Abstract
Background: Minimal residual disease (MRD) has an emerging diagnostic and prognostic role in Diffuse Large B-cell Lymphoma (DLBCL). However, repetitious bone marrow aspirations or lymph node biopsies would bring great pains to patients. At present, deep next-generation sequencing (NGS) based on circulating tumor DNA (ctDNA), allows quantitative mutational analysis with high sensitivity simultaneously, thereby providing us a promising non-invasive and radiation-free approach to monitor MRD and track sophisticated dynamic evolution of tumor clones.
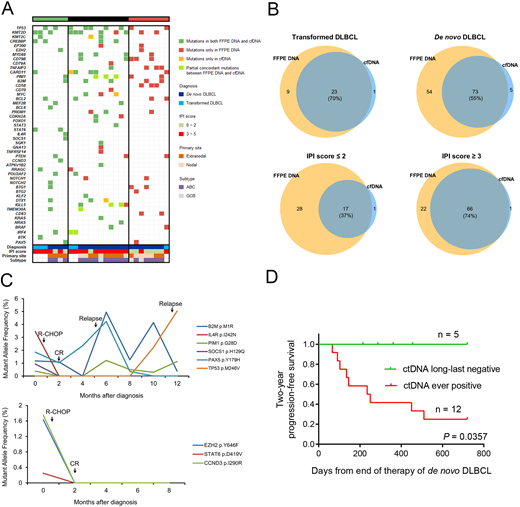
Methods: A total of 21 patients with denovo DLBCL and 6 patients with transformed DLBCL treated at our center were enrolled in this study. Formalin-fixed paraffin-embedded (FFPE) tissues at diagnosis were collected. Peripheral blood samples were obtained at the time of DLBCL diagnosed, remission, relapse, or other specific points in follow-up duration. Genomic DNA (gDNA) and cell-free DNA (cfDNA) were extracted from FFPE specimens and plasma samples, respectively. Targeted sequencing based on Illumina HiSeq 2000 were performed in all samples, which covering the whole coding sequences or hotspot regions in 61 genes known to be significantly associated with DLBCL. Library was prepared using unique molecular identifiers (UMI), which enabled the reduction of most synthesis-based errors. Sequencing depth was performed up to 2000× for FFPE specimens and 25000× for plasma samples, respectively. All mutations detected by cfDNA were validated by droplet digital PCR using Bio-Rad QX200TM. Concordance of filtered variants was determined between FFPE specimens and plasma samples at diagnosis. Correlation between clinical conditions and variant allele frequency (VAF) was also analyzed. This study was approved by Institutional Review Board of Tongji Hospital.
Results: The median sequencing depth of FFPE specimens and cfDNA samples was 2312× and 28211×, respectively. Compared sequencing results of FFPE specimens with paired plasma samples at diagnosis, the number of cases with complete concordance, partial concordance, and complete discordance were 7, 12, and 8. For ultimate filtered variants detected in FFPE specimens and plasma at diagnosis, the consistency was 70% in transformed DLBCL and 55% in de novo DLBCL. Taking International Prognostic Index (IPI) score into consideration, the consistency of detection of denovo DLBCL patients with IPI score ≤ 2 was significant lower than that of patients with IPI score ≥ 3 (37% vs. 74%, P < 0.001). Besides, the presence of mutations in cfDNA also predicted MRD-positivity even though the patients achieved complete remission (CR), and the dynamic change of variant allele frequency is also associated with the evolution of tumor clones in most cases. Finally, long-last negativity of variants in cfDNA after treatment was significantly associated with superior progression-free survival.
Conclusions: Our study indicated that targeted NGS of cfDNA in DLBCL is a promising test that facilitate disease monitoring and individualized therapy. A high level of concordance between FFPE specimens and plasma samples is associated with progression or evolvement that underlies clinical conditions, and ctDNA analysis may be a practical testing method to assess the prognosis of patients, although further investigations based on an extended sequencing panel and study cohort are warranted.
No relevant conflicts of interest to declare.
Author notes
Asterisk with author names denotes non-ASH members.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal