[Background]
Customized gene panel sequencing is commonly used in cancer research to detect point mutations and small insertions or deletions. This technique can also be used to detect copy number alterations (CNAs) in tumor samples based on the comparison of sequence depth of targeted regions between samples and variant allele frequencies of germline single nucleotide polymorphisms (SNPs).
CNAs in TP53 and CDKN2A/2B are frequently found in various types of leukemias. TP53 loss was reported to be a frequent CNA with poor prognosis in adult AML, and CDKN2A/2B alterations were found to be frequent in adult and pediatric acute lymphoblastic leukemia. However, the molecular and clinical profiles of the CNAs of these genes remain unclear in pediatric AML.
[Patients and Methods]
We analyzed clinical samples from 331 pediatric AML patients in the AML-05 trial, which was conducted by the Japanese Pediatric Leukemia/Lymphoma Study Group. Targeted sequencing was performed using a 343-gene custom panel and next-generation sequencer. These 343 genes are reportedly associated with hematopoietic malignancy or solid tumor pathogenesis. Copy number analysis was performed using the in-house pipeline CNACS. Additionally, a SNP array was performed for 40 of the 331 patients, including 34 patients with complex karyotype, to detect CNAs of TP53.
[Results]
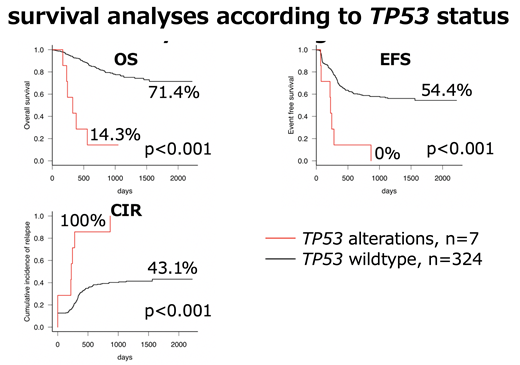
TP53 alterations were identified in 7 (2.1%) of 331 patients. TP53 losses and mutations were detected in 7 and 4 patients, respectively, and all the patients with TP53 mutations concurrently had TP53 losses. Seven patients with TP53 losses were identified by CNACS, although only 4 patients with TP53 losses were found by SNP array. As for the cytogenetics of 7 patients with TP53 losses, six carried a complex karyotype, lacking FAB-M7 morphology. Accordingly, 6 (28.6%) of 21 non-FAB-M7 patients with complex karyotype had TP53 losses. Fusion genes and gene mutations were rare in patients with TP53 losses (NUP98-KDM5A, n=1; KIT, n=1; NRAS, n=1; CEBPA, n=1; ASXL1, n=1), and no patients with core-binding factor AML (CBF-AML) or KMT2A-rearrangements had TP53 losses. In survival analyses, patients with TP53 losses had significantly worse overall survival, event-free survival, and cumulative incidence of relapse rates than patients with wildtype TP53 (14.3% vs 71.4%, p<0.001, 0% vs 54.4%, p<0.001, and 100% vs 43.1%, p<0.001, respectively) (Figure). Among the 7 patients with TP53 losses, 2 had induction failure, and the remaining 5 relapsed. As a result, 6 patients died.
CDKN2A/2B alterations were identified in 10 patients (3.0%) (9 losses and 1 mutation). Three patients with CDKN2A/2B losses had simultaneous TP53 alterations. Unlike TP53 alterations, CDKN2A/2B alterations were found in patients with CBF-AML and those with FAB-M7. In survival analyses, no significant difference was observed between patients with or without CDKN2A/2B alterations.
[Discussion]
TP53 losses were identified in approximately 2% of patients with pediatric AML. However, these alterations were found to be frequent in patients with complex karyotypes, particularly in those with non-FAB-M7 (28.6%). Furthermore, patients with TP53 losses had very poor prognosis. These findings indicate the necessity of risk stratification using TP53 status for pediatric AML.
The efficacy of hematopoietic stem cell transplantation (HSCT) for the treatment of patients with TP53 losses remains unclear because 6 of 7 patients with TP53 losses received HSCT in this study. Moreover, because some of the identified TP53 mutations were possible germline mutations, HSCT may increase the incidence rate of the secondary cancer in patients with TP53 mutations. Thus, the efficacy of new agents (e.g., azacitizine and gemtuzumab ozogamicin) should be evaluated in future clinical trials for the intensification of chemotherapy for these patients.
Meanwhile, the clinical and molecular profiles of CDKN2A/2B alterations were not fully identified in this study. However, the high incidence of CDKN2A/2B alterations in patients with TP53 losses indicates that the correlation between these gene alterations is essential for oncogenesis in a subset of pediatric AML patients.
Ogawa:ChordiaTherapeutics, Inc.: Consultancy, Equity Ownership; Kan Research Laboratory, Inc.: Consultancy; RegCell Corporation: Equity Ownership; Asahi Genomics: Equity Ownership; Qiagen Corporation: Patents & Royalties; Dainippon-Sumitomo Pharmaceutical, Inc.: Research Funding.
Author notes
Asterisk with author names denotes non-ASH members.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal