Abstract
Acute promyelocytic leukemia (APL) is typified by the reciprocal translocation, t(15; 17)(q22; q21), leading to the formation of PML-RARα and RARα-PML fusion genes. We have characterized 7 cases of morphologic APL found to lack the t(15; 17) on conventional cytogenetic assessment. In 6 of 7 cases, cryptic PML-RARα rearrangements were identified by reverse transcriptase-polymerase chain reaction and fluorescent in situ hybridization (FISH); whereas, in the remaining patient, APL was associated with the variant translocation, t(11; 17)(q23; q12-21), leading to the formation of PLZF-RARα and RARα-PLZF fusion genes. In each of the cases with cryptic PML-RARα rearrangements, PML-RARα transcripts were detected in the absence of RARα-PML, consistent with the concept that PML-RARα is the critical oncogenic fusion protein. In 4 of these cases with evaluable metaphase spreads, the occurrence of a nonreciprocal translocation was confirmed by FISH with sole formation of the PML-RARα fusion gene; in 3 cases with morphologically normal chromosomes 15 and 17, RARα was inserted into PML on 15q, whereas in the remaining patient the PML-RARα fusion arose due to insertion of 15q-derived material including PML into RARα on 17q. Immunofluorescence studies were performed using antibodies raised against PML and PIC 1, a ubiquitin-homology domain protein previously identified as an interaction partner of PML. In acute myeloid leukemia (AML) of subtypes other than M3, PIC 1 was localized to the nuclear membrane and colocalized with PML within discrete nuclear bodies. In APL cases with cryptic PML-RARα rearrangements, the characteristic microparticulate pattern of PML staining was detected with partial colocalization with PIC 1, indicative of disruption of the nuclear bodies; whereas in t(11; 17)-associated APL, PML and PIC 1 remained colocalized within discrete nuclear bodies, as observed in non-APL cases. Although deregulation of the putative growth suppressor PML and delocalization of other nuclear body constituents have been advocated to play a key role in the development of t(15; 17)-associated APL, the present study shows that disruption of PML nuclear bodies per se is not a prerequisite for the pathogenesis of APL.
ACUTE PROMYELOCYTIC leukemia (APL) is typified by the reciprocal translocation, t(15; 17)(q22; q21),1 leading to the formation of PML-RARα and RARα-PML fusion genes (reviewed by Grimwade and Solomon2 and references therein). PML-RARα, transcribed from add(15q), retains virtually all the domains considered to be of functional importance to both PML and RARα and has therefore traditionally been considered to play a key role in leukemogenesis, which has recently been confirmed using a transgenic model.3,4 Any role for del(17q)-derived RARα-PML in the development of APL remains unclear, particularly because reciprocal transcripts are detected in only 81% of patients5; nevertheless, a case of APL apparently lacking a PML-RARα fusion gene and in which only RARα-PML transcripts could be detected has recently been reported.6 The molecular pathogenesis of APL is believed to reflect two key processes: leukemic transformation coupled with a block in myeloid differentiation such that the marrow becomes replaced by abnormal promyelocytes.7 In APL cases associated with a PML-RARα rearrangement, this differentiation block may be overcome by retinoids such as all-trans retinoic acid (ATRA).8 To understand these phenomena, much effort has been devoted to the study of the physiologic roles of RARα and PML.
RARα is a member of the steroid hormone nuclear receptor family, serving as a transcription factor mediating the effect of retinoic acid at specific response elements (reviewed by Stunnenberg9 ). In common with vitamin D and thyroid hormone receptors, high-affinity DNA binding of retinoic acid receptors (RARs) requires heterodimerization with a member of the retinoid X receptor family.10,11 Integrity of these retinoid signaling pathways is critical for normal embryogenesis (reviewed by Grimwade and Solomon2 and references therein) and postnatal myeloid differentiation.12 13
In contrast to RARα, the role of PML remains less clearly defined. Initial claims that it also serves as a transcription factor on the basis of N-terminal zinc-binding RING finger and B-box domains14,15 remain unsubstantiated, and indeed more recent work has failed to demonstrate specific DNA-binding for the majority of RING-family members (reviewed by Saurin et al16 ). With the development of appropriate antisera, PML has been found to be predominantly localized to the nucleus within structures known as PML nuclear bodies.17-20 These are composed of several proteins of unknown function, including NDP52,21 Sp100, and Sp140, which were identified as targets for autoantibodies in patients with primary biliary cirrhosis,22,23 and PIC 1, a ubiquitin-homology domain protein, which has been found to interact directly with PML.24 A variety of experimental approaches have implicated PML in immunologic responses (Grimwade and Solomon2 and references therein) and there is some evidence to suggest that PML itself or components of the nuclear bodies are cell-cycle regulated and can mediate growth-suppressor activity.25-28 Cells from a wide range of tissues and blasts from leukemic subtypes other than APL typically demonstrate 10 to 30 discrete nuclear bodies when stained with PML antisera. Whereas, in APL cases associated with the t(15; 17), a microparticulate pattern of PML staining is characteristic,17-19 reflecting disruption of nuclear bodies due to an interaction between PML and PML-RARα.29 This process may promote leukemogenesis by delocalizing the putative growth-suppressor PML and other nuclear body components; this, coupled with an abnormal pattern of retinoid responses also mediated by the fusion protein, compounded by RXR sequestration could account for the block in myeloid differentiation that characterizes the disease. Treatment of such cases with ATRA leads to complete remission by terminal differentiation of the leukemic clone associated with release of inhibitory effects of PML-RARα at retinoid response elements14,30-32 and degradation of the fusion protein33,34 accompanied by normalization of nuclear body architecture.17-20 Rare cases of PML-RARα–mediated APL have been identified that fail to respond to retinoids associated with persistence of the microparticulate PML nuclear staining pattern.19,35 On the basis of these findings it has been suggested that disruption of PML nuclear bodies is critical to the pathogenesis of APL; furthermore, it has been advocated that reconstitution of normal nuclear architecture is essential to permit differentiation in the presence of retinoids.19
Although ATRA therapy is unable to sustain long-term remission in APL,36-38 recent studies have shown that a combined treatment approach using ATRA and chemotherapy confers significant improvements in disease-free survival compared with chemotherapy alone.39,40 Because a favorable response to ATRA appears to be restricted to patients with the t(15; 17),8,41 establishing the presence of this cytogenetic change, or in its absence identification of a PML-RARα rearrangement, is fundamental for optimal treatment of patients and meaningful analysis of APL trials. Although early studies in specialized centers reported the presence of the t(15; 17) in all cases of APL42; it is now clear that some cases with morphologic acute myeloid leukemia (AML) M3 reflect cryptic PML-RARα rearrangements,6,43,44 whereas in others RARα is fused to a partner other than PML. Thus far, three such alternative fusion partners have been identified, including the novel Krüppel-like zinc finger protein PLZF involved in the t(11; 17)(q23; q21)45; nucleophosmin, an RNA processing protein disrupted by the t(5; 17)(q32; q12)46; and most recently NuMA, which is involved in control of mitosis and is rearranged by the t(11; 17)(q13; q21).47 Although such variant translocations are extremely rare, accounting for less than 1% of morphologic APL,48 elucidation of the mechanisms underlying leukemogenesis in these cases is likely to provide considerable insight into the processes involved in the development of PML-RARα–mediated disease and in particular as to whether disruption of PML nuclear bodies is fundamental to the pathogenesis of APL. Cases with variant fusion translocations also afford the opportunity to dissect out mechanisms leading to leukemic transformation from those mediating the block in myeloid differentiation and its reversal by retinoids. In the present study, we have characterized 7 cases of morphologic APL found to lack the t(15; 17) on conventional cytogenetic assessment and considered their implications for the pathogenesis of APL as a whole.
MATERIALS AND METHODS
Patients and cytogenetics. This study considers 7 patients with morphologic APL who were found to lack the t(15; 17) on conventional cytogenetic assessment. Five patients were drawn from the MRC ATRA trial; details of the treatment protocol have been described elsewhere.5 Cytogenetic assessment was undertaken at local centers or by the central UK MRC AML trials cytogenetics service at University College Hospital, London, according to standard methods.49 In each case in which APL was associated with a normal karyotype, preparations were subject to at least 24 hours of culture and a minimum of 20 metaphases were examined.
Reverse transcriptase-polymerase chain reaction (RT-PCR) and sequence analysis. Details of bone marrow and peripheral blood sample preparation, RNA extraction, and RT-PCR protocols to detect PML-RARα and RARα 1-PML fusion transcripts in conjunction with RARα and PML as controls for RNA integrity have been fully described elsewhere.5 50 Using cDNA generated by the same method, PLZF-RARα and RARα-PLZF transcripts were amplified using nested PCR in material derived from a patient with t(11; 17)(q23; q12-21) identified by cytogenetics. Primers used for PLZF-RARα PCR were as follows: PLZF external, 5′-TCCAGAGGGAGCTGTTCAGC-3′; RARα external, 5′-TCTTCTGGATGCTGCGGCGG-3′; PLZF internal, 5′-TCGAGCTTCCTGATAACGAG-3′; and RARα internal, 5′-GGCGCTGACCCCATAGTGGT-3′. Primers for RARα 1-PLZF nested PCR comprised the following: RARα external, 5′-GGCCAGCAACAGCAGCTCCT-3′; PLZF external, 5′-ATGTCAGTGCCAGTATGGGT-3′; RARα internal, 5′-GGTGCCTCCCTACGCCTTCT-3′; and PLZF internal, 5′-CACTGATCACAGACAAAGGC-3′.
PCR was performed in a 50 μL reaction, with 1 μL of the first round PCR products used as template for the second round of PCR, as previously described.51 First and second round PCR reactions comprised 35 cycles, each consisting of 1 minute of denaturation at 95°C, 1 minute of annealing at 57°C, and 1 minute of extension at 72°C, followed by 10 minutes of extension at 72°C (OmniGene apparatus; Hybaid, Teddington, Middlesex, UK). PCR products were size-separated on ethidium bromide-stained 1.5% agarose gels, as previously described.50Bcr 1 (intron 6) and bcr 2 (exon 6) PML breakpoints were distinguished by sequence analysis of PML-RARα PCR products using previously described methods and primers.5PLZF-RARα and RARα-PLZF PCR products were similarly sequenced, using appropriate internal primers, with an automated sequencer (377; ABI, Perkin-Elmer, CA).
Fluorescent in situ hybridization (FISH). FISH using ICRF PML and RARα cosmid probes (15.5 and 121, respectively) was performed to detect PML-RARα fusion genes in patients lacking the t(15; 17), using previously described methods.43 PML cosmid 15.5 encompasses the 5′ region of the gene, including exons 1 and 2, whereas RARα cosmid 121 spans the APL breakpoint region on chromosome 17.43 In APL cases associated with the t(15; 17), a dual signal is detected with these probes on the derivative chromosome 15, identifying the site of the PML-RARα fusion, in addition to single hybridization signals corresponding to the normal PML and RARα loci on 15q and 17q, respectively. FISH was also performed, using PML and RARα probes (Oncor, Gaithersburg, MD), which detect the formation of the reciprocal RARα-PML fusion gene on del(17q) in APL cases associated with the t(15; 17), in addition to single signals relating to the normal PML and RARα loci. To further characterize cases with cryptic PML-RARα fusion genes, whole chromosome paints (WCPs; Vysis, Richmond, Surrey, UK) and biotinylated centromere probes (Oncor) for chromosomes 15 and 17 were used, in accordance with the manufacturer's instructions. Images were captured with a Zeiss Axioskop epifluorescence microscope and cooled CCD camera (Photometrics, AZ), controlled by an Apple Macintosh computer (Apple Computer Inc, Cupertino, CA). Image capture and processing software were obtained from Vysis, UK.
Immunofluorescence. The PML immunofluorescence technique as applied to crude bone marrow or peripheral blood smears using polyclonal PML antisera has been described in detail previously.5 Dual immunofluorescence studies were also undertaken using a polyclonal antiserum raised against the PML nuclear body constituent, PIC 1,24 and a monoclonal antibody (PG-M3; kindly provided by P.G. Pelicci, European Institute of Oncology, Milan, Italy) directed against the amino-terminal of PML, which recognizes both wild-type PML and the PML-RARα fusion protein.52 Bone marrow/peripheral blood smears were fixed in methanol at −20°C for 15 minutes, allowed to air dry, and preblocked with 10% fetal calf serum. Slides were then incubated simultaneously with PG-M3 and PIC 1 antibodies at a dilution of 1/5 and 1/200, respectively, in phosphate-buffered saline (PBS)-Tween 0.5% for 1 hour at room temperature. Slides were subsequently incubated with Texas-Red–coupled antimouse (Dako Ltd, High Wycombe, UK) and fluorescein-coupled antirabbit (Amersham Intl, Amersham, UK) secondary antibodies, each at 1/200 dilution, for 30 minutes at room temperature. All incubations were followed by three washes in PBS followed by a final wash comprising 0.05% Tween in PBS for 10 minutes. Preparations were examined by confocal laser scanning as previously described.5 24
RESULTS
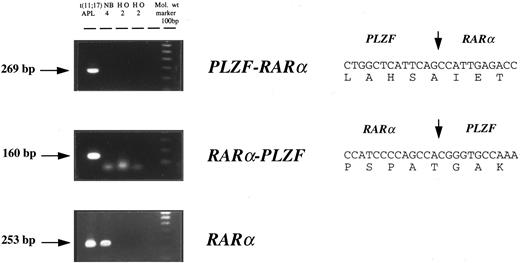
Cytogenetic and molecular characterization of APL cases lacking the t(15; 17). Cytogenetic and molecular findings in 7 patients with morphologic features of APL, but lacking the t(15; 17), are presented in Table 1. In 6 of 7 patients, RT-PCR confirmed the presence of a PML-RARα rearrangement, and in each of these cases PML-RARα transcripts were detected in the absence of RARα-PML. In the remaining patient (case no. 7), APL was associated with t(11; 17) (q23; q12-21); RT-PCR confirmed expression of both PLZF-RARα and reciprocal RARα-PLZF transcripts (Fig 1). Sequencing of PLZF-RARα and RARα-PLZF PCR products was consistent with breakpoints within the second intron of RARα and within the intron separating the exons coding for the second and third zinc fingers of PLZF, as identified in 5 of 6 previously described cases.41 53
Molecular and Cytogenetic Characteristics of Patients With Morphologic APL Lacking the t(15; 17)
| Case No. . | Morphologic Features . | Cytogenetics . | Fusion Transcripts Detected by RT-PCR . | PML Breakpoint . | Rearrangement Detected by FISH . | PML Immunofluorescence . |
|---|---|---|---|---|---|---|
| 1 | Hypergranular M3 | 47,XX,del(5)(q2?3q34),+i(8)(q10) [3] | PML-RARα | bcr 3 | Interstitial insertion of RARα into PML on 15q | ND |
| 45,XX,del(5)(q2?3q34),add(7)(q32),−21 [2] | ||||||
| 92,XXXX,del(5)(q2?3q34) x2 [2] | ||||||
| 46,XX [2] | ||||||
| 2 | Hypergranular M3 | 46,XY | PML-RARα | bcr 1 | Interstitial insertion of RARα into PML on 15q | Microparticulate |
| 3 | Hypogranular M3 variant | 46,XY | PML-RARα | bcr 3 | Interstitial insertion of RARα into PML on 15q | ND |
| 4 | Hypergranular M3 | 46,XY,t(4; 16)(p14; q22),t(9; 12)(q22; q24),dir ins(17; 15)(q21; q15q22) [11] | PML-RARα | bcr 1 | Interstitial insertion of PML into RARα on 17q | ND |
| 46,idem,t(6; 8)(q13; q22) [10] | ||||||
| 46,idem,add(3q),t(6; 14),t(11; 22) [3] | ||||||
| 46,XY [6] | ||||||
| 5 | Hypergranular M3 | 46,XX,del(7)(q22q36) [5] | PML-RARα | bcr 3 | PML-RARα fusion detected by interphase FISH | Microparticulate partial PML and PIC 1 colocalization |
| 46,XX [5] | ||||||
| 6 | Hypergranular M3 | 46,XX | PML-RARα | bcr 3 | PML-RARα fusion detected by interphase FISH | Microparticulate |
| 7 | Hypergranular M3. Marrow replaced by abnormal promyelocytes with basophilic granules. No Auer rods observed. | 46,XY,t(11; 17)(q23; q12-21) [12] | PLZF-RARα, RARα-PLZF | NA | ND | Wild-type PML and PIC 1 colocalized |
| 46,XY [3] |
| Case No. . | Morphologic Features . | Cytogenetics . | Fusion Transcripts Detected by RT-PCR . | PML Breakpoint . | Rearrangement Detected by FISH . | PML Immunofluorescence . |
|---|---|---|---|---|---|---|
| 1 | Hypergranular M3 | 47,XX,del(5)(q2?3q34),+i(8)(q10) [3] | PML-RARα | bcr 3 | Interstitial insertion of RARα into PML on 15q | ND |
| 45,XX,del(5)(q2?3q34),add(7)(q32),−21 [2] | ||||||
| 92,XXXX,del(5)(q2?3q34) x2 [2] | ||||||
| 46,XX [2] | ||||||
| 2 | Hypergranular M3 | 46,XY | PML-RARα | bcr 1 | Interstitial insertion of RARα into PML on 15q | Microparticulate |
| 3 | Hypogranular M3 variant | 46,XY | PML-RARα | bcr 3 | Interstitial insertion of RARα into PML on 15q | ND |
| 4 | Hypergranular M3 | 46,XY,t(4; 16)(p14; q22),t(9; 12)(q22; q24),dir ins(17; 15)(q21; q15q22) [11] | PML-RARα | bcr 1 | Interstitial insertion of PML into RARα on 17q | ND |
| 46,idem,t(6; 8)(q13; q22) [10] | ||||||
| 46,idem,add(3q),t(6; 14),t(11; 22) [3] | ||||||
| 46,XY [6] | ||||||
| 5 | Hypergranular M3 | 46,XX,del(7)(q22q36) [5] | PML-RARα | bcr 3 | PML-RARα fusion detected by interphase FISH | Microparticulate partial PML and PIC 1 colocalization |
| 46,XX [5] | ||||||
| 6 | Hypergranular M3 | 46,XX | PML-RARα | bcr 3 | PML-RARα fusion detected by interphase FISH | Microparticulate |
| 7 | Hypergranular M3. Marrow replaced by abnormal promyelocytes with basophilic granules. No Auer rods observed. | 46,XY,t(11; 17)(q23; q12-21) [12] | PLZF-RARα, RARα-PLZF | NA | ND | Wild-type PML and PIC 1 colocalized |
| 46,XY [3] |
Abbreviations: ND, not determined; NA, not applicable.
Detection of PLZF-RARα and RARα-PLZF transcripts by nested RT-PCR in t(11; 17)(q23; q12-21)-associated APL. PLZF-RARα and RARα-PLZF cDNA sequences are shown on the right; the positions of RARα and PLZF fusion junctions are delineated by vertical arrows.
Detection of PLZF-RARα and RARα-PLZF transcripts by nested RT-PCR in t(11; 17)(q23; q12-21)-associated APL. PLZF-RARα and RARα-PLZF cDNA sequences are shown on the right; the positions of RARα and PLZF fusion junctions are delineated by vertical arrows.
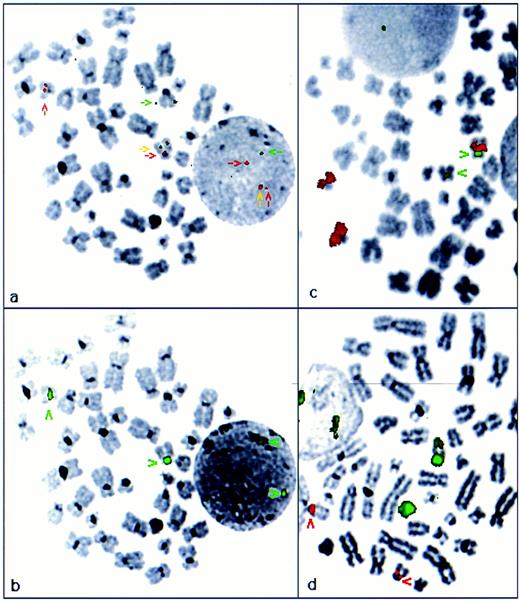
FISH analysis of APL cases with cryptic PML-RARα rearrangements. FISH using ICRF probes, PML cos 15.5 and RARα cos 121, confirmed formation of a PML-RARα fusion in each of the 6 APL cases in which PML-RARα rearrangements were identified by RT-PCR (Table 1). In 4 patients with available diagnostic metaphase spreads, the mechanism underlying the PML-RARα rearrangement was further characterized, using ICRF cosmid probes that specifically detect the PML-RARα fusion gene, in conjunction with centromere probes and WCPs for chromosomes 15 and 17. In 3 patients, in each of whom morphologically normal chromosomes 15 and 17 were identified by conventional cytogenetics (cases no. 1 through 3, Table 1), the PML-RARα fusion gene was localized to chromosome 15q (Fig 2a and b). In each case, 15 and 17 specific paints hybridized solely to their respective chromosomes (Fig 2c and d), consistent with an interstitial insertion of RARα into PML on 15q. Furthermore, in each of these cases, using commercially available probes (Oncor), RARα was found to hybridize to two normal appearing chromosome 17s in addition to forming a fusion signal on 15q, again consistent with formation of a PML-RARα fusion on 15q and absence of the reciprocal RARα-PML fusion gene as suggested by RT-PCR analyses. In the remaining patient with evaluable metaphases, PML 15.5 and RARα 121 probes localized the PML-RARα fusion to 17q (case no. 4; Fig 3a and b). In addition to the PML-RARα fusion signal, a more centromeric RARα hybridization signal was observed. WCPs demonstrated insertion of chromosome 15 material into 17q, such that chromosome 17 appeared abnormally large on conventional cytogenetic assessment (Fig 3c and d). These results indicate that the PML-RARα fusion in this patient reflected insertion of PML with more centromeric chromosome 15-derived material into the genomic region spanned by RARα cos 121; again, this was consistent with detection of PML-RARα in the absence of RARα-PML fusion transcripts by RT-PCR.
Cryptic PML-RARα fusion resulting from interstitial insertion of RARα into PML on 15q (case no. 2; Table 1). (a) FISH analysis using ICRF PML 15.5 (green) and RARα 121 (red) cosmid probes. The RARα probe hybridized to two chromosome 17s of normal appearance, whereas 1 normal PML locus was observed on chromosome 15. The PML-RARα fusion was detected on 15q (yellow arrow). Localization of the fusion gene was confirmed by subsequent hybridization with a chromosome 15 centromere probe, shown in red in (b). (c) Chromosome 15 paint (red); chromosome 17 centromere probe (green). (d) Chromosome 17 paint (green); chromosome 15 centromere probe (red)
Cryptic PML-RARα fusion resulting from interstitial insertion of RARα into PML on 15q (case no. 2; Table 1). (a) FISH analysis using ICRF PML 15.5 (green) and RARα 121 (red) cosmid probes. The RARα probe hybridized to two chromosome 17s of normal appearance, whereas 1 normal PML locus was observed on chromosome 15. The PML-RARα fusion was detected on 15q (yellow arrow). Localization of the fusion gene was confirmed by subsequent hybridization with a chromosome 15 centromere probe, shown in red in (b). (c) Chromosome 15 paint (red); chromosome 17 centromere probe (green). (d) Chromosome 17 paint (green); chromosome 15 centromere probe (red)
PML-RARα fusion resulting from interstitial insertion of PML with associated chromosome 15-derived material into RARα on 17q (case no. 4; Table 1). (a) FISH analysis using ICRF PML (green) and RARα (red) cosmid probes. PML-RARα fusion gene was detected on 17q (yellow arrow), adjacent to RARα hybridization signal (red arrow). Splitting of the RARα- derived signal on der(17q) was indicative of insertion of PML and adjacent sequence into the genomic region covered by RARα cosmid 121. Localization of the PML-RARα fusion gene was confirmed by subsequent hybridization with a chromosome 17 centromere probe shown in green in (b). (c) Chromosome 15 paint (red) and chromosome 17 centromere probe (green), confirming insertion of chromosome 15-derived material into 17q. (d) Chromosome 17 paint (green), chromosome 15 centromere probe (red). Chromosome 17 paint remained localized to 17; der(17q) showed a region of absent signal corresponding to the inserted region of 15q shown in (c).
PML-RARα fusion resulting from interstitial insertion of PML with associated chromosome 15-derived material into RARα on 17q (case no. 4; Table 1). (a) FISH analysis using ICRF PML (green) and RARα (red) cosmid probes. PML-RARα fusion gene was detected on 17q (yellow arrow), adjacent to RARα hybridization signal (red arrow). Splitting of the RARα- derived signal on der(17q) was indicative of insertion of PML and adjacent sequence into the genomic region covered by RARα cosmid 121. Localization of the PML-RARα fusion gene was confirmed by subsequent hybridization with a chromosome 17 centromere probe shown in green in (b). (c) Chromosome 15 paint (red) and chromosome 17 centromere probe (green), confirming insertion of chromosome 15-derived material into 17q. (d) Chromosome 17 paint (green), chromosome 15 centromere probe (red). Chromosome 17 paint remained localized to 17; der(17q) showed a region of absent signal corresponding to the inserted region of 15q shown in (c).
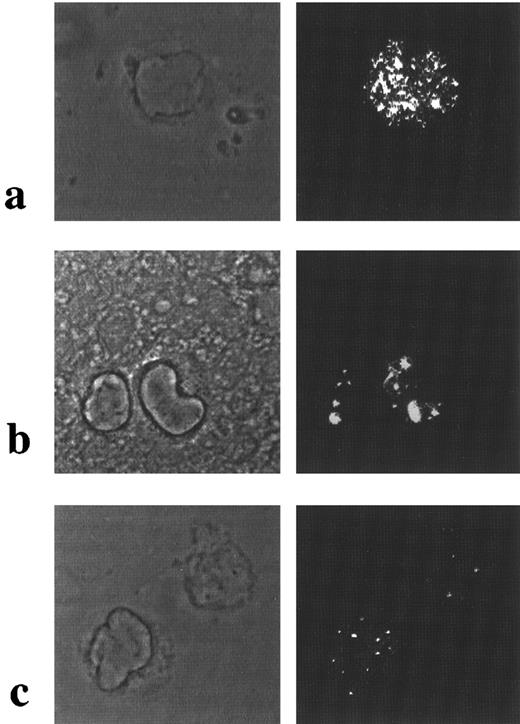
Nuclear architecture in APL cases lacking the t(15; 17). Immunofluorescence studies were performed using polyclonal PML antisera. In 3 patients, cryptic PML-RARα rearrangements detected by FISH and RT-PCR were confirmed by the presence of the characteristic microparticulate nuclear staining pattern in leukemic cells (Table 1 and Fig 4a), identical to that observed in the NB4 cell-line and APL cases associated with the t(15; 17)5; whereas, in the APL case associated with the t(11; 17) leading to a PLZF-RARα rearrangement, a wild-type pattern of PML nuclear staining was observed within the leukemic blasts (Fig 4c), identical to that seen in 2 non-APL AML patients (Fig 4b) and to that previously described for HL60 and U937 cell-lines.5
PML immunofluorescence in AML using polyclonal antisera. Phase contrast is shown in left-hand panels and corresponding PML immunofluorescence on the right. In APL cases with cryptic PML-RARα rearrangements, a microparticulate pattern of PML nuclear staining was observed as shown in (a). In non-APL cases, a wild-type pattern of PML nuclear staining was detected, as shown in AML M2 blasts in (b); similar nuclear staining was observed in t(11; 17)-associated APL (case no. 7; Table 1), shown in (c).
PML immunofluorescence in AML using polyclonal antisera. Phase contrast is shown in left-hand panels and corresponding PML immunofluorescence on the right. In APL cases with cryptic PML-RARα rearrangements, a microparticulate pattern of PML nuclear staining was observed as shown in (a). In non-APL cases, a wild-type pattern of PML nuclear staining was detected, as shown in AML M2 blasts in (b); similar nuclear staining was observed in t(11; 17)-associated APL (case no. 7; Table 1), shown in (c).
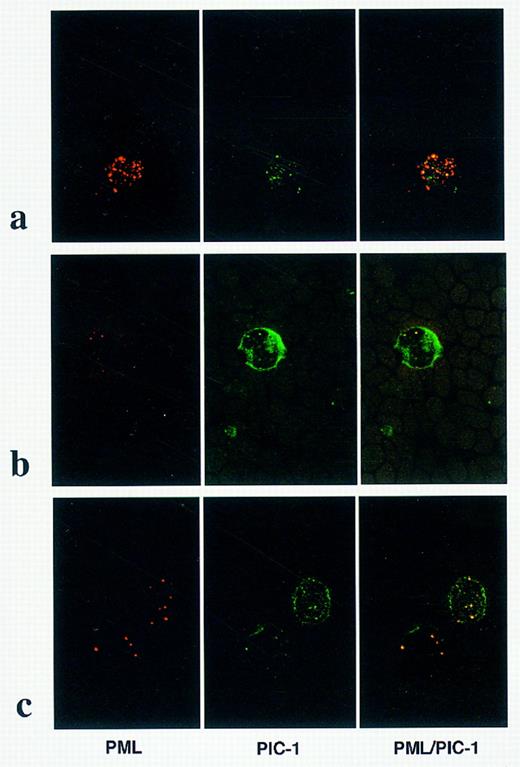
Dual immunofluorescence studies were subsequently undertaken using a PML monoclonal antibody and polyclonal antiserum directed against PIC 1, a newly described constituent of PML nuclear bodies. In 3 non-APL AML cases, PML and PIC 1 were colocalized within discrete nuclear bodies as shown in Fig 5b; as distinct from the pattern detected in a patient with a cryptic PML-RARα rearrangement (case no. 5) in which microparticulate PML staining was observed, with only partial colocalization with PIC 1 (Fig 5a). In the patient with t(11; 17)-associated APL, PML and PIC 1 were colocalized within discrete nuclear bodies (Fig 5c), as observed in non-APL cases. In addition to discrete punctate nuclear staining, PIC 1 was also localized to the nuclear membrane, as clearly shown in Fig 5b and c.
PML and PIC 1 localization in AML. Dual immunofluorescence using PML monoclonal (left-hand panel) and PIC 1 polyclonal (center panel) antibodies. Images are fused in the right-hand panel; yellow signal denotes regions of PML/PIC 1 colocalization. (a) In APL cases associated with cryptic PML-RARα rearrangements, a microparticulate pattern of PML nuclear staining was associated with partial colocalization with PIC 1. In non-APL cases, eg, AML M4, shown in (b) and in t(11; 17)-associated APL (case no. 7; Table 1), shown in (c), PIC 1 was localized to the nuclear membrane and colocalized with PML within discrete nuclear bodies.
PML and PIC 1 localization in AML. Dual immunofluorescence using PML monoclonal (left-hand panel) and PIC 1 polyclonal (center panel) antibodies. Images are fused in the right-hand panel; yellow signal denotes regions of PML/PIC 1 colocalization. (a) In APL cases associated with cryptic PML-RARα rearrangements, a microparticulate pattern of PML nuclear staining was associated with partial colocalization with PIC 1. In non-APL cases, eg, AML M4, shown in (b) and in t(11; 17)-associated APL (case no. 7; Table 1), shown in (c), PIC 1 was localized to the nuclear membrane and colocalized with PML within discrete nuclear bodies.
DISCUSSION
Early studies in specialist cytogenetic centers reported that the t(15; 17) could be detected in all cases of APL.42 In the light of such claims, clinicians encountering AML cases with morphologic features of APL could lose faith in the initial clinical diagnosis if subsequent cytogenetic assessment failed to provide appropriate confirmatory evidence. However, since the characterization of the PML-RARα rearrangement that underlies the t(15; 17) and identification of the rare variant translocations whereby RARα is fused to partners other than PML, it is clear that absence of the t(15; 17) does not preclude a morphologic diagnosis of APL, although it remains the diagnostic hallmark of the disease. Large multicenter studies such as the UK MRC ATRA trial afford the opportunity to determine the frequency of cryptic rearrangements and variant translocations among patients with suspected APL. In this regard, in only 87% of APL patients with molecular evidence of a PML-RARα rearrangement was the t(15; 17) detectable by conventional cytogenetics. In most cases, absence of the t(15; 17) was a reflection of failed cytogenetics; however, 2% of cases were due to cryptic PML-RARα rearrangements.5
In the present study, we have characterized a series of 7 patients with morphologic APL found to lack the t(15; 17) on successful conventional cytogenetic assessment. In 6 patients, of whom 5 had morphologically normal chromosomes 15 and 17, the diagnosis was confirmed by the presence of a PML-RARα rearrangement; in each case, PML-RARα transcripts were detected by RT-PCR in the absence of the reciprocal derived RARα-PML species. FISH analyses in 4 such patients with evaluable metaphase spreads confirmed the presence of a nonreciprocal translocation and in each case were consistent with formation of PML-RARα as the sole fusion gene as a result of an interstitial insertion event, most commonly due to insertion of RARα into PML on 15q. This phenomenon was observed in 3 cases and has been the subject of two previous case reports in which chromosomes 15 and 17 also appeared normal by conventional cytogenetics.6 44 In the remaining patient with evaluable metaphases in the present study, the PML-RARα fusion was found to result from insertion of PML into RARα on 17q, which has not been previously described. Future characterization of the genomic breakpoints of these cases may provide insights not only into mechanisms mediating interstitial insertions but also into mechanisms underlying the more typical classical reciprocal translocation.
Demonstration of PML-RARα as the sole fusion gene formed in each of the APL cases with cryptic PML-RARα rearrangements in this study is consistent with the proposed role of its gene product as a critical mediator of leukemogenesis.2,43,51 This has recently been confirmed in a transgenic model3,4 whereby expression of PML-RARα was associated with impairment of normal myeloid differentiation accompanied by accumulation of primitive precursors and predisposition to the development of an APL-like syndrome responsive to ATRA. Although the presence of a latent period before developing the leukemia argues in favor of a requirement for additional mutational events, as has been suggested in theoretical models of tumorigenesis,54 it could also imply that high-level expression of the transgene did not occur within equivalent progenitors to those forming the targets of leukemic transformation in human APL. Although PML-RARα is clearly established as playing a key role in leukemogenesis and determining the differentiation response to ATRA, any role for RARα-PML in the pathogenesis of the disease remains to be determined. Although a single case of APL in which a RARα-PML fusion gene was apparently formed in isolation has been reported,6 the present study demonstrating a series of cases in which PML-RARα was the sole fusion gene formed argues against a significant role for RARα-PML in the pathogenesis of APL. Furthermore, this study, when considered in conjunction with previous series establishing that RARα-PML transcripts are not detectable in approximately 20% APL patients,5 shows that, at least in a proportion of cases, absence of RARα-PML transcripts reflects the occurrence of PML-RARα rearrangements due to interstitial insertion events rather than the classic reciprocal translocation.
In 1 patient in the present study, APL was associated with t(11; 17)(q23; q12-21), leading to a PLZF-RARα rearrangement. In common with the index case,53 both PLZF-RARα and RARα-PLZF transcripts could be detected by RT-PCR, consistent with the concept that, in contrast to t(15; 17)-mediated APL, both fusion partners are implicated in leukemogenesis.41,55 Whereas PLZF bears no structural similarities to PML, being characterized by an amino-terminal POZ motif, and 9 carboxy-terminal zinc finger domains implicated in its role as a transcription factor41,53,55-58; the two proteins and their respective fusion products share a number of common features. In particular, PLZF is localized to discrete nuclear bodies56,58 whose formation is dependent on the integrity of the POZ domain57 and exhibits growth-suppressor activity in transformation assays.48,59 Furthermore, in common with PML-RARα, PLZF-RARα may sequester RXR and binds retinoid response elements inhibiting transactivation.56,57,60 These processes involving disrupted growth-suppressor function coupled with deregulation of retinoid signaling pathways appear to provide a final common pathway for the pathogenesis of APL. However, in marked contrast to the disease associated with PML-RARα rearrangements, in vitro differentiation assays have shown that blasts derived from t(11; 17)(q23; q21) APL cases are resistant to retinoids.41 This may be accounted for by a number of factors, including differing repertoire and character of response between the respective fusion proteins in the presence of ligand,57,60,61 possibly compounded by the absence of ATRA-induced degradation of PLZF-RARα34 and by upregulation of RARα-PLZF leading to persistent deregulation of the cell cycle.41,55 59
Recent studies suggest that there is at least partial colocalization of PML and PLZF within the nucleus,61,62 raising the possibilities that their growth-suppressor activities might be interrelated and that fusion proteins associated with variant APL translocations could promote leukemogenesis by disruption of PML nuclear bodies. Therefore, in the context of such a model, one might expect ATRA-resistant t(11; 17) cases to maintain a disrupted pattern of PML nuclear bodies, reminiscent of that reported in retinoid-resistant PML-RARα–mediated cases.19,35 However, the present study refutes such a hypothesis; disruption of PML nuclear bodies was only observed in APL cases associated with cryptic PML-RARα rearrangements in which PML nuclear staining patterns were identical to t(15; 17)-positive cases, whereas a wild-type pattern of PML nuclear staining was detected in blasts derived from t(11; 17)(q23; q21)-associated APL, reminiscent of that observed in other subtypes of AML. Furthermore, using dual immunofluorescence techniques, we were able to show that PIC 1 (GMP 1,63 SUMO 164), a novel-ubiquitin homology domain protein identified as an interaction partner of PML in a yeast-two hybrid screen,24 interacts with PML in vivo in AML blasts as well as in nonhemopoietic cell lines, as previously described.24 In t(11; 17)-associated APL and in non-APL cases, in addition to perfect colocalization with PML within discrete nuclear bodies, a perinuclear pattern of PIC 1 staining was observed consistent with its reported interaction with RanGap1, targeting it to the nuclear pore complex.63,64 In contrast, in APL cases associated with cryptic PML-RARα rearrangements, only partial colocalization of PML and PIC 1 was observed, confirming results previously obtained with the APL cell-line NB4,24 reflecting disruption of PML nuclear bodies in the presence of the PML-RARα fusion protein. Recent work has established that the second heptad repeat of the coiled-coil domain of PML-RARα is critical for nuclear body disruption, which does not appear to be necessary either for the block in differentiation or its reversal by retinoids; both of these effects are dependent on the integrity of the first heptad repeat of the coiled-coil region.29 Whether this perturbation of nuclear architecture is involved in the process of leukemic transformation is still undetermined. It remains a possibility that disruption of the nuclear bodies is merely a secondary phenomenon reflecting an interaction between the PML-RARα fusion protein and wild-type PML and is of no importance to leukemogenesis. Nevertheless, the present study underlines the fact that disruption of PML nuclear bodies provides a valuable marker for the PML-RARα fusion protein in cases lacking the t(15; 17) and confirms PML immunofluorescence as a suitable technique for rapid identification of the subgroup of APL patients likely to benefit from retinoids.5 65 In conclusion, although deregulation of the putative growth-suppressor PML and delocalization of other nuclear body constituents have been advocated to play a key role in the development of t(15; 17)-associated APL, the present study shows that disruption of PML nuclear bodies per se is not a prerequisite for the pathogenesis of APL.
NOTE ADDED IN PROOF
A wild-type PML nuclear localization pattern has also recently been reported in an APL case with the variant translocation t(11;17)(q13;q21) leading to a NUMA/RARα rearrangement.47PIC 1 has been designated UBL 1 by the Gene Nomenclature Committee.
ACKNOWLEDGMENT
The authors are grateful to all the clinicians who entered patients into the MRC ATRA trial and forwarded material for molecular and cytogenetic analyses, particularly Dr Steve Kelsey. We thank Steve Chatters and Joanne Rogers in the Cytogenetics Laboratory at University College Hospital, London, and the cytogeneticists involved in karyotyping these patients who provided material for FISH analyses, particularly Debra Lillington, Michael Neat, and David Stevenson. We are grateful to Peter Jordan for assistance with immunofluorescence studies, to the photographic department at ICRF, Lincoln's Inn Fields, and to Iain Goldsmith and the oligonucleotide synthesis service at ICRF, Clare Hall. We also thank Hans Nicolai, Dr Melissa Brown, Dr Aurélie Catteau, and Dr Chun-Fang Xu for helpful discussions and assistance with sequence analyses.
D.G. was supported by an MRC clinical training fellowship. E.S. and K.H. were supported by EEC grants BIOMED-CT92-0755 and Biotech BI02-CT-930450. E.D. was supported by an EC TMR fellowship. D.G. and K.H. are currently supported by ICRF. S.L. and DNA/RNA banking facilities at University College Hospital, London are currently supported by the Kay Kendall Leukaemia Fund.
Address reprint requests to Dr David Grimwade, Cancer Genetics Laboratory, UMDS, 8th Floor, Guy's Tower, Guy's Hospital, London SE1 9RT, UK.


This feature is available to Subscribers Only
Sign In or Create an Account Close Modal