Abstract
The murine double minute 2 (MDM2) protein facilitates G1 to S phase transition by activation of E2F-1 and can enhance cell survival by suppressing wild-type p53 (wtp53) function. In this study, we examined MDM2 expression and function in multiple myeloma (MM) cells. MDM2 is strongly and constitutively expressed in MM cell lines (ARH-77, RPMI 8226, and OCI-My5) and in the cells of plasma cell leukemia (PCL) patients, but is not expressed in normal bone marrow mononuclear cells (BM MNCs). Treatment of MM cells with MDM2 antisense, but not sense, nonsense, or scrambled, oligodeoxyribonucleotides (ODNs) decreased DNA synthesis and cell viability; it also induced G1 growth arrest, as evidenced by propidium iodide (PI) staining and induction of retinoblastoma protein (pRB) to E2F-1 binding. Moreover, inhibition of MDM2 using antisense ODNs also triggered MM cell apoptosis as evidenced by acridine orange–ethidium bromide staining. We next studied the association of MDM2 with wtp53 and/or mutant p53 (mtp53), E2F-1, CDK4, and p21. MDM2 constitutively binds to E2F-1 in all MM cells, to both wtp53 and mtp53, and to p21 in tumor cells lacking p53. These data suggest that MDM2 may enhance cell-cycle progression in MM cells both by activating E2F-1 and by downregulating cell-cycle inhibitory proteins (wtp53 and p21). Overexpression of MDM2 may therefore contribute to both growth and survival of MM cells, suggesting the potential utility of treatment strategies targeting MDM2 in MM.
DOUBLE MINUTES (DMs) are small, acentromeric, extrachromosomal nuclear bodies.1-3 Amplified DNA sequences in the form of such DMs were previously found in a spontaneously transforming, highly tumorigenic derivative of the murine Balb/c cell line,4,5 so-called 3T3DM cells. DMs can be stably maintained in these cells during long-term growth both in vitro and in vivo, suggesting the ability of one or more genes to confer some growth advantage.6 Studies involving three of these murine DM (mdm) genes (mdm1, mdm2, and mdm3) showed that mdm2-bearing 3T3DM cells maintained tumorigenicity when introduced subcutaneously into athymic nude mice, whereas mdm1- and mdm3-bearing cells failed to generate any tumors.5,6 The non-DM chromosomal mdm2 gene is found on murine chromosome 10, region C1-3,5 whereas MDM2, its human homologue, is located at the 12q13-14 locus of human chromosome 12.7,8 Since MDM2 possesses an acidic domain and zinc-finger motifs, it may function to transit cells from the G1 to S phase of the cell cycle by direct DNA binding to the promoters of genes activated upon entry into the cell cycle.9,10 It can also facilitate the shift into S phase by activating E2F-1.11 E2F-1 binds to dephosphorylated retinoblastoma protein (pRB), and the complex directly binds DNA to induce G1 growth arrest12; conversely, activation of E2F-1 triggers dissociation of E2F-1–pRB complexes, thereby removing the block from the G1 to S phase. Finally, MDM2 also inhibits wild-type p53 (wtp53)-mediated apoptosis.13 Therefore, MDM2 can function both to induce cell proliferation and to enhance cell survival.
Murine DM2 (MDM2) protein and/or MDM2 mRNA are overexpressed in a variety of neoplasms, including acute leukemias,14-17 myelodysplastic syndrome,14 chronic lymphocytic leukemia and lymphomas (including Sézary syndrome),18-20 multiple myelomas (MMs),15 sarcomas,7,21-23 thyroid carcinomas,24 breast carcinomas,25-28 non–small-cell lung carcinomas (NSCLCs),29 Lewis lung carcinoma,30 a pancreatic carcinoid tumor,31 glioblastomas,32 cervical cancer,33 transitional cell bladder carcinomas,34,35 testicular germ cell tumors,36 and choriocarcinomas.37 Whereas MDM2 gene amplifications among overexpressing neoplasms are frequent in bony and soft-tissue sarcomas,7,11,13,38,39 breast carcinomas,23,25 NSCLC,26 and testicular germ cell tumors,33 they are rare in human leukemias and lymphomas,14,15,17,36,40 bladder carcinoma cell lines,32 and thyroid carcinomas.21 In addition to gene amplifications, enhanced mRNA transcription, prolonged protein half-life, and enhanced translation34 can also lead to MDM2 overexpression. Finally, overexpression of MDM2 is correlated with advanced disease,18,30 higher-grade neoplasms,19 progressive disease,13 poor response to chemotherapy,15 poor prognosis and/or survival,12,15 and, in the case of breast carcinoma, estrogen receptor immunoreactivity.15,22 23 However, to date, the functional significance of MDM2 overexpression in human cancers is not well understood.
In the present study, we characterized the expression and function of MDM2 in MM cell lines and plasma cell leukemia (PCL) patients. We demonstrate that MDM2 protein is overexpressed in these tumor cells. Our studies using antisense oligodeoxyribonucleotide (ODN) to specifically inhibit MDM2 function in MM cells show that MDM2 promotes both entry into the cell cycle and tumor cell survival, and suggest the utility of treatments targeting MDM2 in MM.
MATERIALS AND METHODS
MM cell lines and PCL patient samples.The human MM-derived cell lines used were ARH-77 (American Type Culture Collection [ATCC] CRL-1621, Rockville, MD), RPMI 8226 (ATCC CCL-155), U266 (ATCC TIB-196), and OCI-My5 (a gift from Dr H.A. Messner, Ontario Cancer Institute, Toronto, Canada). ARH-77 and RPMI-8226 MM cells were cultured in RPMI 1640 with L-glutamine medium (Cellgro; GIBCO-BRL, Gaithersburg, MD) supplemented with 10% heat-treated fetal bovine serum (FBS) (PAA Laboratories Inc, Newport Beach, CA), 25 IU/mL penicillin and 25 mg/mL streptomycin (Pen-Strep; GIBCO-BRL), and 5 mmol/L L-glutamine (GIBCO-BRL). U266 MM cells were cultured in RPMI-1640 with L-glutamine medium supplemented with 15% FBS, Pen-Strep, and 5 mmol/L L-glutamine. OCI-My5 MM cells were cultured in Iscove's modified Dulbecco's medium (Sigma Diagnostics, St Louis, MO) supplemented with 10% FBS, Pen-Strep, and 5 mmol/L L-glutamine. Saos-2 (ATCC HTB-85), a human osteosarcoma cell line without detectable p53 protein, was cultured in McCoy's 5A (modified) medium with L-glutamine (GIBCO-BRL) supplemented with 15% FBS, Pen-Strep, and 5 mmol/L L-glutamine. All cell lines were grown at 37°C in a humidified 5% CO2 atmosphere.
Mononuclear cells (MNCs) were obtained by Ficoll-Paque (Pharmacia Biotech, Uppsala, Sweden) separation from peripheral blood samples of two patients with PCL (PCL1 and PCL2). The MNC fraction in both PCL1 and PCL2 included more than 95% CD38+ CD45RA− tumor cells. Bone marrow (BM) MNCs were similarly obtained from freshly donated BM from normal donors after provision of appropriate informed consent.
Increasing wtp53 protein expression in BM MNCs by γ-irradiation.Normal BM MNCs were divided into three fractions: the first fraction was not irradiated; the second was lysed after 1 hour γ-irradiation (XRT) exposure (2.7 Gy); and the third after 4 hour XRT (2.7 Gy). Increased wtp53 protein expression was confirmed by Western blotting (WB) (description follows).
Antibodies.The following primary monoclonal antibodies (MoAbs) were used: SMP14 anti-MDM2 MoAb recognizing amino acid residues 154 to 167 of human MDM2, DO-1 horseradish peroxidase (hrp)-conjugated anti-p53 MoAb recognizing pantropic p53 (ptp53), and C-20 anti–E2F-1 MoAb (all Santa Cruz Biotechnology Inc, Santa Cruz, CA); and Ab-5 anti-p53 MoAb recognizing wtp53, Ab-3 anti-p53 MoAb recognizing mutant p53 (mtp53), Ab-6 anti-p53 MoAb recognizing ptp53, Ab-1 C36 anti-pRB MoAb, Ab-6 AF11 anti-pRB MoAb, Ab-1 anti-p21WAF1 MoAb, and AB-1 anti-Actin MoAb (all from Oncogene Science Inc, Cambridge, MA). The following primary polyclonal antibodies (pAbs) were used: H-22 anti-CDK4 pAb and C-19 anti-p21WAF1/CIP1 pAb (both Santa Cruz Biotechnology Inc). Hrp-conjugated sheep anti-mouse Ig MoAb and hrp-conjugated donkey anti-rabbit Ig MoAb (both Amersham Life Science, Buckinghamshire, UK) were used as secondary MoAbs.
Western blotting and immunoprecipitation analysis.Cells were grown to 70% to 80% confluency, harvested, and pelleted by centrifugation at 3,000g for 5 minutes at room temperature. Each cell pellet was washed thrice with 50 mL phosphate-buffered saline (PBS) (Sigma Diagnostics) for MM cell lines or Tris-buffered saline (TBS) for PCL cells, and then lysed on ice for 30 minutes in lysis buffer (containing 50 mmol/L Tris, pH 8.0, 150 mmol/L NaCl, 0.1% Nonidet P-40, 1.0 mmol/L EDTA, 50 mmol/L sodium fluoride, 1.0 mmol/L sodium orthovanadate, 2.0 μg/mL aprotinin, 2.0 μg/mL leupeptin, and 5.0 μg/mL phenylmethylsulfonyl fluoride) with frequent vortexing. Cell lysates were centrifuged at 6,000 × g for 15 minutes at 4°C, and the supernatants were divided into 1-mL aliquots and transferred to 1.5-mL Eppendorf tubes. Each aliquot was precleared by incubating with 1 μL rabbit anti-mouse ascites fluid and 100 μL 10% (vol/vol) Protein-A Sepharose CL-4B beads (Pharmacia Biotech) for 1 hour at 4°C with continuous rocking. The Sepharose beads were then precipitated by centrifugation at 10,000g for 30 seconds, and the supernatants were collected and assayed for total protein content by Bradford's microtiter plate method (Biorad, Hercules, CA).
For Western blotting, 100 μg whole cell lysate was equally loaded into each well of a sodium dodecyl sulfate (SDS)-polyacrylamide gel. For immunoprecipitation, 500 μg whole cell lysate was first incubated with 1.0 μg of the antibody of interest for 1 hour at 4°C with continuous rocking. Thereafter, 100 μL 10% (vol/vol) Protein-A Sepharose CL-4B beads were added and incubated overnight at 4°C with continuous rocking. The Sepharose beads were precipitated the next day by centrifugation at 10,000g for 30 seconds and the supernatant was discarded. The beads were washed thrice with lysis buffer and pelleted; the final supernatant was discarded. The remaining lysis buffer was removed by aspiration, and the pellet was resuspended in 25 μL 1 × sample buffer (containing 2% SDS, 60 mmol/L Tris, pH 6.8, 10% 2-mercaptoethanol, 20% glycerol, and 0.001% bromophenol blue). After a brief vortex mixing, the sample was incubated in a water bath at 85°C for 10 minutes, and then vortexed for 30 seconds and centrifuged at 16,000g for 15 minutes. Twenty-five microliters of the immunoprecipitated sample was equally loaded into each well of 6% (pRB), 8% (MDM2, E2F-1, and p53), 10% (CDK4), and 12% (p21) SDS-polyacrylamide gels.
After both whole cell lysates and immunoprecipitated samples were separated by SDS-polyacrylamide gel electrophoresis (SDS-PAGE), proteins were transferred onto polyvinylidine difluoride (PVDF ) membranes (Immobilon-P; Millipore, Bedford, MA) by a semidry method. The membranes were next blocked by incubating overnight at room temperature with continuous rocking in blotto (containing 5% skim milk [Mix & Drink; Saco Foods Inc, Middleton, WI], 0.2% Tween-20 (Sigma Diagnostics), and 0.02% sodium azide) and then washed in Tris-buffered saline with Tween-20 (TBST) (containing 20 mmol/L Tris, pH 7.6, 150 mmol/L NaCl, and 0.05% Tween-20) for 45 minutes with continuous rocking and frequent changes of TBST. Membranes were next incubated for 1 hour at room temperature with the appropriate primary Abs diluted in TBST to a final concentration of 1 μg/mL. Washing was then performed as already described, and when required, membranes were incubated for 45 minutes at room temperature with secondary hrp-conjugated antibodies diluted 1:1,000 in TBST. After the final wash, the hrp chemiluminescence detection was performed as directed by the manufacturer (ECL, Amersham Life Science), followed by exposure to photographic film (Biomax MR; Eastman Kodak Co, Rochester, NY) for 5 to 60 seconds.
MDM2 antisense, sense, and nonsense ODN assays.RPMI 8226 MM cells were cultured in 96-well tissue culture plates at an initial cell density of 1.7 × 106 cells/mL for cell proliferation and cell density analyses, and in 24-well tissue culture plates at an initial cell density of 0.5 × 106/mL for cell-cycle and apoptosis analyses. Twenty-molar HEPES buffer (Intergen, Purchase, NY) was added to wells receiving ODNs; control cells were cultured in media without addition of ODNs. The MDM2 ODN sequences were as follows: 15-base antisense ODN (A15) (5′-dGACATGTTGGTATTG-3′); 20-base antisense ODN (A20) (5′-dGACATGTTGGTATTGCACAT-3′)41; 15-base sense ODN (S15) (5′-dATGTGCAATACCAAC-3′); 20-base sense ODN (S20) (5′dATGTGCAATACCAACATGTC-3′); 15-base nonsense ODN (N15) (5′-dGTGTGTGTGTGTGTG-3′); 20-base nonsense ODN (N20) (5′-dGTGTGTGTGTGTGTGTGTGT-3′); 15-base scrambled ODN (X15) (5′-dGTTGATCACTAGTTG-3′); and 20-base scrambled ODN (X20) (5′-dTGATAGTCGTATGCTGATAC-3′). ODNs were used at a concentration of 0.1, 1.0, and 2.0 μmol/L to assay for effects on the kinetics of proliferation; a cell-cycle profile, apoptosis assay, and immunoblotting were performed in cells cultured with ODNs at 1.0 μmol/L.
For cell proliferation analyses, cells were cultured for periods of up to 24 hours with each ODN (antisense [A15 and A20], sense [S15 and S20], nonsense [N15 and N20], and scrambled [X15 and X20]), and DNA synthesis was assayed by tritiated (3H) thymidine (TdR) uptake. In some cultures, ODNs were replenished every 4 hours. Briefly, 0.5 μCI 3H-TdR (Dupont NEN, Boston, MA) was added to each culture well and incubated for 4 hours before harvesting onto glass filters using the HARVESTAR 96 MACH II (Tomtec Inc, Orange, CT) and counting on the 1205 BETAPLATE liquid scintillation counter (Wallac, Gaithersburg, MD). Cell density was simultaneously determined from an aliquot (10 μL) of each well by trypan blue (GIBCO-BRL) exclusion.
For cell-cycle and apoptosis analyses, cells were similarly cultured for periods of up to 24 hours with ODN, washed thrice in ice-cold PBS, and pelleted. The cell pellet was resuspended in 0.5 mL ice-cold propidium iodide ([PI] 15 μg/mL; Sigma Diagnostics) in 0.1% sodium citrate and 0.1% Nonidet P-40, and incubated for 30 minutes at 4°C. The cell-cycle distribution of these cells was analyzed by flow cytometry (Coulter Corp, Miami, FL). The percentage of apoptotic cells was simultaneously determined from an aliquot (10 μL) of each well obtained before PI staining by acridine orange (100 μg/mL) and ethidium bromide (100 μg/mL) (both Sigma Diagnostics) staining. Apoptotic cells were enumerated by fluorescence microscopy (Microstar IV; Reichert-Jung, Buffalo, NY) at 490 nm excitation wavelength; the mean percentage of apoptotic cells was determined for three samples (200 cells per sample).
MDM2 half-life determination by 35S-methionine pulse chase.RPMI 8226 MM cells were cultured for 4 hours at a cell density of 1.0 × 106/mL in methionine-free RPMI-1640 with L-glutamine medium supplemented with 10% dialyzed FBS, followed by culturing for 2 hours in the same medium supplemented with 50 μCi/mL 35S-methionine. Cells were next cultured in RPMI-1640 with L-glutamine medium supplemented with 10% FBS, and samples were withdrawn at 0, 10, 15, 20, 25, 30, 40, 50, and 60 minutes. Expression of labeled MDM2 was determined by immunoprecipitating cell lysates with 1.0 μg SMP14 anti-MDM2 MoAb followed by SDS-PAGE on an 8% gel, transfer to a PVDF membrane, and autoradiography over 10 days without an image-intensifying screen.
RESULTS
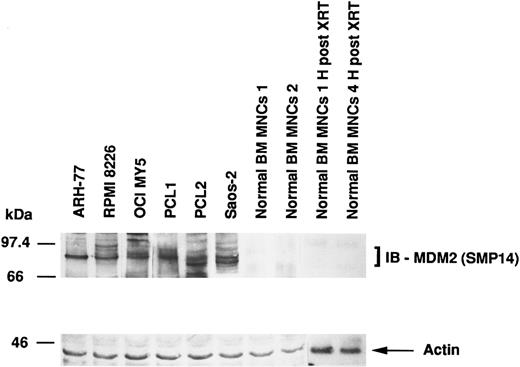
Expression of MDM2 protein in MM-derived cell lines, cells from PCL patients, Saos-2 osteosarcoma cells, and normal BM MNCs.MDM2 protein expression in MM cell lines and cells from PCL patients was compared with the expression in p53-deficient Saos-2 osteosarcoma cells and normal BM MNCs. Figure 1 shows that MM-derived cell lines and PCL cells, but not normal BM MNCs, constitutively express MDM2 protein. Saos-2 osteosarcoma cells, which strongly express MDM2, served as positive controls. In addition, we also examined γ-irradiated BM MNCs, since x-ray therapy (XRT) has been reported to increase MDM2 expression.42 However, we were unable to increase MDM2 expression in normal BM MNCs. Immunoblotting with antiactin MoAb confirms equal protein loading.
Expression of MDM2 protein in MM-derived cell lines. PCL patients cells, Saos-2 osteosarcoma cells, and normal BM MNCs. Total cell lysates of MM cell lines (ARH-77, RPMI 8226, and OCI-My5), cells from PCL patients (PCL1 and PCL2), Saos-2 p53-deficient osteosarcoma cells, and normal BM MNCs (without and 1 and 4 hours post-XRT) were immunoprecipitated with SMP14 anti-MDM2 MoAb followed by immunoblotting with the same Ab. Immunoprecipitation and immunoblotting with AB-1 anti-actin MoAb confirmed equal protein loading.
Expression of MDM2 protein in MM-derived cell lines. PCL patients cells, Saos-2 osteosarcoma cells, and normal BM MNCs. Total cell lysates of MM cell lines (ARH-77, RPMI 8226, and OCI-My5), cells from PCL patients (PCL1 and PCL2), Saos-2 p53-deficient osteosarcoma cells, and normal BM MNCs (without and 1 and 4 hours post-XRT) were immunoprecipitated with SMP14 anti-MDM2 MoAb followed by immunoblotting with the same Ab. Immunoprecipitation and immunoblotting with AB-1 anti-actin MoAb confirmed equal protein loading.
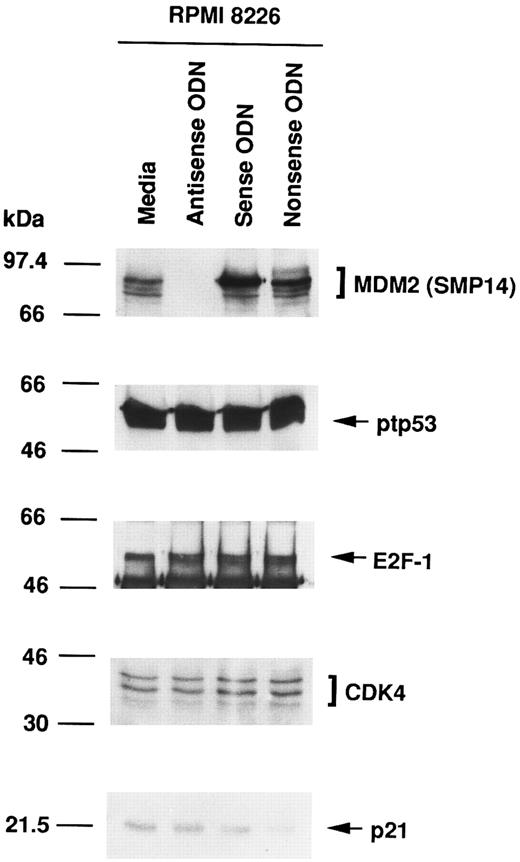
Effect of culture with MDM2 antisense ODN on expression of MDM2, p53, E2F-1, CDK4, and p21 proteins in RPMI 8226 MM cells.To assess the function of MDM2 in MM cells, we next attempted to block MDM2 expression using specific antisense ODN (A20). Figure 2 shows that MDM2 protein expression in RPMI 8226 cells was inhibited in cells cultured with MDM2 antisense ODN, but not in cultures with media, MDM2 sense ODN (S20), or MDM2 nonsense ODN (N20). In contrast, no significant changes were evident in the constitutive expression of p53, E2F-1, CDK4, and p21 in RPMI 8226 cells treated with MDM2 antisense ODN compared with cells cultured with media, MDM2 sense ODN, or MDM2 nonsense ODN, confirming the specificity of MDM2 antisense ODN action.
Effect of culture with MDM2 antisense ODN on expression of MDM2, p53, E2F-1, CDK4, and p21 proteins in RPMI 8226 MM cells. RPMI 8226 MM cells were cultured in media alone or with MDM2 antisense, sense, or nonsense ODNs for 4 hours. Total cell lysates were immunoprecipitated and immunoblotted with SMP14 anti-MDM2 MoAb, AB-6 anti-ptp53 MoAb, C-20 anti–E2F-1 MoAb, H-22 anti-CDK4 pAb, and C-19 anti-p21 pAb.
Effect of culture with MDM2 antisense ODN on expression of MDM2, p53, E2F-1, CDK4, and p21 proteins in RPMI 8226 MM cells. RPMI 8226 MM cells were cultured in media alone or with MDM2 antisense, sense, or nonsense ODNs for 4 hours. Total cell lysates were immunoprecipitated and immunoblotted with SMP14 anti-MDM2 MoAb, AB-6 anti-ptp53 MoAb, C-20 anti–E2F-1 MoAb, H-22 anti-CDK4 pAb, and C-19 anti-p21 pAb.
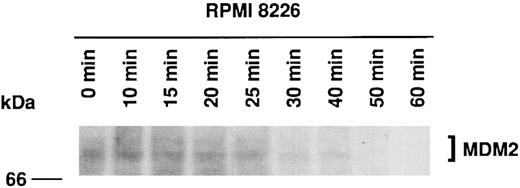
Half-life of MDM2 in RPMI 8226 MM cells.Three previous reports have indicated that MDM2 has a half-life of 15 to 30 minutes in rat embryo fibroblasts,13 3T3DM cells,43 and choriocarcinoma cells (JAR and JEG-3).37 We next determined the half-life of MDM2 in RPMI 8226 MM cells using 35S-methionine pulse-chase experiments. Figure 3 shows that MDM2 expression is markedly reduced at 30 minutes, suggesting that the half-life of MDM2 in RPMI 8226 cells is approximately 25 minutes.
Half-life of MDM2 in RPMI 8226 MM cells. RPMI 8226 MM cells (1.0 × 106 cells/mL) were cultured for 4 hours in methionine-free RPMI-1640 with L-glutamine medium supplemented with 10% dialyzed FBS, followed by culturing for 2 hours in the same medium supplemented with 50 μCi/mL 35S-methionine. Cells were next cultured in RPMI 1640 with L-glutamine medium supplemented with 10% FBS, and samples of equal volume were withdrawn at 0, 10, 15, 20, 25, 30, 40, 50, and 60 minutes. Expression of labeled MDM2 was determined by immunoprecipitation of cell lysates with 1.0 μg SMP14 anti-MDM2 MoAb followed by SDS-PAGE on an 8% gel, transfer to a PVDF membrane, and autoradiography over 10 days without an image-intensifying screen. The half-life of MDM2 was 25 minutes.
Half-life of MDM2 in RPMI 8226 MM cells. RPMI 8226 MM cells (1.0 × 106 cells/mL) were cultured for 4 hours in methionine-free RPMI-1640 with L-glutamine medium supplemented with 10% dialyzed FBS, followed by culturing for 2 hours in the same medium supplemented with 50 μCi/mL 35S-methionine. Cells were next cultured in RPMI 1640 with L-glutamine medium supplemented with 10% FBS, and samples of equal volume were withdrawn at 0, 10, 15, 20, 25, 30, 40, 50, and 60 minutes. Expression of labeled MDM2 was determined by immunoprecipitation of cell lysates with 1.0 μg SMP14 anti-MDM2 MoAb followed by SDS-PAGE on an 8% gel, transfer to a PVDF membrane, and autoradiography over 10 days without an image-intensifying screen. The half-life of MDM2 was 25 minutes.
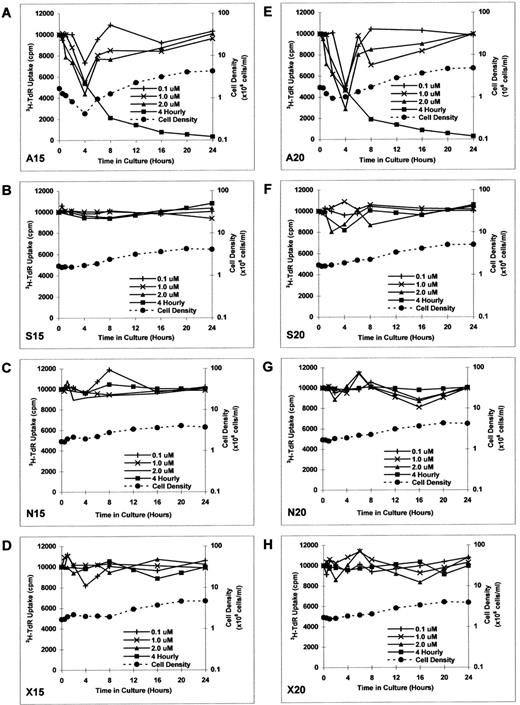
Effect of culture with MDM2 antisense ODN on proliferation in RPMI 8226 MM cells.To determine the kinetics of MDM2 antisense ODN on proliferation of RPMI 8226 MM cells, we constructed additional MDM2 antisense, sense, nonsense, and scrambled ODNs. RPMI 8226 MM cells were cultured in media with MDM2 antisense ODN (A15 and A20), MDM2 sense ODN (S15 and S20), MDM2 nonsense ODN (N15 and N20), or MDM2 scrambled ODN (X15 and X20) at concentrations of 0.1, 1.0, and 2.0 μmol/L for up to 24 hours without replenishment of ODNs. Both A15 and A20 MDM2 antisense ODNs maximally inhibited proliferation at 4 hours, with recovery thereafter to baseline levels of DNA synthesis (Fig 4A and E). Culture with S15, S20, N15, N20, X15, and X20 ODNs did not significantly affect proliferation (Fig 4B to D and F to H).
Effect of culture with MDM2 antisense ODN on proliferation in RPMI 8226 MM cells. RPMI 8226 MM cells were cultured in media with MDM2 antisense ODN (A15 and A20, A and E), MDM2 sense ODN (S15 and S20, B and F ), MDM2 nonsense ODN (N15 and N20, C and G), or MDM2 scrambled ODN (X15 and X20, D and H) at concentrations of 0.1, 1.0, and 2.0 μmol/L for up to 24 hours, without replenishment of ODNs. In addition, RPMI 8226 MM cells were cultured with 1.0 μmol/L of each ODN, with replenishment of ODN every 4 hours. Proliferation was measured by 3H-TdR uptake at 0, 0.5, 1, 2, 4, 6, 8, 12, 16, 20, and 24 hours. Viable cell density at 0, 0.5, 1, 2, 4, 6, 8, 16, and 24 hours was also determined by trypan blue exclusion for RPMI 8226 MM cells cultured in 1.0 μmol/L of each ODN, without replenishment of ODN.
Effect of culture with MDM2 antisense ODN on proliferation in RPMI 8226 MM cells. RPMI 8226 MM cells were cultured in media with MDM2 antisense ODN (A15 and A20, A and E), MDM2 sense ODN (S15 and S20, B and F ), MDM2 nonsense ODN (N15 and N20, C and G), or MDM2 scrambled ODN (X15 and X20, D and H) at concentrations of 0.1, 1.0, and 2.0 μmol/L for up to 24 hours, without replenishment of ODNs. In addition, RPMI 8226 MM cells were cultured with 1.0 μmol/L of each ODN, with replenishment of ODN every 4 hours. Proliferation was measured by 3H-TdR uptake at 0, 0.5, 1, 2, 4, 6, 8, 12, 16, 20, and 24 hours. Viable cell density at 0, 0.5, 1, 2, 4, 6, 8, 16, and 24 hours was also determined by trypan blue exclusion for RPMI 8226 MM cells cultured in 1.0 μmol/L of each ODN, without replenishment of ODN.
RPMI 8226 MM cells were also cultured with 1.0 μmol/L of each ODN, with replenishment of the respective ODNs every 4 hours. In this case, continued inhibition of proliferation was noted to 24 hours in cultures with A15 and A20 (Fig 4A and E); again, S15, S20, N15, N20, X15, and X20 ODNs did not similarly affect DNA synthesis (Fig 4B to D and F to H).
Viable cell density at 0, 0.5, 1, 2, 4, 6, 8, 16, and 24 hours was assessed by trypan blue exclusion of RPMI 8226 MM cells cultured with 1.0 μmol/L of each ODN (without ODN replenishment). MDM2 antisense ODNs (A15 and A20) maximally inhibited cell density at 2 to 4 hours (Fig 4A and E). In contrast, RPMI 8226 MM cells were not inhibited when cultured with S15, S20, N15, N20, X15, or X20 ODNs (Fig 4B to D and F to H); cell density in these cultures quadrupled over 24 hours, suggesting that the doubling time of these MM cells was approximately 12 hours. During the recovery phase of A15 and A20 ODN cultures, tumor cells demonstrated similar doubling times (Fig 4A and E), suggesting that recovery from transient growth inhibition with MDM2 antisense ODN was likely to be complete.
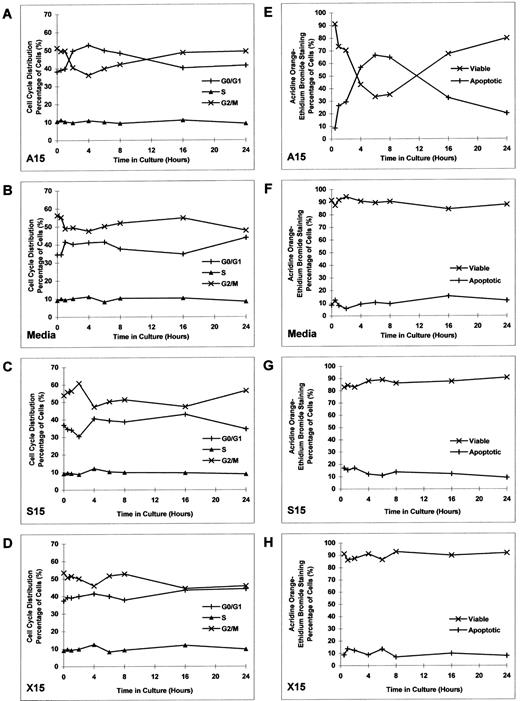
Effect of culture with MDM2 antisense ODN on cell-cycle distribution and apoptosis in RPMI 8226 MM cells.Having established the inhibitory effect of MDM2 antisense ODN on proliferation of RPMI 8226 MM cells, we next assayed the effects of MDM2 antisense ODN on cell-cycle distribution and apoptosis. RPMI 8226 cells were cultured with MDM2 antisense ODN (A15), media alone, MDM2 sense ODN (S15), or MDM2 scrambled ODN (X15) at concentrations of 1.0 μmol/L for up to 24 hours, without replenishment of ODNs. Cell-cycle distribution was defined by PI staining and flow cytometric analysis. Culture with A15 MDM2 antisense ODN triggered transient G1 growth arrest, manifested by an increase in G0/G1 and a corresponding decrease in G2/M (Fig 5A). No similar effects were observed in cells cultured with media, S15 MDM2 sense ODN, or X15 MDM2 scrambled ODN (Fig 5B to D).
Effect of culture with MDM2 antisense ODN on cell-cycle distribution and apoptosis in RPMI 8226 MM cells. RPMI 8226 MM cells were cultured with MDM2 antisense ODN (A15), media alone, MDM2 sense ODN (S15), or MDM2 scrambled ODN (X15) at concentrations of 1.0 μmol/L for up to 24 hours, without replenishment of ODNs. Cell-cycle distribution (A to D) was defined by PI staining and flow cytometric analysis. The percentage of apoptotic cells (E to H) was determined by acridine orange–ethidium bromide staining.
Effect of culture with MDM2 antisense ODN on cell-cycle distribution and apoptosis in RPMI 8226 MM cells. RPMI 8226 MM cells were cultured with MDM2 antisense ODN (A15), media alone, MDM2 sense ODN (S15), or MDM2 scrambled ODN (X15) at concentrations of 1.0 μmol/L for up to 24 hours, without replenishment of ODNs. Cell-cycle distribution (A to D) was defined by PI staining and flow cytometric analysis. The percentage of apoptotic cells (E to H) was determined by acridine orange–ethidium bromide staining.
Since we observed an increase in G0/G1 induced by A15 ODN, we next examined the percentage of apoptotic cells using acridine orange–ethidium bromide staining. RPMI 8226 cells were cultured with MDM2 antisense ODN (A15), media alone, MDM2 sense ODN (S15), or MDM2 scrambled ODN (X15) at concentrations of 1.0 μmol/L for up to 24 hours, without replenishment of ODNs. A15 MDM2 antisense ODN induced apoptosis of RPMI 8226 cells, which peaked at 6 to 8 hours (Fig 5E). In contrast, no similar changes were noted in cultures with media alone, S15 MDM2 sense ODN, or X15 MDM2 scrambled ODN (Fig 5F to H).
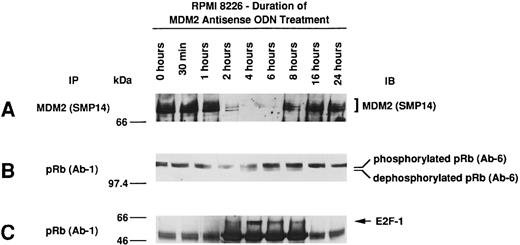
Effect of culture with MDM2 antisense ODN on expression of MDM2, phosphorylation of pRB, and pRB–E2F-1 binding in RPMI 8226 MM cells.Given the observed effects of MDM2 antisense ODN on proliferation, the cell-cycle profile, and apoptosis, we next determined the effect of MDM2 antisense ODN on MDM2 expression, pRB phosphorylation, and pRB–E2F-1 binding. Culture of RPMI 8226 MM cells with 1.0 μmol/L MDM2 antisense ODN (A15) maximally inhibited MDM2 expression at 4 to 6 hours, with recovery thereafter to baseline levels by 16 hours (Fig 6A). pRB is predominantly phosphorylated in RPMI 8226 cells; treatment with MDM2 antisense ODN induced dephosphorylation of pRB at 4 to 8 hours (Fig 6B), and maximal pRB binding to E2F-1 occurred at 2 to 8 hours (Fig 6C).
Effect of culture with MDM2 antisense ODN on expression of MDM2, phosphorylation of pRB, and pRB–E2F-1 binding in RPMI 8226 MM cells. RPMI 8226 MM cells were cultured with 1.0 μmol/L MDM2 antisense ODN (A15) for up to 24 hours. Cell lysates prepared before and at 0.5, 1, 2, 4, 6, 8, 16, and 24 hours were immunoprecipitated with SMP-14 anti-MDM2 MoAb and blotted with the same Ab (A). Cell lysates were immunoprecipitated with Ab-1 C36 anti-pRB MoAb, followed by immunoblotting with Ab-6 AF11 anti-pRB MoAb (B) or C-20 anti–E2F-1 MoAb (C).
Effect of culture with MDM2 antisense ODN on expression of MDM2, phosphorylation of pRB, and pRB–E2F-1 binding in RPMI 8226 MM cells. RPMI 8226 MM cells were cultured with 1.0 μmol/L MDM2 antisense ODN (A15) for up to 24 hours. Cell lysates prepared before and at 0.5, 1, 2, 4, 6, 8, 16, and 24 hours were immunoprecipitated with SMP-14 anti-MDM2 MoAb and blotted with the same Ab (A). Cell lysates were immunoprecipitated with Ab-1 C36 anti-pRB MoAb, followed by immunoblotting with Ab-6 AF11 anti-pRB MoAb (B) or C-20 anti–E2F-1 MoAb (C).
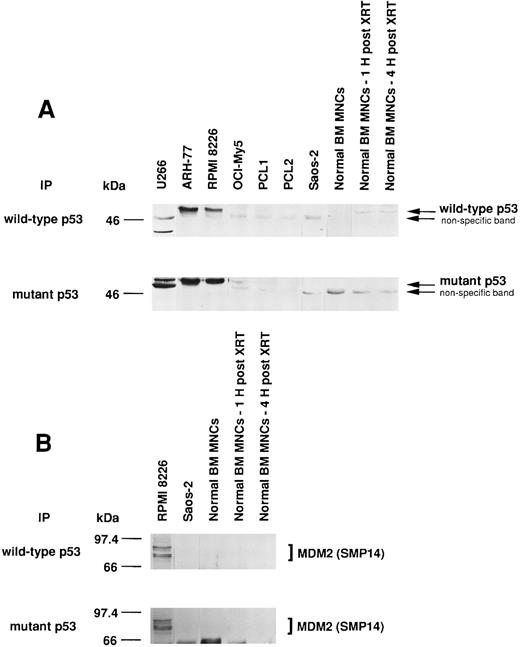
Expression of wtp53 and mtp53, as well as p53-MDM2 binding, in MM-derived cell lines, cells from PCL patient, Saos-2 osteosarcoma cells, and normal BM MNCs.Since MDM2 inhibits p53-mediated apoptosis, we next tested for wtp53 and mtp53 expression and p53 binding to MDM2 in MM cell lines, cells from PCL patients, Saos-2 p53-deficient osteosarcoma cells, and normal BM MNCs. We also examined γ-irradiated BM MNCs, since XRT has been reported to increase wtp53 protein expression.44 U266 MM cells express only mtp53, whereas ARH-77 and RPMI 8226 MM cells express both wtp53 and mtp53 protein (Fig 7A). OCI-My5 MM cells and cells from patient PCL1 weakly express both wtp53 and mtp53, and cells from patient PCL2 weakly express only mtp53. Normal BM MNCs weakly express wtp53 but not mtp53, and expression can be increased after 1 hour and 4 hours by XRT.
Expression of wtp53 and mtp53, as well as p53-MDM2 binding, in MM-derived cell lines, PCL patient cells, Saos-2 osteosarcoma cells, and normal BM MNCs. Total cell lysates of MM cell lines (U266, ARH-77, RPMI 8226, and OCI-My5), PCL patient cells (PCL1 and PCL2), Saos-2 p53-deficient osteosarcoma cells, and normal BM MNCs (without or 1 or 4 hours post-XRT) were immunoprecipitated with Ab-5 anti-wtp53 MoAb or Ab-3 anti-mtp53 MoAb and immunoblotted with DO-1 hrp anti-ptp53 MoAb. Normal BM MNCs served as a positive control for wtp53; U266 MM cells served as a positive control for mtp53; and p53-deficient Saos-2 cells served as a negative control for both wtp53 and mtp53 (A). Total cell lysates of RPMI 8226, Saos-2 osteosarcoma cells, and normal BM MNCs (without or 1 or 4 hours post-XRT) were immunoprecipitated with Ab-5 anti-wtp53 MoAb or Ab-3 anti-mtp53 MoAb, followed by immunoblotting with SMP14 anti-MDM2 MoAb (B).
Expression of wtp53 and mtp53, as well as p53-MDM2 binding, in MM-derived cell lines, PCL patient cells, Saos-2 osteosarcoma cells, and normal BM MNCs. Total cell lysates of MM cell lines (U266, ARH-77, RPMI 8226, and OCI-My5), PCL patient cells (PCL1 and PCL2), Saos-2 p53-deficient osteosarcoma cells, and normal BM MNCs (without or 1 or 4 hours post-XRT) were immunoprecipitated with Ab-5 anti-wtp53 MoAb or Ab-3 anti-mtp53 MoAb and immunoblotted with DO-1 hrp anti-ptp53 MoAb. Normal BM MNCs served as a positive control for wtp53; U266 MM cells served as a positive control for mtp53; and p53-deficient Saos-2 cells served as a negative control for both wtp53 and mtp53 (A). Total cell lysates of RPMI 8226, Saos-2 osteosarcoma cells, and normal BM MNCs (without or 1 or 4 hours post-XRT) were immunoprecipitated with Ab-5 anti-wtp53 MoAb or Ab-3 anti-mtp53 MoAb, followed by immunoblotting with SMP14 anti-MDM2 MoAb (B).
We next examined whether MDM2 was binding to wtp53 and/or mtp53 in RPMI 8226 MM cells. Both wtp53 and mtp53 were associated with MDM2 in these tumor cells (Fig 7B). In contrast, no wtp53- or mtp53-MDM2 protein complexes were detected in Saos-2 cells, which lack p53 protein, or in untreated or irradiated normal BM MNCs, which lack MDM2 expression.
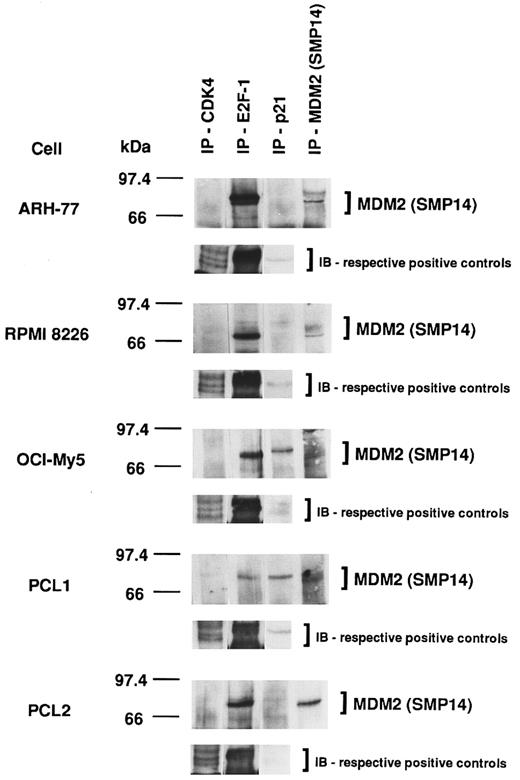
Association of MDM2 with cell-cycle regulatory proteins in p53-positive and p53-negative MM-derived cell lines and PCL patient cells.Given the heterogeneity of p53 expression in MM cell lines and PCL patient cells demonstrated already, we next wanted to determine the impact of p53 expression on the association of MDM2 with other cell-cycle regulatory proteins. MDM2 protein was constitutively associated with CDK4, E2F-1, and p21 in all MM cell lines and PCL patient cells; however, constitutive binding of MDM2 to CDK4 was weak in all tumor cells, whereas MDM2–E2F-1 binding was strong in all tumor cells (Fig 8). Furthermore, cells that weakly express p53 (OCI-My5 and PCL1) demonstrate more MDM2-p21 complex formation compared with cells that strongly express p53 (ARH-77 and RPMI 8226), which demonstrate less MDM2-p21 binding. Immunoprecipitation and immunoblotting of these cell lysates using the same respective Abs served as positive controls.
Association of MDM2 with cell-cycle regulatory proteins in p53-positive and p53-negative MM-derived cell lines and PCL patient cells. Total cell lysates of MM cell lines (ARH-77, RPMI 8226, and OCI-My5) and PCL patient cells (PCL1 and PCL2) were immunoprecipitated with H-22 anti-CDK4 pAb, C-20 anti–E2F-1 MoAb, C-19 anti-p21 pAb, or SMP14 anti-MDM2 MoAb, followed by immunoblotting with SMP14 anti-MDM2 MoAb. These cell lysates were immunoprecipitated and immunoblotted using the same Abs as positive controls.
Association of MDM2 with cell-cycle regulatory proteins in p53-positive and p53-negative MM-derived cell lines and PCL patient cells. Total cell lysates of MM cell lines (ARH-77, RPMI 8226, and OCI-My5) and PCL patient cells (PCL1 and PCL2) were immunoprecipitated with H-22 anti-CDK4 pAb, C-20 anti–E2F-1 MoAb, C-19 anti-p21 pAb, or SMP14 anti-MDM2 MoAb, followed by immunoblotting with SMP14 anti-MDM2 MoAb. These cell lysates were immunoprecipitated and immunoblotted using the same Abs as positive controls.
DISCUSSION
In this study, we have shown that MDM2 protein is overexpressed in MM cell lines and PCL patient cells as compared with normal BM MNCs. To assay for the biologic function of MDM2 overexpression in MM cells, we inhibited MDM2 activity using specific antisense ODN and examined changes in DNA synthesis, cell viability, cell-cycle analysis, and apoptosis. In addition, since MDM2 and p53 are functionally related and overexpression of MDM2 can abrogate wtp53 tumor suppressive and apoptotic function,13 we also examined the expression of both wtp53 and mtp53 protein, as well as the binding of wtp53 and mtp53 to MDM2 in tumor cells. Furthermore, MDM2 association with other cell-cycle regulatory proteins, E2F-1, CDK4, and p21, was also studied, since binding of these proteins to MDM2 could also affect proliferation and apoptosis of these MM cells. Our studies suggest that inhibition of MDM2 induces both G1 growth arrest and apoptosis in MM cells.
In studying the biologic function of MDM2 overexpression, we first confirmed the specificity of MDM2 antisense ODNs used in these studies, evidenced by abrogation of MDM2 protein expression in these tumor cells without associated alteration in the expression of p53, E2F-1, CDK4, or p21. In contrast to a previous report using U87-MG human glioblastoma cells,41 RPMI 8226 MM cells clearly demonstrate large amounts of MDM2 and both wtp53 and mtp53 protein constitutively, and MDM2 expression was inhibited by MDM2 antisense ODN at an ODN concentration of 1.0 to 2.0 μmol/L within 2 to 6 hours. We also confirmed that the half-life of MDM2 was approximately 25 minutes in RPMI 8226 cells, supporting the observed rapid and marked inhibition of MDM2 expression by MDM2 antisense ODN in these MM cells. These differences between results in U87-MG human glioblastoma cells41 and our results in RPMI 8226 MM cells suggest that the effects of MDM2 antisense ODN may be modulated by other factors, such as p53 status, in different cell lines.
We next showed that treatment with MDM2 antisense ODN, but not with MDM2 sense, nonsense, or scrambled ODN, was able to transiently decrease cell proliferation and viability at 2 to 4 hours, indicating specific biologic sequelae of the antisense ODN. Moreover, if antisense ODN was not replenished, recovery of proliferation and viability to baseline occurred within 8 to 16 hours, consistent with the short observed half-life of MDM2 in these MM cells. Our subsequent experiments replenished MDM2 antisense ODN every 4 hours and resulted in sustained enhanced antiproliferative effects, further demonstrating the specificity of biologic sequelae related to the anti-MDM2 antisense ODNs.
MDM2 antisense ODNs also induced G1 growth arrest and triggered apoptosis, assessed by PI staining and flow cytometric analysis. pRB was predominantly phosphorylated and not bound to E2F-1 in MM cells, allowing for G1 to S phase transition. However, binding of E2F-1 to pRB was induced by MDM2 antisense ODN, restoring G1 growth arrest. The observation that MDM2 sense, MDM2 nonsense, and MDM2 scrambled ODN cells continued to progress from G1 to S phase further supports the specific role of MDM2 antisense ODN in G1/S blockade. Finally, acridine orange–ethidium bromide staining also indicated a significant increase in apoptotic MM cells following MDM2 antisense ODN treatment, suggesting that abrogation of MDM2 may also facilitate wtp53-mediated apoptosis. These studies therefore confirm that MDM2 plays a role in the regulation of both growth and survival in MM cells.
Overexpression of MDM2 has been demonstrated in a variety of tumors of solid organs and hematologic malignancies, including two of four cases of MM.10-36 As noted earlier, we found MDM2 to be overexpressed in all MM cell lines and PCL patients studied. In contrast, p53 expression was heterogeneous in MM cells: wtp53 and mtp53 were strongly expressed in ARH-77 and RPMI 8226 MM cells and were less evident in OCI-My5 cells and patient PCL1; only low-level mtp53 was expressed in patient PCL2. Normal BM MNCs, which express only wtp53, and U266 MM cells, which express only mtp53, served as positive controls for wtp53 and mtp53, respectively. Both wtp53 and mtp53 bind MDM2,45 confirmed in RPMI 8226 MM cells in this study, suggesting competition for a common p53 binding site. It has been shown that MDM2 binding to wtp53 inhibits wtp53 action on genes bearing specific p53 response elements (REs),46 and that the MDM2 gene has two tandem p53 REs in its intronic promoter.47 Binding of wtp53 can induce MDM2 gene transcription through activation of its p53 REs.43 This creates an autoregulatory feedback loop, whereby wtp53 enhances MDM2 expression and MDM2 then binds and inhibits wtp53. Hence, the presence of both wtp53 and mtp53 protein expression in the setting of MDM2 overexpression may interfere with this autoregulatory mechanism and is potentially tumorigenic: mtp53 binds to MDM2 and prevents wtp53-MDM2 complex formation, thereby allowing unopposed wtp53 effects. Our studies demonstrate coexpression of wtp53 and mtp53 in the setting of MDM2 overexpression in RPMI 8226 MM cells, highlighting the potential importance of the expression and function of these proteins in the pathogenesis of MM.
Finally, MDM2 is also known to associate with cell-cycle regulatory proteins other than p53, including pRB48 and the E2F-1/DP-1 complex.11 In our study, MDM2 expressed in MM cell lines and PCL patient cells binds E2F-1; in MM cells that lack p53, MDM2 also binds p21. p21 is activated by wtp53 and potentiates wtp53 tumor-suppressor function by strongly inhibiting cell-cycle regulatory proteins such as cyclin E and CDK2.49-51 The binding of MDM2 to p21, especially in MM cells lacking p53, may abrogate the inhibitory function of p21 and thereby facilitate MM cell growth. Our studies therefore suggest that inhibition of MDM2 may serve as a useful treatment strategy in MM.
ACKNOWLEDGMENT
We wish to thank John Daley for his help in the cell-cycle analysis, and Kristin Kimbrough for help in pulse chase experiments.
Supported by National Institutes of Health Grant No. CA50947; the Kraft Family Research Fund; the Ministry of Health, Singapore; Health Manpower Development Plan; and the National Medical Research Council-Shaw Medical Research Fellowship, Singapore.
Address reprint requests to Kenneth C. Anderson, MD, Division of Hematologic Malignancies, Dana-Farber Cancer Institute, 44 Binney St, Boston, MA 02115.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal