignancy Diagnostic Service, St James’s University Hospital, Beckett St, Leeds LS9 7TF, United Kingdom; e-mail: catherine.cargo@nhs.net.


This feature is available to Subscribers Only
Sign In or Create an Account Close Modal