Key Points
Anti-PF4 antibodies as detected by some PF4/heparin enzyme immunoassays are not specific for VITT.
We provide a functional whole-blood flow cytometry test for detection of platelet-activating anti-PF4 antibodies in VITT.
Abstract
Vaccination is crucial in combatting the severe acute respiratory syndrome coronavirus 2 pandemic. The rare complication of thrombocytopenia and thrombotic complications at unusual sites after ChAdOx1 nCov-19 vaccination is caused by platelet-activating antibodies directed against platelet factor 4 (PF4). We present a widely applicable whole-blood standard flow cytometric assay to identify the pathogenic antibodies associated with vaccine-induced immune-mediated thrombotic thrombocytopenia (VITT) after ChAdOx1 nCov-19 vaccination. This assay will enable rapid diagnosis by many laboratories. This trial was registered at www.clinicaltrials.gov as #NCT04370119.
Introduction
Coronavirus disease 2019 (COVID-19) is caused by the single-stranded RNA virus severe acute respiratory syndrome coronavirus2 (SARS-CoV-2). Currently, 4 different vaccines to prevent symptomatic COVID-19 have been approved by the European Medicines Agency.1
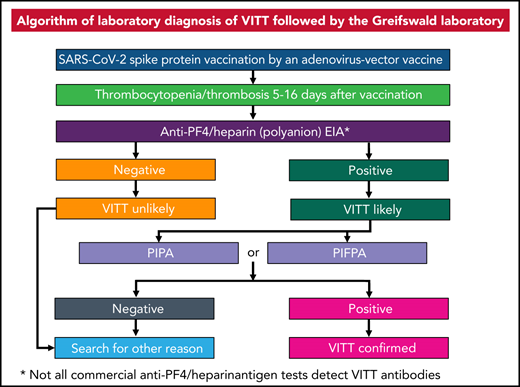
Recently, several cases of thrombosis combined with moderate to severe thrombocytopenia were observed in patients vaccinated with the ChAdOx1 nCov-19 (AstraZeneca) vaccine2‐5 and in patients vaccinated with the Johnson & Johnson vaccine.6,7 We have identified immunoglobulin G antibodies directed against platelet factor 4 (PF4)-activating platelets via FcγRIIa to be the likely cause of vaccine-induced immune-mediated thrombotic thrombocytopenia (VITT).5 VITT shows striking similarities with autoimmune heparin-induced thrombocytopenia (HIT).8,9 In contrast to HIT, platelet activation in VITT occurs in the presence of PF4 rather than low heparin concentrations. The antibodies associated with recent ChAdOx1 nCov-19 vaccination can be detected by some PF4/heparin enzyme immunoassays (EIAs),10 which are widely available to diagnose HIT. However, these assays may not be specific for VITT-related antibodies. Not all antibodies binding to PF4/heparin complexes by EIAs are functionally active and others may be typical HIT antibodies. We modified the functional heparin-induced platelet activation test,11,12 a washed platelet assay, to detect vaccine-related antibodies and to differentiate them from HIT antibodies. We have named the modified assay the PF4-induced platelet activation (PIPA) test (for details, see supplemental Methods, available on the Blood Web site). As washed platelet assays are restricted to specialized laboratories, we developed a flow cytometric assay using whole blood to detect PF4-dependent platelet-activating antibodies in ChAdOx1 nCov-19–vaccinated patients. By analogy to the washed platelet test, we refer to the modified assay as the PF4-induced flow cytometry–based platelet activation (PIFPA) test.
Study design
For the PIFPA test, citrated whole blood was obtained from healthy donors. Whole blood was supplied with 54 U/mL (final) hirudin (Canyon Pharmaceuticals; if hirudin is not available, then other thrombin inhibitors like d-Phe-Pro-Arg-chloromethylketone [PPACK] can be used), and incubated with 0, 5, or 20 µg/mL PF4 (final) for 20 minutes at 37°C. Afterward, heat-inactivated serum (56°C, 30 minutes) from the following donors was added in a whole blood-to-serum ratio of 2:1 and incubated for 20 minutes at 37°C: patients with VITT, asymptomatic vaccinated donors being positive or negative in the anti-PF4/heparin EIA, unvaccinated donors, or patients with HIT. Samples were stained using CD61-phycoerythrin (PE) (clone SZ21; Beckman Coulter) and CD62P-PE-Cy5 (Becton Dickinson) antibodies for 10 minutes (room temperature) in the dark. Stained samples were fixed with 2% paraformaldehyde (Morphisto) for 20 minutes, washed in phosphate-buffered saline (pH 7.4; Pan Biotech), centrifuged (650g, 7 minutes, room temperature), and resuspended in 1× fluorescence-activated cell sorter lysing solution (Becton Dickinson). Samples were measured in a Cytomics FC500 flow cytometer (Beckman Coulter). Platelets were positively gated using CD61-PE. Activation was determined by granule release measured using CD62P-PE-Cy5 and given as mean fluorescence intensity (MFI) of the CD62P+ gated events multiplied by the percentage of gated platelets. Platelet activation for each serum was given as median activation from the 4 whole-blood samples. Further details of thetest method are given in supplemental Methods.
The use of whole blood and platelets from healthy donors and serum from patients was approved by the Ethics Board at University Medicine Greifswald and was conducted in accordance with the Declaration of Helsinki.
Results and discussion
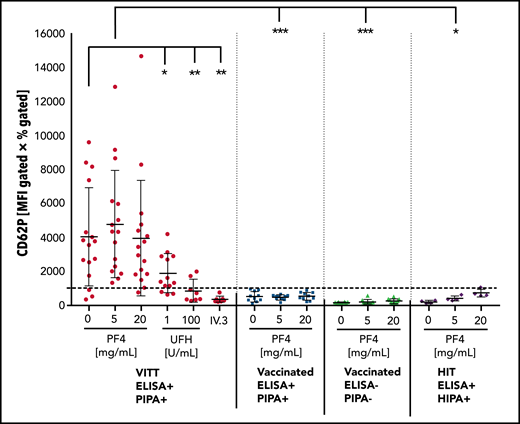
We tested sera from individuals who developed VITT with thrombosis and thrombocytopenia developing 5 to 16 days after vaccination with ChAdOx1 nCov-19 as well as postvaccination serum samples from University Hospital Greifswald health care workers vaccinated with ChAdOx1 nCov-19 (Screening for COVID-19 and Monitoring of Serological Responses to SARS-CoV-2 in Health care Workers [SeCo] study; approved by the Greifswald Ethics Committee, no. BB068/20). Sera were pretested in PF4/heparin EIAs and the functional PIPA test. The flow cytometry–based PIFPA test was performed with 16 VITT samples (14 sera, 2 citrated plasma) and all available EIA+/PIPA− sera, as well as 10 representative EIA−/PIPA− sera and 4 sera of patients with HIT who had a positive EIA and positive HIPA result. Cutoff was determined with sera of 13 unvaccinated healthy controls incubated with 5 µg/mL PF4 as mean plus 2× the standard deviation, with samples above the cutoff regarded as positive. The readout of the PIFPA is granule release of platelets measured by CD62P expression. As platelet activation via FcγRIIa shows variability among donors, the PIFPA was always performed with blood from 4 unselected healthy donors, and the median of CD62P expression is shown for each serum in Figure 1. Serum from patients with VITT had significantly higher median CD62P expression compared with vaccinated controls without thrombotic complications and without functionally relevant antibodies (EIA+/PIPA− [P = .0009]; EIA−/PIPA− [P = .0003]). However, 3 of the 16 VITT samples remained below the cutoff. By adding 5 µg/mL PF4, all VITT samples tested positive and were clearly distinguished from sera of vaccinated, clinically healthy, asymptomatic individuals with positive EIA but a negative PIPA test (P = .0003). All 10 sera that tested EIA− and PIPA− also tested negative in the PIFPA, and only 1 of the 4 tested HIT sera was slightly above the cutoff when incubated with 20 µg/mL PF4. A low dose of unfractionated heparin (1 U/mL) decreased CD62P expression (P = .018) and high doses of heparin further reduced the signal (P = .0058). Inhibition of FcγRIIA by the monoclonal antibody IV.3 inhibited platelet activation by 6 tested VITT samples (P = .0032), supporting the important role of FcγRIIa in the pathogenesis of this prothrombotic complication of ChAdOx1 nCov-19 vaccination.
Platelet activation in the PIFPA test. Platelet activation induced by (heat-inactivated; 56°C, 30 minutes) sera and PF4 is shown by CD62P expression and given by the MFI multiplied by percentage of gated events. Data are shown for 16 samples from patients with VITT (14 sera, 2 plasmas), 10 sera from patients after vaccination with positive EIA but negative PIPA result, 10 sera from patients after vaccination with negative EIA and PIPA, and 4 sera from patients with typical HIT. One data point for each serum represents median activation from 4 different whole-blood samples. Cutoff was determined with sera of 13 unvaccinated healthy controls incubated with 5 µg/mL PF4 as the mean plus 2 standard deviations (SD). With the addition of 5 µg/mL PF4, sera from patients with VITT can be readily discriminated from vaccinated donors with no functionally relevant antibodies (P < .0003). Statistical significance was calculated by the unpaired Student t test. *P < .05; **P < .01; ***P < .001. HIPA, heparin-induced platelet aggregation; UFH, unfractionated heparin.
Platelet activation in the PIFPA test. Platelet activation induced by (heat-inactivated; 56°C, 30 minutes) sera and PF4 is shown by CD62P expression and given by the MFI multiplied by percentage of gated events. Data are shown for 16 samples from patients with VITT (14 sera, 2 plasmas), 10 sera from patients after vaccination with positive EIA but negative PIPA result, 10 sera from patients after vaccination with negative EIA and PIPA, and 4 sera from patients with typical HIT. One data point for each serum represents median activation from 4 different whole-blood samples. Cutoff was determined with sera of 13 unvaccinated healthy controls incubated with 5 µg/mL PF4 as the mean plus 2 standard deviations (SD). With the addition of 5 µg/mL PF4, sera from patients with VITT can be readily discriminated from vaccinated donors with no functionally relevant antibodies (P < .0003). Statistical significance was calculated by the unpaired Student t test. *P < .05; **P < .01; ***P < .001. HIPA, heparin-induced platelet aggregation; UFH, unfractionated heparin.
The PIFPA test shows very comparable results to the PIPA test (Table 1) and can be applied to confirm or rule out platelet-activating PF4-dependent antibodies, which occur 5 to 16 days after ChAdOx1 nCov-19 vaccination based on available data, without the need for washed platelets. The exact mechanism whereby vaccination with ChAdOx1 nCov-19 is associated with these platelet-activating anti-PF4 antibodies is still unknown. The disorder appears analogous to autoimmune HIT, where antibodies are capable of inducing strong platelet activation despite the absence of heparin, in contrast to typical HIT sera, which require the addition of heparin. Although the PIFPA seems to be specific, we cannot rule out that patients with less severe complications (eg, isolated thrombocytopenia, which have not been identified up to now) may have more weakly reacting antibodies induced by ChAdOx1 nCov-19 for which an additional cofactor might be required (by analogy to HIT, akin to heparin) to produce platelet activation by patient serum. The scientific community should undertake all efforts to screen for such a cofactor (or cofactors). In addition, laboratory tests for antibodies causing HIT using washed platelets are always more sensitive and probably even more specific than assays using whole blood.13 We therefore recommend that patients with the typical clinical presentation of VITT, but testing negative in the PIFPA, should be further assessed in the PIPA until more information is available.
Comparison of the diagnostic tests
| . | PF4/heparin EIA (OD) . | PIPA . | PIFPA . |
|---|---|---|---|
| VITT | |||
| 1 | + (3.18) | + | + |
| 2 | + (3.40) | + | + |
| 3 | + (3.50) | + | + |
| 4 | + (3.0) | + | + |
| 5 | + (3.16) | + | + |
| 6 | + (2.02) | + | + |
| 7 | + (3.05) | + | + |
| 8 | + (3.18) | + | + |
| 9 | + (3.17) | + | + |
| 10 | + (3.18) | + | + |
| 11 | + (3.17) | + | + |
| 12 | + (3.20) | + | + |
| 13 | + (3.26) | + | + |
| 14 | + (3.78) | + | + |
| 15 | + (3.27) | + | + |
| 16 | + (2.82) | + | + |
| Vaccinated asymptomatic | |||
| 01 | + (0.52) | Negative | Negative |
| 02 | + (0.77) | Negative | Negative |
| 03 | + (0.89) | Negative | Negative |
| 04 | + (2.15) | Negative | Negative |
| 05 | + (0.85) | Negative | Negative |
| 06 | + (0.60) | Negative | Negative |
| 07 | + (0.73) | Negative | Negative |
| 08 | + (0.55) | Negative | Negative |
| 09 | + (0.67) | Negative | Negative |
| 10 | + (0.78) | Negative | Negative |
| 11 | Negative (0.15) | Negative | Negative |
| 12 | Negative (0.12) | Negative | Negative |
| 13 | Negative (0.10) | Negative | Negative |
| 14 | Negative (0.06) | Negative | Negative |
| 15 | Negative (0.16) | Negative | Negative |
| 16 | Negative (0.18) | Negative | Negative |
| 17 | Negative (0.21) | Negative | Negative |
| 18 | Negative (0.11) | Negative | Negative |
| 19 | Negative (0.11) | Negative | Negative |
| 20 | Negative (0.12) | Negative | Negative |
| . | PF4/heparin EIA (OD) . | PIPA . | PIFPA . |
|---|---|---|---|
| VITT | |||
| 1 | + (3.18) | + | + |
| 2 | + (3.40) | + | + |
| 3 | + (3.50) | + | + |
| 4 | + (3.0) | + | + |
| 5 | + (3.16) | + | + |
| 6 | + (2.02) | + | + |
| 7 | + (3.05) | + | + |
| 8 | + (3.18) | + | + |
| 9 | + (3.17) | + | + |
| 10 | + (3.18) | + | + |
| 11 | + (3.17) | + | + |
| 12 | + (3.20) | + | + |
| 13 | + (3.26) | + | + |
| 14 | + (3.78) | + | + |
| 15 | + (3.27) | + | + |
| 16 | + (2.82) | + | + |
| Vaccinated asymptomatic | |||
| 01 | + (0.52) | Negative | Negative |
| 02 | + (0.77) | Negative | Negative |
| 03 | + (0.89) | Negative | Negative |
| 04 | + (2.15) | Negative | Negative |
| 05 | + (0.85) | Negative | Negative |
| 06 | + (0.60) | Negative | Negative |
| 07 | + (0.73) | Negative | Negative |
| 08 | + (0.55) | Negative | Negative |
| 09 | + (0.67) | Negative | Negative |
| 10 | + (0.78) | Negative | Negative |
| 11 | Negative (0.15) | Negative | Negative |
| 12 | Negative (0.12) | Negative | Negative |
| 13 | Negative (0.10) | Negative | Negative |
| 14 | Negative (0.06) | Negative | Negative |
| 15 | Negative (0.16) | Negative | Negative |
| 16 | Negative (0.18) | Negative | Negative |
| 17 | Negative (0.21) | Negative | Negative |
| 18 | Negative (0.11) | Negative | Negative |
| 19 | Negative (0.11) | Negative | Negative |
| 20 | Negative (0.12) | Negative | Negative |
Results of the PF4/heparin EIA and the washed platelet PIPA test were compared with the flow cytometry–based PIFPA test. Results of the PF4/heparin EIAs are given in parentheses.
+ indicates that a test was rated positive; OD, optical density.
In summary, we present an assay to detect platelet-activating antibodies that seem highly specific for VITT after vaccination with ChAdOx1 nCov-19. The pathogenesis of this severe adverse effect, which threatens the vaccination program in the SARS-CoV-2 pandemic, may be further clarified by rapidly identifying the real incidence of VITT caused by platelet-activating antibodies.
The data supporting the findings of this study are available from the corresponding author upon reasonable request.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank Ulrike Strobel, Ricarda Raschke, Ines Warning, Carmen Freyer, Katrin Stein, Jessica Fuhrmann, and Julia Klauke for excellent technical support.
This work was supported by the Deutsche Forschungsgemeinschaft (DFG; German Research Foundation) project number 374031971–TRR 240.
Authorship
Contribution: S.H., M.W., C.Z., and J.W. performed the experiments, analyzed the data, and wrote the manuscript; C.Z., L.S., and T.T. coordinated patient samples and the study protocol; L.U., N.-O.H., and K.B. coordinated the SeCo study and provided the samples; K.A. discussed the results and reviewed the manuscript; and A.G. developed the PIPA and the PIFPA concept, supervised the study and experiments, and wrote the manuscript.
Conflict-of-interest disclosure: A.G. reports grants and nonfinancial support from Aspen, Boehringer Ingelheim, Merck Sharp & Dohme (MSD), Bristol Myers Squibb (BMS), Paringenix, Bayer Healthcare, Gore Inc, Rovi, Sagent, and Biomarin/Prosensa; personal fees from Aspen, Boehringer Ingelheim, MSD, Macopharma, BMS, Chromatec, and Instrumentation Laboratory; and nonfinancial support from Boehringer Ingelheim, Portola, Ergomed, and Gesellschaft für Thrombose und Hämostaseforschung (GTH) eV, outside of the submitted work. T.T. reports personal fees and educational support from BMS, Pfizer, and Chugai Pharma; personal fees from Bayer and Novartis;and educational support from Novo Nordisk and Daiichi Sankyo, outside of the submitted work. The remainingauthors declare no competing financial interests.
Correspondence: Andreas Greinacher, Institut für Immunologie und Transfusionsmedizin, University Medicine Greifswald, Sauerbruchstr, 17487 Greifswald, Germany; e-mail: andreas.greinacher@med.uni-greifswald.de.
REFERENCES
Author notes
S.H. and M.W. contributed equally to this study.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal