Key Points
PD-L1 expression is increased in 46/1 haplotype carriers.
3D chromatin structure differs between JAK2 haplotypes, and noncoding elements in the JAK2 loci regulate both JAK2 and PD-L1 expression.
Visual Abstract
Although described more than a decade ago, the mechanism by which the JAK2 46/1 haplotype increases the risk of developing JAK2-mutated myeloproliferative neoplasms (MPNs) remains unexplained. Inflammation and immunity are linked to MPN development and thus could be relevant to the mechanism by which 46/1 mediates its effect. Here, we show that programmed death-1 receptor ligand (PD-L1) expression is elevated in 46/1 haplotype, both in healthy carriers and in CD34+ cells from patients with MPN. Using circular chromosome conformation capture, we observed that PD-L1 and the neighboring PD-L2 loci physically interact with JAK2 in a manner that differs between 46/1 and nonrisk haplotypes. CRISPR/Cas9 genome editing identified a region within JAK2 intron 2 that influences both JAK2 and PD-L1 expression. We suggest that increased PD-L1 expression may be relevant to the mechanism by which 46/1 leads to an increased inherited risk of developing MPN.
Introduction
Classic Philadelphia-negative myeloproliferative neoplasms (MPNs) are clonal hematopoietic stem cell disorders characterized by the proliferation of ≥1 cell lineage.1 A key component in their pathogenesis is aberrant cytokine pathway signaling, caused in most cases by coding mutations in JAK2, CALR, and MPL.1
The great majority of MPN cases are sporadic (with no familial aggregation); however, several common inherited genetic variants have been associated with an increased inherited risk of developing an MPN.2,3 Of these, the JAK2 46/1 haplotype, a >200 kb region at 9p24.1, which includes the JAK2, INSL4, and INSL6 genes, is probably the most relevant, because it explains ∼28% of the population-attributable risk of developing an MPN.4 This common low penetrance haplotype accounts for a quarter of JAK2 alleles in the general population but as many as 70% to 80% of mutated JAK2 alleles in MPNs. 46/1 was described more than a decade ago but, despite its obvious importance and clinical associations (eg, association with increased JAK2 V617F allele burden in polycythemia vera and lower constitutional symptoms/better survival in myelofibrosis),5,6 the mechanism by which it exerts its effects remains unclear.4,7,8 Two nonexclusive hypotheses have been proposed: the hypermutability of JAK2 on the 46/1 haplotype; and the “fertile ground” hypothesis, in which JAK2 V617F is proposed to have a selective advantage if it is acquired on 46/1.3,9
Although not part of the 46/1 haplotype, the gene encoding CD274/programmed death-1 receptor ligand (PD-L1; hereafter referred to as PD-L1) is also located at 9p24.1, ∼320 kb proximal to JAK2. This is of interest because PD-L1 is overexpressed in JAK2 V617F-mutated MPN cases, potentially leading to immune escape.10 Furthermore, PD-L1 is expressed on disease-initiating MPN stem cells and the degree of PD-L1 expression in CD34+ cells has been linked to JAK2 V617F mutational burden.11,12 Because both the presence and mutational burden of JAK2 V617F is strongly associated with 46/1,4,7,8,13 we aimed to explore whether there was any relationship between 46/1 and PD-L1 expression, which could, for example, be driven by long-range cis-regulatory elements within the region or structural alterations to chromatin, independent of JAK2 V617F status.
Study design
Patients and samples
Peripheral blood was collected from healthy blood donors after informed consent was obtained. Frozen CD34+ cells from JAK2 V617F-positive patients with MPN were obtained from the Hospital Universitario 12 de Octubre Biobank. This study was conducted according to the biomedical research guidelines of the Declaration of Helsinki and approved by the Ethics Committee of Hospital Universitario 12 de Octubre (no. 16/096). TaqMan single nucleotide polymorphism (SNP) genotyping analysis (C_31941696_10; Thermo Fisher) was used for haplotyping (see the supplemental Methods, available on the Blood website).
PD-L1 protein levels
PD-L1 expression in leukocyte populations and cell lines (K562 and HEL) was studied by flow cytometry. For CD34+ cells from patients with MPN, PD-L1 enzyme-linked immunosorbent assay was used (Thermo Scientific; catalog no. BMS2327). For further details, see the supplemental Methods.
4C-Seq
Circular chromosome conformation capture (4C) technology allows for the identification of interactions that take place between a defined genomic region (viewpoint [VP]) and the rest of the genome. The protocol we used is described in the supplemental Methods; in brief, we designed primers to study 7 VP around and within the 46/1 JAK2 haplotype, selected by the presence of markers of regulatory activity. 4C followed by deep sequencing (4C-Seq) was performed, as previously described,14 on 107 granulocytes purified from peripheral blood by Ficoll gradient (Ficoll-Paque PLUS; GE Healthcare; 2 nullizygous for the 46/1 haplotype and 2 homozygous for the 46/1 haplotype). Quantification of the interactions between each VP and the PD-L1 and PD-L2 regions (chr9:5447958-5470872 and chr9:5507588-5570246, respectively) were performed by calculating the sum of the normalized reads counts for each sample within the regions of interest. To study significant contacts, the frequency of captured sites per window was used to fit a distance decreasing monotone function, and z scores were calculated from its residuals using a modified version of 4C-Seq.15 Significant contacts were considered in cases in which the z score was >2 in both replicates and deviated significantly (adjusted P < .05) from its normal cumulative distribution in at least one of the replicates. After data analysis, processed reads and interactions were visualized using the WashU Epigenome Browser (https://epigenomegateway.wustl.edu/).
Cell culture and transfection and CRISPR experiments
K562 and HEL cell lines were cultured as described in the supplemental Methods. For genome editing, K562 cells were transfected with the CRISPR/Cas9 gene-editing tool as described.16 A total of 5 × 105 K562 cells were plated in 6 multiwell plates (MW6) and transfected with 1.25 μg of each plasmid using lipofectamine 3000, following the manufacturer’s instructions. For HEL cells, 5 × 106 cells were electroporated with 35 μg of each plasmid using the Bio-Rad Gene Pulser II with the following conditions: 975 μF, 264 mV, and 22 milliseconds. Forty-eight hours after transfection, single GFP+ cells were sorted using a FACSAria Fusion (BSC II). Clones were grown, and DNA was extracted and genotyped using QuickExtract Solution (Lucigen). For HEL cells, 2 rounds of electroporation were performed to obtain high deletion efficiency. K562 cells were treated with 10 ng/mL of human interferon gamma overnight (PeproTech; catalog no. 300-02). Guides and primers used for genotyping are described in supplemental Table 4. For quantitative polymerase chain reaction methods, see the supplemental Methods.
Statistics
Student t test or 1-way analysis of variance (GraphPad PRISM 8.4.3) were used for independent samples, depending on the experimental design.
Results and discussion
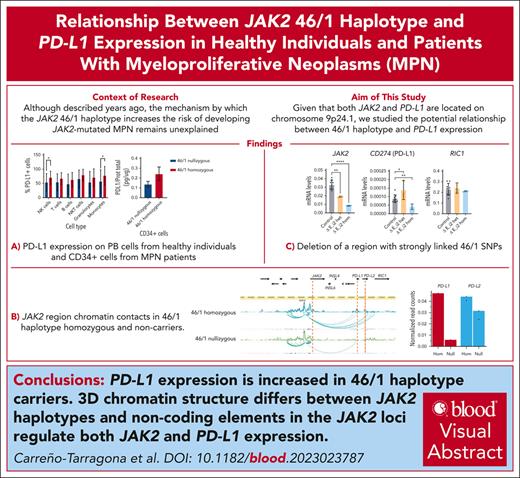
To explore the possibility that PD-L1 expression is influenced by JAK2 haplotype status, we performed flow cytometry to measure PD-L1 levels in different peripheral blood cell populations from a cohort of healthy individuals selected as either homozygous or nullizygous for 46/1 (n = 37 each). As shown in Figure 1A, there is a clear tendency toward overexpression of PD-L1 in all cell types in 46/1 homozygotes, which was statistically significant for natural killer lymphocytes and monocytes. We also studied PD-L1 levels in CD34+ cells from JAK2 V617F-positive patients with MPN (nullizygous, n = 11; homozygous, n = 9) and observe a clear tendency toward higher expression in 46/1 homozygotes as shown previously (Figure 1B).12
JAK2 46/1 haplotype influences PD-L1 expression levels. (A) The percentage of PD-L1–expressing cells, measured by flow cytometry, in different peripheral blood cell populations from healthy donors (46/1 nullizygous [blue] and 46/1 homozygous [red]; n = 37 each). ∗P < .05 (nonparametric Kruskal-Wallis test). (B) PD-L1 protein levels, determined by enzyme-linked immunosorbent assay and expressed as PD-L1/total protein (pg/μg) in CD34+ cells obtained from bone marrow of JAK2-positive patients with MPN (46/1 nullizygous [blue], n = 11; 46/1 homozygous [red], n = 9). Error bars indicate standard error of the mean. B, B lymphocytes; NK, natural killer cells; NKT, natural killer T cells; T, T lymphocytes.
JAK2 46/1 haplotype influences PD-L1 expression levels. (A) The percentage of PD-L1–expressing cells, measured by flow cytometry, in different peripheral blood cell populations from healthy donors (46/1 nullizygous [blue] and 46/1 homozygous [red]; n = 37 each). ∗P < .05 (nonparametric Kruskal-Wallis test). (B) PD-L1 protein levels, determined by enzyme-linked immunosorbent assay and expressed as PD-L1/total protein (pg/μg) in CD34+ cells obtained from bone marrow of JAK2-positive patients with MPN (46/1 nullizygous [blue], n = 11; 46/1 homozygous [red], n = 9). Error bars indicate standard error of the mean. B, B lymphocytes; NK, natural killer cells; NKT, natural killer T cells; T, T lymphocytes.
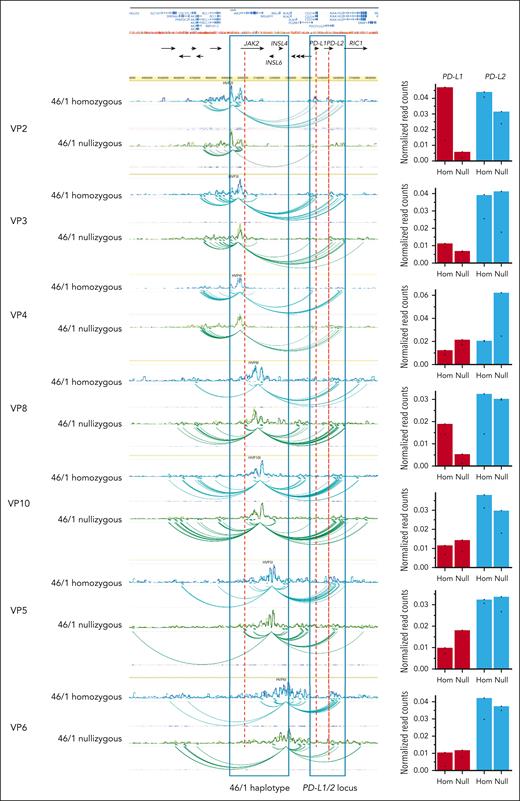
Because the PD-L1 locus lies in close proximity to the JAK2 gene and the 46/1 haplotype block, we hypothesized that transcriptional regulatory elements located within the haplotype might physically interact with PD-L1 and influence its expression. Therefore, we selected 7 regions (VPs) within or in the immediate vicinity of the haplotype (see the supplemental Methods for details) to map long-distance chromatin interactions through 4C-Seq. Using neutrophils from 46/1 nullizygous (n = 2) and 46/1 homozygous (n = 2) healthy donors, we found that the JAK2 locus has contacts with both the PD-L1 and PD-L2 loci. Furthermore, the three-dimensional (3D) chromatin interactions between the JAK2 and PDL1/2 regions differed between 46/1 homozygotes and 46/1 nullizygotes (Figure 2).
Chromatin contacts of JAK2 with the PD-L1 locus differ between the nonrisk and the 46/1 haplotype. Visualization of a 1.6 Mb region from human chromosome 9, spanning the JAK2 and PDL1/2 loci (hg19 chr9:4300000-5900000). Chromatin interactions established from the 7 distinct genomic regions or viewpoints (VP2, VP3, VP4, VP8, VP10, VP5, and VP6; genomic coordinates in supplemental Table 2) are shown as spider plots. Healthy individuals who were 46/1 nullizygous (green and light green, n = 2) or 46/1 homozygous (blue and light blue, n = 2) are shown for each VP, with differences between them around PD-L1/2 indicated. Regions corresponding to the 46/1 haplotype and the PDL-1/2 loci are boxed. Black horizontal arrows below the University of California Santa Cruz browser view indicate the position of all genes in the region, and the names of those referred to in this study are included. On the right of each spider plot, normalized read counts (or contacts) are shown for both PD-L1 (red) and PD-L2 (blue). As indicated, the left column for each locus represents 46/1 homozygous (Hom) and right column 46/1 nullizygous (Null). As an example, there is a marked reduction in normalized read counts between VP2 and PD-L1 on comparison of 46/1 homozygotes with 46/1 nullizygotes. There is also a reduction in the interaction between VP2 and PD-L2, but this is much more modest.
Chromatin contacts of JAK2 with the PD-L1 locus differ between the nonrisk and the 46/1 haplotype. Visualization of a 1.6 Mb region from human chromosome 9, spanning the JAK2 and PDL1/2 loci (hg19 chr9:4300000-5900000). Chromatin interactions established from the 7 distinct genomic regions or viewpoints (VP2, VP3, VP4, VP8, VP10, VP5, and VP6; genomic coordinates in supplemental Table 2) are shown as spider plots. Healthy individuals who were 46/1 nullizygous (green and light green, n = 2) or 46/1 homozygous (blue and light blue, n = 2) are shown for each VP, with differences between them around PD-L1/2 indicated. Regions corresponding to the 46/1 haplotype and the PDL-1/2 loci are boxed. Black horizontal arrows below the University of California Santa Cruz browser view indicate the position of all genes in the region, and the names of those referred to in this study are included. On the right of each spider plot, normalized read counts (or contacts) are shown for both PD-L1 (red) and PD-L2 (blue). As indicated, the left column for each locus represents 46/1 homozygous (Hom) and right column 46/1 nullizygous (Null). As an example, there is a marked reduction in normalized read counts between VP2 and PD-L1 on comparison of 46/1 homozygotes with 46/1 nullizygotes. There is also a reduction in the interaction between VP2 and PD-L2, but this is much more modest.
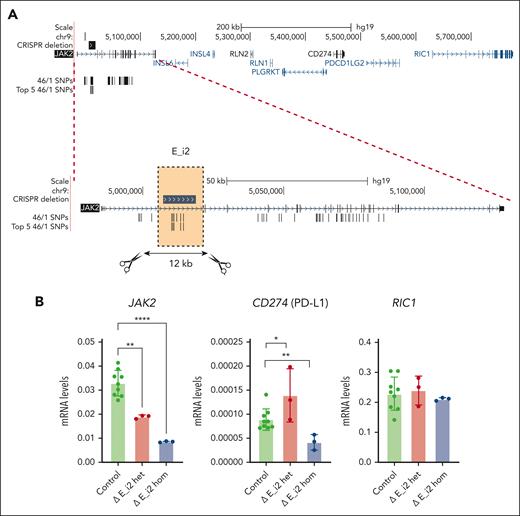
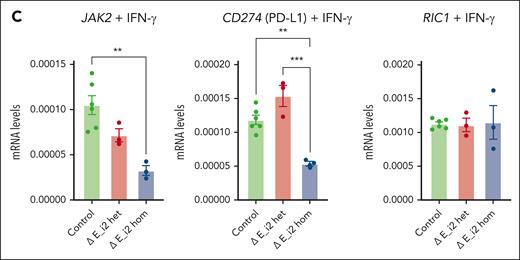
Finally, we aimed to identify putative transcriptional cis-regulatory elements present in the JAK2 haplotype that could be involved in the regulation of PD-L1. We selected a 12 kb region located in JAK2 intron 2, based on the location of strongly linked 46/1 SNPs (E_i2; Figure 3A; supplemental Methods), and deleted it with CRISPR/Cas9 in the myeloid cell line K562. Correctly deleted cells, both heterozygous and homozygous, together with nondeleted controls (supplemental Figure 3A), were isolated, and the expression of JAK2, PD-L1, and RIC1 (a gene neighboring the PD-L1/2 locus with minimal JAK2 contacts; Figure 2) was measured. Expression of PD-L2 is barely detectable in myeloid cells, so we did not include it in the analysis. Deletion of E_i2, either 1 or 2 copies, led to a strong reduction of JAK2 messenger RNA levels, and PD-L1 expression was also reduced but only when both alleles were deleted (Figure 3B). RIC1 expression did not change in either heterozygous or homozygous deleted clones. Because the expression of PD-L1 in K562 cells is low, we increased its levels by stimulating cells with interferon gamma17,18 (supplemental Figure 3B), observing the same reduction in JAK2 and PD-L1 expression in the deleted cells (Figure 3C). In addition, analysis of protein levels by flow cytometry confirmed a decrease of PD-L1 in the homozygous clone (supplemental Figure 3C). In addition, we studied the effect of E_i2 deletion in HEL cells, an MPN line that contains multiple copies of JAK2 V617F.19 We quantified the degree of deletion of JAK2 in multiple clones (supplemental Figure 3D-E) and observed that edited HEL cells have lower levels of PD-L1 protein (supplemental Figure 3F). Thus, our data identified a regulatory element located in the JAK2 haplotype that influences PD-L1 expression.
JAK2 haplotype regulates PD-L1 expression as shown by CRISPR/Cas9 deletion. (A) Genomic locus of JAK2, indicating the region of intron 2 that was deleted to test its regulatory activity (hg19 chr9:5006961-5018796) based on the location of strongly linked 46/1 SNPs (see the supplemental Methods; supplemental Table 3). (B) Expression of JAK2 (left), PD-L1 (middle), and RIC1 (right) in control (green), heterozygous (red), and homozygous (blue) intron 2 element (E_i2) deleted K562 clones. (C) Expression of JAK2 (left), PD-L1 (middle), and RIC1 (right) in control (green), heterozygous (red), and homozygous (blue) intron 2 element (E_i2) deleted K562 clones treated overnight with 10 ng/mL of interferon gamma. In panels B-C, mRNA levels were normalized using GAPDH as an endogenous control, and data were analyzed by 1-way analysis of variance followed by Tukey multiple comparison test; ∗P < .05; ∗∗P < .01; ∗∗∗P < .005; ∗∗∗∗P < .001. Het, hetrorozygous; Hom, homozygous; IFN-γ, interferon gamma; mRNA, messenger RNA.
JAK2 haplotype regulates PD-L1 expression as shown by CRISPR/Cas9 deletion. (A) Genomic locus of JAK2, indicating the region of intron 2 that was deleted to test its regulatory activity (hg19 chr9:5006961-5018796) based on the location of strongly linked 46/1 SNPs (see the supplemental Methods; supplemental Table 3). (B) Expression of JAK2 (left), PD-L1 (middle), and RIC1 (right) in control (green), heterozygous (red), and homozygous (blue) intron 2 element (E_i2) deleted K562 clones. (C) Expression of JAK2 (left), PD-L1 (middle), and RIC1 (right) in control (green), heterozygous (red), and homozygous (blue) intron 2 element (E_i2) deleted K562 clones treated overnight with 10 ng/mL of interferon gamma. In panels B-C, mRNA levels were normalized using GAPDH as an endogenous control, and data were analyzed by 1-way analysis of variance followed by Tukey multiple comparison test; ∗P < .05; ∗∗P < .01; ∗∗∗P < .005; ∗∗∗∗P < .001. Het, hetrorozygous; Hom, homozygous; IFN-γ, interferon gamma; mRNA, messenger RNA.
It has been shown that only a minority of patients with clonal hematopoiesis of indeterminate potential associated with the JAK2 V617F mutation develop an MPN and that inflammation/immunity as well as constitutional genetics plays a central role in MPN progression.20-23 In this context, the PD1/PD-L1 axis has been implicated in playing a key role in MPN development after acquisition of JAK2 V617F.10 We have shown that the JAK2 46/1 haplotype influences PD-L1 expression levels, and this may be mediated by interacting transcriptional regulatory elements within the JAK2 locus. Further investigations are needed to confirm our hypothesis that increased PD-L1 expression is part of the mechanisms by which 46/1 leads to an increased inherited risk of developing MPN.
Acknowledgments
The authors thank Javier Traba (Centro de Biología Molecular [CBM]) for assistance with gamma interferon experiments.
This study was funded by the Asociación Española Contra el Cáncer (AECC) Ideas Semilla AECC 2017 grants PID2020-115755GB-I00 and PID2023-151742NB-I00 (funded by Ministerio de Ciencia, Innovación y Universidades /Agencia Estatal de Investigación [AEI]/10.13039/501100011033); by the Subdirección General de Investigación Sanitaria (Instituto de Salud Carlos III, Spain) grant PI19/01518; by the Cancer Research Innovation in Science (CRIS) Against Cancer Foundation grant 2018/001; and by the Instituto de Investigación Hospital 12 de Octubre (IMAS12). CBM is supported by an institutional grant from the Fundación Ramón Areces and Severo Ochoa Center of Excellence (grant CEX2021-001154-S, funded by Ministerio de Ciencia e Innovación /AEI/10.13039/501100011033).
Authorship
Contribution: G.C.-T. designed the study, performed experiments, analyzed and interpreted the data, and wrote the manuscript with the support of M.M., J.M.-L., R.A., and N.C.P.C.; R.R. participated in circular chromosome conformation capture experiments and analysis; W.J.T. performed the analysis to identify 46/1 SNPs; A.L. assisted with sample processing and fluorescence-activated cell sorter studies; J.V., M.T., A.J.C., and A.M. participated in the molecular characterization of JAK2 haplotype interactions with PD-L1 expression; and R.G.-V. performed and analyzed PD-L1 enzyme-linked immunosorbent assay.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
The current affiliation for R.R. is Department of Genetic Medicine and Development, Faculty of Medicine, and Institute of Genetics and Genomics in Geneva (iGE3), University of Geneva, Geneva, Switzerland.
Correspondence: Gonzalo Carreño-Tarragona, Hematology Department, I+12, Hospital Universitario 12 de Octubre, Avda Córdoba S/N, 28041 Madrid, Spain; email: gonzalo.carreno@salud.madrid.org.
References
Author notes
J.M.-L. and M.M. are joint senior authors.
Original data are available on request from the corresponding author, Gonzalo Carreño-Tarragona (gonzalo.carreno@salud.madrid.org).
The online version of this article contains a data supplement.
There is a Blood Commentary on this article in this issue.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.

![JAK2 46/1 haplotype influences PD-L1 expression levels. (A) The percentage of PD-L1–expressing cells, measured by flow cytometry, in different peripheral blood cell populations from healthy donors (46/1 nullizygous [blue] and 46/1 homozygous [red]; n = 37 each). ∗P < .05 (nonparametric Kruskal-Wallis test). (B) PD-L1 protein levels, determined by enzyme-linked immunosorbent assay and expressed as PD-L1/total protein (pg/μg) in CD34+ cells obtained from bone marrow of JAK2-positive patients with MPN (46/1 nullizygous [blue], n = 11; 46/1 homozygous [red], n = 9). Error bars indicate standard error of the mean. B, B lymphocytes; NK, natural killer cells; NKT, natural killer T cells; T, T lymphocytes.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/145/19/10.1182_blood.2023023787/1/m_blood_bld-2023-023787-gr1.jpeg?Expires=1769082656&Signature=zR7Hrj63vhPpgXU6NAC78ez4voFhETdaHWHQtb1yOoa~rLBtiqpquhHsLu5N~MOCtGw1VcQ3NFYPJ8ex8C3gw0wBhe6~tcZkYqtJvtwveXSbE6IdaUSAlT2Hnk6fHYofq9Xe62nwI7kmKpOpqxyqAMQbzxzPQj3wgr6Rq895TkjHhOEGUGU5rlP6ifI0mgDd7fDyidC0AHdUnOuIt-xPGKRhJBo277-vfAX591qtkG5s3SyQ74O2W7a1wLY0s~wmdlxyDnsliKLDcROsenxApyHj027R6TruFjldy19qYla0jNFRkdd0VgzOjdGgPKxzEkwcqHY9N2IMwMf71kysYQ__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
This feature is available to Subscribers Only
Sign In or Create an Account Close Modal