In this issue of Blood, Huang et al1 show that HIC2 messenger RNA (mRNA) is a direct target of let-7 microRNA (miRNA), providing a crucial link between LIN28B/let-7 and active transcription of the fetal hemoglobin (HBG) genes through silencing BCL11A.
Elevation of fetal HBG transcription in adults, reversing the perinatal switch to adult HBB, has been a therapeutic goal for decades to moderate the symptoms of sickle cell disease and β-thalassemia. In adult erythroid cells, the repressor BCL11A binds to the promoters of the HBG genes and interferes with their long-range activation by the locus control region, making reduction of BCL11A highly desirable.2,3BCL11A is regulated at the transcriptional level by a complex intronic enhancer.4 Elevation of fetal hemoglobin (HbF) in adult erythroid cells was successfully achieved by genome editing to delete the BCL11A enhancer in mobilized patient hematopoietic stem cells ex vivo, which was recently approved in the UK for treatment of sickle cell disease and β-thalassemia and by the US Food and Drug Administration for sickle cell disease.
BCL11A mRNA and protein levels are higher in adult compared with fetal erythroblasts. Efforts to understand this differential level of expression were given a significant boost by the discovery that the repressor HIC2 binds within the BCL11A enhancer evicting GATA1 and decommissioning enhancer activity.5 HIC2 is a fetal-stage-expressed factor, explaining why BCL11A is reduced at the fetal stage but begging the question how HIC2 is repressed in adults.
Using careful quantitation of nascent RNA, mRNA, and protein in adult and fetal erythroblasts, Huang et al concluded that HIC2 was regulated posttranscriptionally. They then depleted DICER, the miRNA processing enzyme, to globally block miRNA biogenesis, a pathway for mRNA degradation.6 In adult-type erythroid HUDEP2 cells, HIC2 protein was strongly increased by DICER depletion, accompanied by reduced BCL11A and induction of HBG. However, codepletion of DICER and HIC2 only partially reversed HBG induction, leaving open the possibility that miRNAs may contribute to HBG silencing by additional HIC2-unrelated mechanisms.
miRNAs have substantially different expression patterns in adult and fetal erythroblasts. Notably, the Miller group had proposed7 a central role for small RNAs in globin gene switching. They reported that overexpression of LIN28B in cultured erythroblasts reduced let-7 miRNA, one of its major targets, suppressed BCL11A and upregulated HBG. However, the mechanistic connection between let-7 and BCL11A remained unclear. To identify specific miRNAs that might be involved in HIC2 regulation, Huang et al focused on those more abundant in adult than in fetal cells that also had predicted binding sites in the HIC2 3′ untranslated region (3′ UTR), through which mRNA degradation is mediated. Seven predominately adult expressed miRNAs were obtained that were, remarkably, all members of the let-7 miRNA family.
The authors next overexpressed let-7 in fetal-type erythroid HUDEP1 cells. This required bypassing inhibition of let-7 processing by LIN28B, which binds to the let-7 loop sequence and is abundant in fetal erythroblasts. To this end, they produced a chimeric miRNA with the mature sequence of let-7a-1, to enable HIC2 3′ UTR targeting, and the loop structure of miR-21, an unrelated miRNA not inhibited by LIN28B. Overexpression of the chimeric miRNA indeed downregulated HIC2 mRNA and protein and induced BCL11A primary transcripts. Drilling down further, the authors fused the HIC2 3′ UTR to luciferase. Mutation of each of 4 predicted let-7 binding sites in the 3′ UTR had little effect but deletion of all 4 sites strongly increased the luciferase signal. The results support that let-7 targets HIC2 mRNA for degradation through its 3′ UTR and that HIC2 reduction is sufficient to upregulate BCL11A.
In reciprocal experiments, Huang et al repressed let-7 in adult erythroid HUDEP2 cells using a decoy containing lentiviral vector. Decreasing the levels of highly expressed let-7a/f together increased HIC2 mature mRNA and protein levels but not primary RNA. HIC2 protein increased more than mRNA. This suggests let7a/f influences both mRNA turnover and translation. Inhibition of let-7a/f also reduced BCL11A primary and mature transcripts and protein, and HBG transcription was strongly induced. Thus, let-7 represses HIC2 and contributes to BCL11A and HBG regulation.
In xenotransplantation studies, Huang et al engrafted mice with healthy human donor CD34+ cells that had been genetically modified with lentiviral vectors to knock down let-7a/f. Human erythroid progenitors sorted from bone marrow after 16 weeks showed 50% reduction of BCL11A primary transcripts and mature mRNA and strongly increased HBG and HBE1 expression. Sensitive quantitation indicated HBG represented 45% of total β-like globin transcripts, a 90-fold increase. Mechanistic studies ex vivo in human CD34+ cells confirmed let7a/f inhibition increased HBG mRNA and protein and significantly increased the percentage of HbF expressing cells. HIC2 was strongly increased in genetically modified cells and BCL11A was strongly reduced.
let-7a/f inhibition might have had more global consequences than HIC2 reduction. However, RNA sequencing revealed few (125) differentially expressed genes (DEGs). The let-7a/f up/down DEGs correlated significantly with high/low HIC2. Taken together, the data support a regulatory pathway connecting let-7 and HIC2 translation and mRNA stability with BCL11A and HBG transcriptional regulation (see figure).
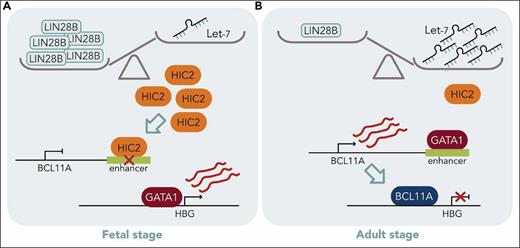
HIC2 links LIN28B/let-7 to HBG transcription. (A) The balance depicts abundance of RNA binding protein LIN28B in fetal cells resulting in blocked let-7 miRNA biogenesis. HIC2 can then inactivate the BCL11A enhancer, allowing transcription of HBG and HbF production. (B) In adult cells, LIN28B is low and let-7 is abundant, binding to the 3′ UTR of HIC2 and leading to its degradation. Now, GATA1 can occupy the BCL11A enhancer, BCL11A is activated and then the protein silences HBG. Professional illustration by Somersault18:24.
HIC2 links LIN28B/let-7 to HBG transcription. (A) The balance depicts abundance of RNA binding protein LIN28B in fetal cells resulting in blocked let-7 miRNA biogenesis. HIC2 can then inactivate the BCL11A enhancer, allowing transcription of HBG and HbF production. (B) In adult cells, LIN28B is low and let-7 is abundant, binding to the 3′ UTR of HIC2 and leading to its degradation. Now, GATA1 can occupy the BCL11A enhancer, BCL11A is activated and then the protein silences HBG. Professional illustration by Somersault18:24.
Is HIC2 the only relevant let-7a/f target in the pathway to HBG transcriptional control? Chromatin structure analyses are supportive of this hypothesis. In agreement with the few gene expression changes in the let-7 inhibition model, there were relatively few changes observed using assay for transposase-accessible chromatin (ATAC)-sequencing. Interestingly, HIC2 and GATA binding motifs were enriched among the peaks that lost accessibility, consistent with common targets. Globally, the sites that lost accessibility correlated with increased HIC2 signal and decreased GATA1 signal. The BCL11A enhancer exemplifies this exchange. Increased HIC2 signal and decreased GATA1 signal at the enhancer correlated with decreased long-range interactions with the BCL11A promoter and decreased transcription.
In a final experiment to clinch that HIC2 critically connects let-7 to HBG, Huang et al depleted HIC2 in let7a/f-inhibited HUDEP2 cells. BCL11A was restored to levels in let-7a/f-inhibited cells alone and HBG was resilenced. The present report connects the dots between let-7 miRNA and HBG, showing that HIC2 is targeted for degradation by let-7, and supplies a satisfying explanation to the proposal of the Miller group that small RNAs have a central role in globin gene switching. The miRNA pathway provides a so far underexplored regulatory option for augmenting fetal hemoglobin.
Conflict-of-interest disclosure: A.D. declares no competing financial interests.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal