Abstract
Retinoic acid (RA) induces differentiation, followed by apoptosis in acute promyelocytic leukemia (APL) cells, both in vitro and in patients. One problem in understanding these mechanisms is to distinguish molecular events leading to differentiation from those leading to apoptosis. We have identified a leukemic cell line, PLB-985, where RA directly induces apoptosis with no morphologic, genetic, or cell-surface marker evidence of differentiation. These cells differentiate following dimethyl sulfoxide (DMSO), but not RA, treatment. Two-color flow cytometry showed no alteration of the cell cycle after RA treatment, and cell-surface marker analysis of CD11a, CD11b, and CD13 showed no modulation typical of differentiating cells. RNA expression of myeloblastin and transglutaminase, genes regulated by RA-induced differentiation in NB4 cells, was unchanged by RA treatment. Instead, RA induced apoptosis, as shown by typical apoptotic morphological features, genomic DNA laddering, and positive labeling in the TUNEL assay. We found that induction of apoptosis in this model requires a different pattern of retinoid receptor binding and transcriptional activation than is seen in APL cells. As previously described, treatment with retinoid receptor-selective ligands showed that stimulation of RAR alone is sufficient to induce differentiation and apoptosis in NB4 cells, and that stimulation of RXR has no effect on the parameters analyzed. In PLB-985 cells, on the other hand, apoptosis was induced only upon costimulation of both RAR and RXR. Stimulation of either receptor alone had no effect on the cells. Consistent with these findings, bcl-2 RNA and protein levels were downregulated after stimulation of both RAR and RXR, but not with an RAR-specific ligand alone, as in NB4 cells. The expression of several other bcl-2 family members (bcl-X, ich-1, bax, bag, and bak ) and retinoid receptors (RARα, RXRα, and RXRβ) was not affected by treatment with RAR- and/or RXR-activating retinoids; RARβ RNA was undetectable before and after retinoid treatment. Thus, our cell model provides a useful tool in determining the genetic events mediating apoptosis as a response to RA, unobscured by events implicated in differentiation.
RETINOIDS ARE natural and synthetic derivatives of vitamin A (retinol) that have been shown to exert dramatic effects on cell death and differentiation in vertebrate embryogenesis, and, more recently, in human tumor cells.1 These compounds inhibit proliferation and induce differentiation and apoptosis in some human leukemia cells in vivo and in vitro. This has been shown in leukemic cells in patients treated with all-trans retinoic acid (ATRA), as well as in the human acute promyelocytic leukemia (APL) cell line, NB4.2-5 Other isomers of ATRA (9-cis- and 13-cis-RA) have been shown to give similar results. However, retinoids have not been successfully used to induce differentiation or apoptosis in most non-APL cell lines or in patients with other subtypes of leukemia. Furthermore, the molecular mechanisms by which retinoids induce both differentiation and apoptosis are not well described. An understanding at the molecular level of why and how retinoids are effective against some leukemia cells but not others is essential to broadening their therapeutic use.
In the retinoid-responsive myeloid leukemic cell lines NB4 and HL-60, retinoid-induced differentiation and apoptosis appear to occur in a sequential order, whereby apoptosis follows differentiation and growth arrest. NB4 cells appear to be blocked at a precursory stage of myelocytic differentiation, likely due to the presence of a dominant hybrid PML/RARα protein,6-9 resulting from a translocation between chromosomes 15 and 17, fusing the 5′ promyelocytic leukemia (PML) gene on chromosome 15 to the 3′ RARα gene on chromosome 17. This mutation is also a hallmark of APL.10,11 Treatment with RA releases this block in differentiation,12-14 apparently by inducing a specific proteolytic degradation of the fusion PML/RARα protein.15,16 The cells proceed to differentiate into granulocytes and subsequently die by apoptosis. In HL-60 cells, which lack the characteristic t(15; 17) and do not synthesize the fusion PML/RARα protein, RA also induces differentiation and apoptosis, evidently by a different mechanism.17
In these cellular models, apoptotic death is the ultimate fate of RA-responsive leukemic cells. However, it is not clear whether apoptosis occurs as a consequence of the cells reaching a terminally differentiated state, or is directly induced by retinoids. It is difficult to distinguish the molecular events involved in apoptosis from those required for differentiation. For example, members of the bcl-2 family of genes and their binding partners are involved in the activation and suppression of apoptosis.18-20 Many of these genes are also regulated during differentiation, making it difficult to dissect precisely which genes are associated with each pathway. There are recent data that the process of differentiation in HL-60 cells can be separated from the induction of apoptosis depending on the type of RA receptor that is activated. RA mediates its effects by binding two types of receptors, RARs and RXRs,21,22 and members of both families are expressed by NB4 and HL-60 cells.23 In recent experiments, stimulation of RARα in HL-60 cells induced differentiation whereas stimulation of RXRα induced apoptosis.5,24 Further, retinoid-resistant HL-60 cells bearing a mutated dominant negative RARα but overexpressing RXR were reported to undergo apoptosis in response to RA.24 We and others have developed NB4 subclones that do not undergo differentiation, but such clones described to date also have been resistant to retinoid-induced apoptosis.
We have characterized a naturally occurring myeloid cell line, PLB-985,25 that undergoes differentiation and apoptosis in response to one agent, but apoptosis without differentiation in response to retinoids. Using two-color flow cytometry, we show that the induction of apoptosis in these cells does not affect the cell cycle nor the expression of cell-surface markers of differentiation. bcl-2 transcription, but not that of other family members, is downregulated within 1 hour after the addition of RA. Furthermore, by using receptor-selective ligands we have shown that the induction of apoptosis requires the activation of both RAR and RXR, in contrast to NB4 cells, where the activation of RAR alone suffices to induce differentiation and apoptosis. Thus, we propose that the PLB-985 cell line provides a useful tool to dissect the molecular pathways involved in RA-induced apoptosis.
MATERIALS AND METHODS
Cell culture and cell morphology. PLB-985 and NB4 cells were maintained in RPMI 1640 medium (GIBCO-BRL, Burlington, Ontario, Canada) supplemented with 10% fetal bovine serum (FBS; Upstate Biotechnology Inc, Lake Placid, NY) in a humidified incubator at 37°C with 5% CO2 . Retinoid-treated cells were incubated in the presence of 10−6 or 10−7 mol/L retinoids (9-cis-, 13-cis-, or all-trans-RA, or selective ligands 100153 and 100272 [TTNPB]). Dimethyl sulfoxide (DMSO)-treated cells were grown in the presence of 1.25% DMSO. The medium for all cells was changed after 3 days of incubation, and fresh retinoids or DMSO were added to the medium. Logarithmically growing cells were diluted threefold at this time point. Cytospins from control and treated cells were stained with Wright's stain for morphological characterization. Cell viability was assessed by trypan blue (GIBCO, Grand Island, NY) exclusion using a hemocytometer.
DNA laddering analysis and TUNEL (TdT) assay. Total genomic DNA was extracted according to standard phenol/chloroform protocol.26 Five micrograms of undigested DNA was separated on a 1.5% TBE/agarose gel, stained with ethidium bromide, and photographed.
Genomic DNA strand breaks characteristic of apoptosis were labeled in situ by terminal deoxynycleotidyl transferase (TdT) using an in situ cell death detection kit (Boehringer Mannheim, Laval, Québec, Canada) according to the manufacturer's instructions. Briefly, cytospins containing 50,000 cells were fixed in 4% paraformaldehyde/phosphate-buffered saline (PBS) and permeabilized in 0.1% Triton X-100 and 0.1% sodium citrate. The cells were then exposed to the TUNEL reaction mixture containing the TdT enzyme and fluorescein-labeled nucleotides, washed, and photographed under a fluorescence microscope.
Cell surface marker analysis. Control and treated cells were washed twice in PBS and incubated with anti-CD11b, anti-CD11c and anti-CD13 antibodies (Coulter Electronics, Inc, Miami Lakes, FL). Fluorimetric analysis of at least 5,000 cells was performed on an Epics XL-MCL cytometer using the manufacturer's suggested protocol (Coulter Electronics).
Transfections and CAT assay for ligand binding specificity. PLB-985 and NB4 cells were transiently transfected by electroporation as previously described.27 βREtkCAT was used as a reporter plasmid containing a typical DR5 RAR-response element driving the CAT gene (a generous gift from H. Sucov, La Jolla, CA). CMVβgal, a plasmid containing the β-gal gene constitutively expressed from a CMV promotor (Clonetech Laboratories, Palo Alto, CA) was used as an internal transfection control and was cotransfected together with the CAT plasmid. CAT activity was measured using a modified organic diffusion protocol as described.27
RNA extraction and analysis. Total cellular RNA was isolated using the guanidinium thiocyanate method as previously described,28 separated on a 1% formaldehyde agarose gel, and blotted on a Hybond N (Amersham Life Science, Arlington Heights, IL) or a Magna (MSI, Westborough, MA) nylon membrane. Membranes were UV-cross-linked and hybridized according to a modified protocol.29 Briefly, membranes were wetted in 0.1× SSC (20× SSC is 3 mol/L sodium chloride and 0.3 mol/L sodium citrate, pH 7.0) and prehybridized for 1 hour at 65°C in a rotating oven in 0.05 mL/cm2 solution I (0.6 mol/L NaCl, 120 mmol/L Tris-base, 8 mmol/L EDTA, 0.1% sodium pyrophosphate, 0.2% sodium dodecyl sulfate [SDS], and 0.05% heparin [sodium salt], adjusted to pH 7.4 with HCl). After prehybridization, solution I was replaced with solution II (solution I + 10% dextran sulfate, sodium salt), and the heat-denatured radioactive probe was added. Hybridization was performed overnight at 65°C in a rotating oven. The filters were then washed once in 2× SSC for 5 minutes at room temperature, then at 65°C for 1 hour in 0.1× SSC, 0.1% SDS followed by a quick rinse in 0.1× SSC. The membranes were exposed against an autoradiography film. Ribonuclease protection analysis was performed on total RNA as previously described.30
Probes used in Northern blots. The FL-1 fragment was used as a probe for human bcl-2 mRNA (a kind gift from M. Cleary, Stanford University Medical Center, Stanford, CA).31 Probes for bcl-X, ich-1, bax, bag, and bak were generated by reverse transcriptasepolymerase chain reaction (RT-PCR) using left and right primers (synthesized by the Biotechnology Research Institute, MRC, Montréal, Québec, Canada) as follows (numbers denote the first 5′ base-pair): bcl-X: 98-GGACAATGGACTGGTTGAGC and 633-CGATCCGACTCACCAATACC32; ich-1: 54-GCATGCATCCTCATCATCAG and 847-CTCTTGGAGCTGGAGCAGTT33; bax: 38-CCACCAGCTCTGAGCAGATC and 407-ATGGTTCTGATCAGTTCCGG34; bag: 130-CAGACTCAGCGTGATCGTCA and 751-CACTGCACTTCATTCAGCCA (mouse sequence)35; bak: 264-GCTTCTGAGGAGCAGGTAGC and 696-AGTGATGCAGCATGAAGTCG.36 The myeloblastin probe used was a full-length cDNA fragment.37
Protein extraction and Western blotting. Total protein was extracted from cells and separated on SDS-polyacrylamide gel electrophoresis (SDS-PAGE) as previously described.27 Membranes were blocked overnight in PBS-5% skim milk solution and hybridized for 4 hours at room temperature with a 1:1,000 dilution of anti–bcl-2 antibody (no. 15131A; PharMingen, San Diego, CA) in a PBS-5% skim milk solution on a gyrating platform. Membranes were washed twice for 15 minutes in PBS and hybridized with a horseradish peroxidase (HRP)-coupled secondary antibody for 1 hour at room temperature, after which they were washed again as before. Protein detection was done using an ECL chemiluminescence system (Amersham Life Science, Arlington Heights, IL).
RESULTS
Cell growth response to retinoid treatment. We initially studied the effect of different retinoids on the growth pattern of the myelomonoblastic cell line PLB-985 and compared those with the known effects of retinoids on the APL cell line NB4. Retinoids induce NB4 cells to differentiate into mature granulocytes, which undergo cell cycle arrest and later die by apoptosis.2 38
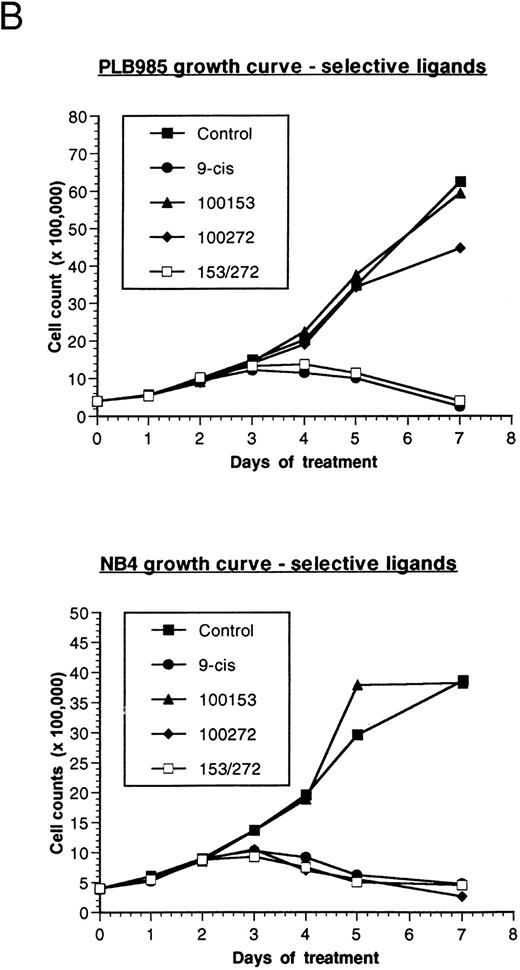
Effect of retinoids on the growth of PLB-985 and NB4 cells. (A) PLB-985 cells were grown in the presence of 10−7 mol/L 9-cis-RA, 13-cis-RA, or all-trans-RA, and viable cells were counted at 1, 2, 4, and 7 days after the addition of retinoids. (B) PLB-985 and NB4 cells were grown in the presence of 10−7 mol/L 9-cis-RA or the selective ligands, and cell counts were made at days 1, 2, 3, 4, 5, and 7. Fresh medium and retinoids were added at day 3 and cell numbers were adjusted as described in Materials and Methods.
Effect of retinoids on the growth of PLB-985 and NB4 cells. (A) PLB-985 cells were grown in the presence of 10−7 mol/L 9-cis-RA, 13-cis-RA, or all-trans-RA, and viable cells were counted at 1, 2, 4, and 7 days after the addition of retinoids. (B) PLB-985 and NB4 cells were grown in the presence of 10−7 mol/L 9-cis-RA or the selective ligands, and cell counts were made at days 1, 2, 3, 4, 5, and 7. Fresh medium and retinoids were added at day 3 and cell numbers were adjusted as described in Materials and Methods.
Addition of either 9-cis- (9-cis-RA), all-trans- (ATRA), or 13-cis-retinoic acid (13-cis-RA) at a concentration of 10−7 mol/L resulted in growth inhibition of both PLB-985 and NB4 cells. Cell counts levelled off by the third day of culture with all three retinoids, and few viable cells remained after 7 days of treatment (Fig 1A; NB4 data not shown). Growth inhibition in PLB-985 cells was most pronounced with 9-cis-RA. Higher concentrations of retinoids amplified this effect, and no viable cells remained after 7 days of treatment with 10−6 mol/L 9-cis-RA (data not shown).
9-cis-RA binds and activates both classes of nuclear retinoid receptors, RAR and RXR, whereas ATRA binds preferentially RAR's.21,22,39 However, stereoisomerization may permit ATRA or 13-cis-RA to bind RXR's, in addition to RAR's, albeit at a lower efficiency.21,22 To determine whether the induction of the observed cellular growth arrest and eventual cell death required activation of RAR, RXR, or both, we obtained RAR- and RXR-selective ligands (a gift from Richard Heyman, Ligand Pharmaceuticals, San Diego, CA). Ligand 100272 (TTNPB) selectively binds the RAR family receptors and ligand 100153 selectively binds RXR's. Both ligands bind their respective receptors with high affinity and with no apparent cross-reactivity at the concentrations used in this study (R. Heyman, personal communication, March 1995). We verified the specificity of the selective ligands by transfecting a CAT (chloramphenicol acetyl transferase) reporter plasmid into PLB-985 and NB4 cells, and assaying CAT activity after the addition of the selective ligands. The plasmid βREtkCAT,40 containing a typical RAR-responsive element driving the CAT reported gene, was activated in both cell lines only by the RAR-selective ligand 100272, whereas the RXR-selective ligand 100153 did not induce any CAT activity (data not shown). We also analyzed by RNase protection analysis the activation of CD38, an RAR-inducible myeloid antigen.41-43 As expected, 9-cis-RA and the RAR-selective ligand 100272 induced the transcription of CD38, whereas the RXR-selective ligand 100153 did not significantly affect it (data not shown). Thus, the selective ligands were specific for their respective receptors at 10−7 mol/L concentration.
PLB-985 and NB4 cells were treated separately with each of these selective ligands at a concentration of 10−7 mol/L, or with both selective ligands mixed together, and cell growth was assessed. NB4 cells grew normally with the RXR-selective ligand (100153) but were sensitive to the RAR-selective ligand (100272). This RAR-selective ligand alone inhibited NB4 cell growth to the same extent as 9-cis-RA (Fig 1B, bottom). As expected, combined treatment with RAR- and RXR-selective ligands also inhibited NB4 cell growth.
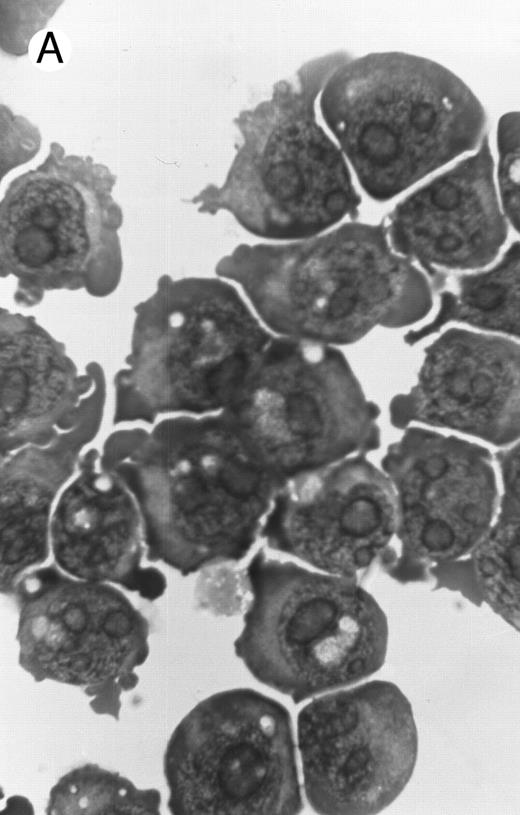
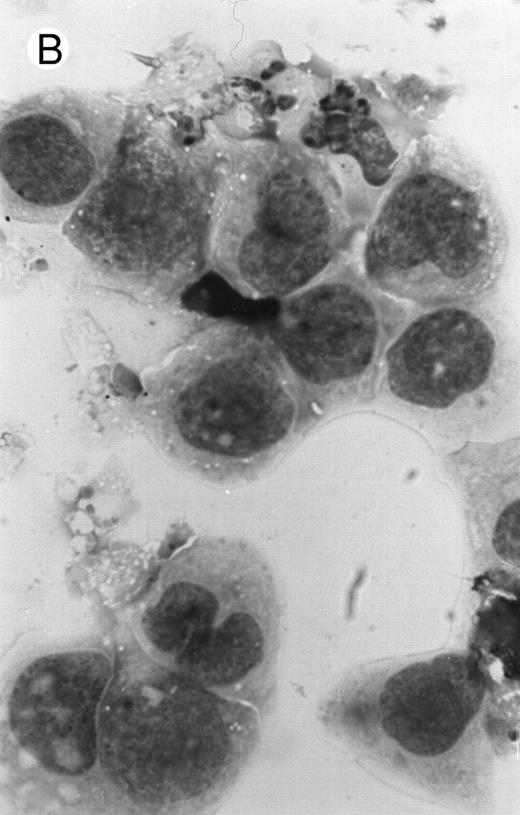
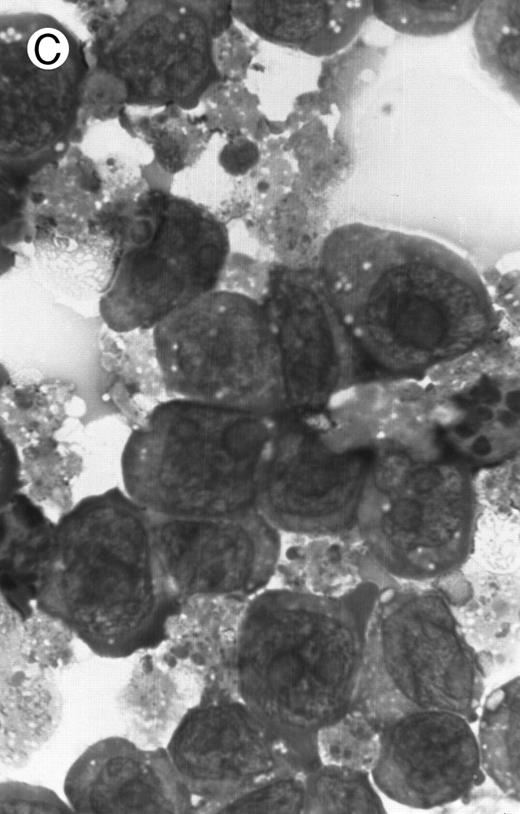
Morphology of PLB-985 cells (A) before treatment and after a 4-day treatment with (B) 1.25% DMSO (differentiation) or (C) 10−7 mol/L 9-cis-RA (apoptosis) (original magnification × 1,000).
Morphology of PLB-985 cells (A) before treatment and after a 4-day treatment with (B) 1.25% DMSO (differentiation) or (C) 10−7 mol/L 9-cis-RA (apoptosis) (original magnification × 1,000).
In contrast to NB4 cells, neither RAR nor RXR activation was sufficient by itself to affect cell growth in PLB-985 cells. However, when both selective ligands were present together in the same culture, cell growth was inhibited to the same extent as with 9-cis-RA (Fig 1B, top). Together, these results indicated that growth arrest and subsequent cell death in PLB-985 cells has a requirement for the activation of both RAR and RXR, whereas in NB4 cells the stimulation of RAR alone is sufficient to induce growth arrest, differentiation, and cell death.
Cell differentiation response to retinoid treatment. PLB-985 cells are myelomonoblastic with a potential to differentiate either into granulocytes upon treatment with 1.25% DMSO or into monocytes in response to PMA.25 NB4 cells, on the other hand, differentiate into granulocytes upon treatment with retinoids.
We have shown above that activation of both RAR and RXR by 9-cis-RA or by the combination of selective ligands inhibits cell growth and induces cell death in PLB-985 cells. We wished to verify whether these retinoids also induced differentiation of PLB-985 cells before the onset of cell death. Microscopic examination of cell morphology after 4 days of treatment with 9-cis-RA demonstrated no differentiated phenotype (Fig 2C). On the other hand, DMSO-treated PLB-985 cells had a typical differentiated morphology (Fig 2B). We then examined alterations in several cell surface markers, especially those specific for differentiated granulocytes. We contrasted the known differentiation effects of retinoids on NB4 with PLB-985 cells. NB4 and PLB-985 cells express low levels of CD11b and no detectable CD11c, but high levels of CD13.2 44 As previously reported, treatment of NB4 cells with 9-cis-RA increases expression of CD11b and CD11c, and significantly reduces expression of CD13 (Fig 3, bottom), consistent with the differentiation of NB4 cells into granulocytes. Also consistent with their effects on cell growth, the RXR-selective ligand 100153 did not have any effect on the surface marker expression in NB4 cells, but the RAR-selective ligand 100272 induced the same differentiation-specific changes as 9-cis-RA. In PLB-985 cells, DMSO-induced granulocytic differentiation stimulates the surface expression of CD11b44 (and data not shown). In our experiments PLB-985 cells treated with 9-cis-RA did not show any increase in CD11b and CD11c levels. These cells also continued to express high levels of CD13, even at day 4, when the cell counts began to decrease (Fig 3, top). As expected, the selective ligands, either alone or in combination, also did not have any effect on the expression of these markers in PLB-985 cells. These results show that stimulation of both RAR and RXR inhibits growth and induces cell death without differentiation in PLB-985 cells.
Cell differentiation surface marker expression on PLB-985 (top) and NB4 (bottom) cells. Cells were treated with 10−7 mol/L 9-cis-RA or the selective ligands, and the cell-surface expression of CD11b, CD11c, and CD13 antigens was assessed by flow cytometry at 1, 2, 3, and 4 days after treatment. Graphs indicate percent of positive cells.
Cell differentiation surface marker expression on PLB-985 (top) and NB4 (bottom) cells. Cells were treated with 10−7 mol/L 9-cis-RA or the selective ligands, and the cell-surface expression of CD11b, CD11c, and CD13 antigens was assessed by flow cytometry at 1, 2, 3, and 4 days after treatment. Graphs indicate percent of positive cells.
Induction of apoptosis by 9-cis-RA. Microscopic observation of retinoid-treated PLB-985 cells showed typical characteristics of apoptosis, including cell shrinkage, membrane blebbing, and nuclear fragmentation (Fig 2C). Furthermore, apoptotic cells were observed as early as 2 days after the addition of retinoids, and increased in numbers with the progression of treatment (data not shown). A typical molecular characteristic of apoptosis is genomic DNA cleavage into internucleosomal segments attributed to the specific activation of an endogenous endonuclease.45 Several cell lines (MOLT-3, T-cell ALL; CEM, T-cell ALL; PLB-985; Raji, Burkitt lymphoma; HUT 78, cutaneous T-cell lymphoma) were treated for 4 days with 10−6 mol/L 9-cis-RA. DNA was isolated from these cultures, separated on an agarose gel, and visualized by ethidium bromide staining. Only RA-treated PLB-985 cells exhibited a typical apoptotic “DNA laddering” pattern (data not shown). Similar results were obtained after treatment with all-trans-RA and 13-cis-RA (data not shown); however, this effect was most pronounced with 9-cis-RA.
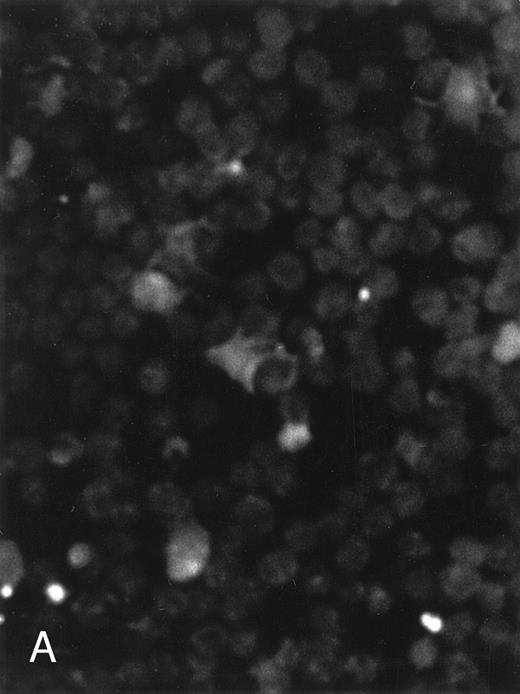
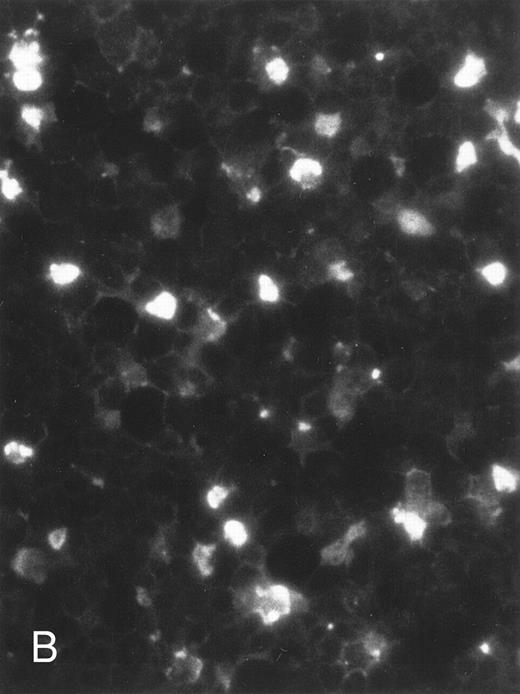
The apoptosis-specific genomic DNA cleavage was also assessed by the TUNEL (TdT-mediated dUTP nick end labelling) assay (In situ Cell Death Detection Kit, AP; Boehringer Mannheim). PLB-985 cells were treated with 10−6 mol/L 9-cis-RA for 3 days and assayed for DNA strand breakage. Control cells growing exponentially showed very limited fluorescent labeling, whereas RA-treated cells showed numerous positive cells (Fig 4). These results confirmed that RA induced specific DNA strand breakage in PLB-985 cells, indicative of the apoptotic process.
Induction of genomic DNA-nicking in apoptotic PLB-985 cells treated with 10−6 mol/L 9-cis-RA for 3 days. Cytospins were prepared from (A) normal and (B) RA-treated cells, and the TUNEL assay was performed, labeling the apoptosis-specific DNA strand breaks with fluorescent label as described in Materials and Methods.
Induction of genomic DNA-nicking in apoptotic PLB-985 cells treated with 10−6 mol/L 9-cis-RA for 3 days. Cytospins were prepared from (A) normal and (B) RA-treated cells, and the TUNEL assay was performed, labeling the apoptosis-specific DNA strand breaks with fluorescent label as described in Materials and Methods.
We also wished to determine whether PLB-985 cells arrested or accumulated at a specific cell cycle stage before the onset of apoptosis. This was determined by analyzing the percentage of cells in each stage at various times after the beginning of treatment with 9-cis-RA or with the combination of RAR- and RXR-selective ligands. We could not detect any significant differences in the cell-cycle pattern between logarithmically growing control PLB-985 cells and those treated with the retinoids (data not shown). PLB-985 cells did not accumulate at any particular cycle stage, suggesting a direct entry into apoptosis without growth arrest.
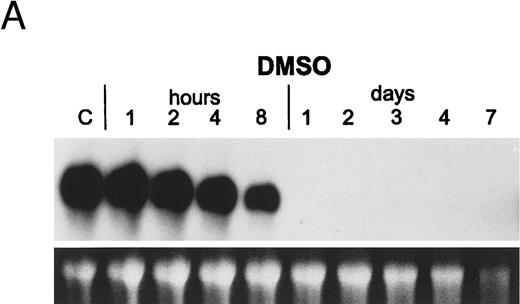
Regulation of gene expression in PLB-985 cells. Myeloblastin (MBN), a serine protease, is downregulated upon growth arrest and differentiation of myeloid leukemia cells.37,46 PLB-985 cells induced to differentiate with 1.25% DMSO showed a marked reduction in MBN mRNA within 8 hours, and no MBN transcript was detected after 24 hours of treatment (Fig 5A). However, treatment with 9-cis-RA had no effect on MBN expression, even after several days of treatment (Fig 5B). The transcriptional induction of transglutaminase (TGase), a protein cross-linking enzyme, has also been shown to be an indicator of differentiation in the myelomonocytic cell line HL-6046-48 and in NB4.49 We studied the effect of retinoids on the expression of TGase in PLB-985 cells and found that it was not induced upon RA treatment (data not shown). These results show that transcriptional regulation of genes specific for myeloid differentiation is not affected by RA in PLB-985 cells, confirming the absence of differentiation in these cells.
Regulation of myeloblastin gene expression in PLB-985 and NB4 cells. (A) Northern blot showing downregulation of myeloblastin RNA in PLB-985 cells induced to differentiate with 1.25% DMSO. Cells were treated from 1 hour to 4 days with DMSO. Five micrograms of total cell RNA from each sample was separated on agarose/formaldehyde gel, transferred to a nylon membrane, and probed with a myeloblastin cDNA probe. Photograph of EtBr-stained 28S rRNA band is shown, verifying integrity and equal loading of RNA. (B) PLB-985 cells were treated with 9-cis-RA or the selective ligands for 1, 2, 3, 4, and 7 days. Lane C contains control normal cell RNA.
Regulation of myeloblastin gene expression in PLB-985 and NB4 cells. (A) Northern blot showing downregulation of myeloblastin RNA in PLB-985 cells induced to differentiate with 1.25% DMSO. Cells were treated from 1 hour to 4 days with DMSO. Five micrograms of total cell RNA from each sample was separated on agarose/formaldehyde gel, transferred to a nylon membrane, and probed with a myeloblastin cDNA probe. Photograph of EtBr-stained 28S rRNA band is shown, verifying integrity and equal loading of RNA. (B) PLB-985 cells were treated with 9-cis-RA or the selective ligands for 1, 2, 3, 4, and 7 days. Lane C contains control normal cell RNA.
Since the level of expression of the various retinoid receptors may be critical to the phenotypic response, we determined the expression pattern of RARα, RARβ, RXRα, and RXRβ in PLB-985 cells by RNase protection. RARα was expressed at higher levels than RXRα or RXRβ, whereas RARβ was not expressed at all (data not shown). In contrast to the effects of retinoids on NB4 cells,27 the addition of either 9-cis-RA or any of the selective ligands did not significantly modify the expression of these receptors.
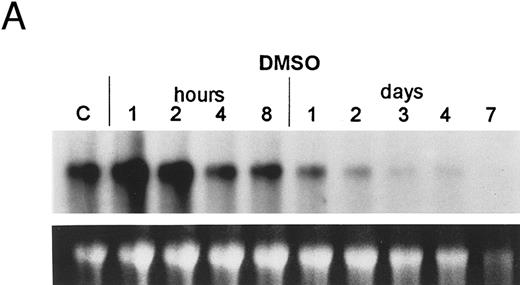
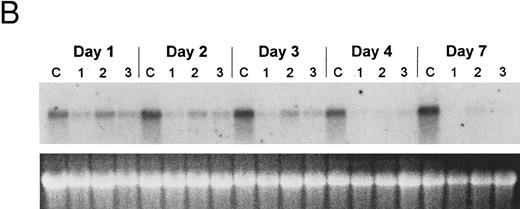
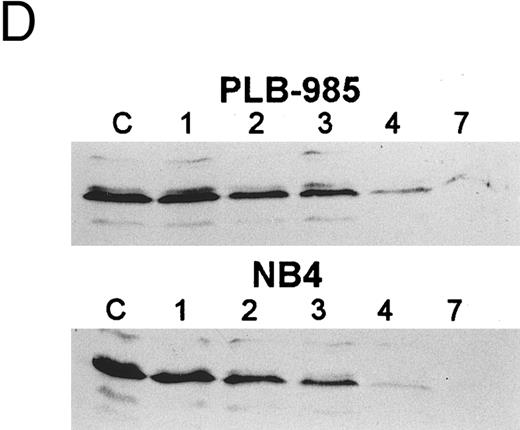
The bcl-2 family of genes has been implicated in the induction of apoptosis in several systems. Thus, initially we tested by Northern blot analysis the expression of bcl-2, bcl-X, ich-1, baxL , baxS , bag, and bak expression after 9-cis-RA treatment in PLB-985 cells. Only bcl-2 showed modulation of its expression after treatment. Untreated cells express a relatively high level of bcl-2 mRNA, and DMSO-induced differentiation downregulated this expression (Fig 6A). Treatment with 9-cis-, 13-cis-, or all-trans-RA also caused downregulation of bcl-2 mRNA levels (Fig 6B). Of the three retinoids tested, 9-cis-RA had the most pronounced inhibitory effect on bcl-2 expression (compare lane 1 with lanes 2 and 3 in each set). This downregulation was apparent as early as 2 hours after the addition of the retinoid (Fig 6C and data not shown). By day 4 bcl-2 mRNA levels were practically undetectable in the treated PLB-985 cells (Fig 6B). We also verified the levels of bcl-2 protein in these samples by Western blots. The levels of bcl-2 protein parallelled those of the mRNA on the Northern blots (Fig 6D).
Regulation of bcl-2 expression in PLB-985 cells. Northern blots of: (A) cells treated with 1.25% DMSO for 1 hour to 7 days; (B) cells treated with either 9-cis- (lanes 1), 13-cis-(lanes 2), or all-trans-RA (lanes 3) for 1, 2, 3, 4, and 7 days; and (C) cells treated with 9-cis-RA for 1 to 48 hours. Lane C contains control normal cell RNA. (D) Western blot showing downregulation of bcl-2 protein in PLB-985 and NB4 cells treated with 9-cis-RA for 1, 2, 3, 4, and 7 days. Ten micrograms of protein extract was separated on a 15% SDS-PAGE, transferred to a membrane, and probed with an α-bcl-2 antibody using ECL, described in Materials and Methods. Lane C contains control normal cell protein.
Regulation of bcl-2 expression in PLB-985 cells. Northern blots of: (A) cells treated with 1.25% DMSO for 1 hour to 7 days; (B) cells treated with either 9-cis- (lanes 1), 13-cis-(lanes 2), or all-trans-RA (lanes 3) for 1, 2, 3, 4, and 7 days; and (C) cells treated with 9-cis-RA for 1 to 48 hours. Lane C contains control normal cell RNA. (D) Western blot showing downregulation of bcl-2 protein in PLB-985 and NB4 cells treated with 9-cis-RA for 1, 2, 3, 4, and 7 days. Ten micrograms of protein extract was separated on a 15% SDS-PAGE, transferred to a membrane, and probed with an α-bcl-2 antibody using ECL, described in Materials and Methods. Lane C contains control normal cell protein.
Receptor specificity of gene regulation. PLB-985 cells expressed relatively high basal levels of MBN mRNA (Fig 5A), but treatment with the selective ligands individually or in combination did not affect the level of the MBN transcript (Fig 5B). The addition of the selective ligands individually also had no effect on the expression of bcl-2 mRNA in PLB-985 cells, but the combination of both RAR- and RXR-selective ligands caused a reduction in bcl-2 levels (Fig 7), similar to the results with 9-cis-RA. However, NB4 cells did not require both RAR- and RXR-selective ligands to downregulate bcl-2 expression: the RAR-selective ligand (100272) was sufficient to downregulate bcl-2 expression by itself, whereas the RXR-selective ligand (100153) had no effect on bcl-2 RNA levels (Fig 7). Both ligands together gave the same results as with the RAR-selective ligand alone. These results show that retinoid downregulation of bcl-2 in PLB-985 cells requires the activation of both RAR and RXR receptors, whereas in NB4 cells only the activation of RAR is sufficient for bcl-2 downregulation.
Northern blot comparing regulation of bcl-2 mRNA in PLB-985 and NB4 cells after treatment with 9-cis-RA or the selective ligands for 1, 2, 3, 4, and 7 days. Lane C contains control normal cell RNA. The Northern blot was prepared as in Fig 5.
Northern blot comparing regulation of bcl-2 mRNA in PLB-985 and NB4 cells after treatment with 9-cis-RA or the selective ligands for 1, 2, 3, 4, and 7 days. Lane C contains control normal cell RNA. The Northern blot was prepared as in Fig 5.
DISCUSSION
We have characterized a myelomonoblastic cell line PLB-985 that undergoes apoptosis without cell differentiation upon RA stimulation. Examples of apoptosis induction without differentiation and cell-cycle arrest exist in other cell types, including keratinocytes,50 erythroid progenitors,51 and neuronal cells.52 PLB-985 cells could be induced to differentiate along the granulocytic pathway with 1.25% DMSO,25 (and our results). However, RA treatment did not affect cell morphology, nor expression of any differentiation-specific surface marker or gene. Although some morphological features of differentiation were previously seen in PLB-985 cells when treated with 1 μmol/L RA,25 our extensive analysis, including surface marker, myeloblastin, and transglutaminase expression, has clearly demonstrated the lack of granulocytic differentiation upon RA treatment.
We have found that the RA-induced apoptosis in PLB-985 cells requires the stimulation of both the RAR and RXR. This is in contrast to the APL cell line NB4, where RAR stimulation alone leads to differentiation and apoptosis, and specific ligand binding to RXR is not needed. Two recent studies5 24 presented evidence that differentiation in HL-60 cells involves RARα (as in NB4 cells), and apoptosis depends on RXRα stimulation. In contrast to NB4 cells, HL-60 subclones were derived in which each process could be induced independently from one another. Cellular models with distinct phenotypes induced by activation of RAR, RXR, or both may be useful in finding the specific molecular events in the induction of differentiation and apoptosis of leukemic cells.
The downregulation of bcl-2 expression is widely observed in many cell systems that are stimulated to differentiate and undergo apoptosis, and its ability to suppress apoptosis is well documented (reviewed by Reed53 ). Overexpression of bcl-2 has been shown to inhibit the onset of apoptosis,54,55 whereas inhibition of its expression by antisense oligonucleotides promotes apoptosis.56,57 In PLB-985 cells, we have shown that bcl-2 is downregulated during both DMSO-induced differentiation and RA-induced apoptosis. In HL-60 cells, bcl-2 is also downregulated during DMSO- or RA-induced differentiation.58 Thus, bcl-2 downregulation may be a common effector of both differentiation and apoptosis. Alternatively, bcl-2 could be an early genetic event specific only to apoptosis, but since differentiation ultimately leads to apoptosis, bcl-2 downregulation would also be apparent during differentiation. Our observations that bcl-2 is downregulated within a few hours after the addition of RA, thus making it an early event, are consistent with the former hypothesis. However, in HL-60 cells, overexpression of exogenous bcl-2 was reported to markedly prolong in vitro life span without altering their maturation potential.59
Bcl-2 interacts with several other members of its own family of proteins, such as Bax, Bcl-XL , and Bak, forming homodimers or heterodimers with different, if not opposite, effects on cell death. One model proposes that Bax and Bcl-2 compete for one another, maintaining a balance in the form of heterodimers.34 Thus, an increase in one molecule may tilt the balance toward an abundance of one homodimer species over the other, with Bcl-2 homodimers promoting cell survival and Bax homodimers promoting apoptosis.60 In PLB-985 cells, we did not find any modulation of expression in any of these bcl-2–related genes. This suggests another regulatory mechanism for these proteins in PLB-985 apoptosis, perhaps involving a modification such as phosphorylation, since phosphorylation of Bcl-261 and Bad62 63 has been shown to modulate their activity and affect cell survival.
Bcl-2 expression also appears to inhibit the proteolytic activation of a key apoptotic enzyme, CPP32β/Yama/prICE/caspase 3,64-67 a member of the ICE/CED-3 family of proteases. Several lines of evidence indicate that bcl-2 acts upstream of CPP32β to inhibit its activation.64,67 68 Our preliminary results show that CPP32β appears to be activated within 2 days after RA treatment of PLB-985 cells (Monczak et al, unpublished results, March 1997), corresponding to the time frame of bcl-2 downregulation.
The expression of RARβ appears to be involved in the mediation of normal cellular differentiation in several cell types. In epithelial cells RARβ is induced in differentiating and adult lung tissue, and about half of human lung cancer cell lines and primary tumors show reduced or absent RARβ expression.69-71 Transgenic expression of RARβ can induce differentiation in malignant epithelial cells72,73 and differentiation and growth arrest without apoptosis in human RARβ-negative breast cancer cell lines.74 APL cells do not normally express RARβ, but treatment with ATRA induces its expression.30 75 Since PLB-985 cells do not express RARβ even after retinoid treatment, but undergo apoptosis without differentiation, we speculate that the lack of RARβ expression in RA-treated PLB-985 cells may uncouple the differentiation pathway from apoptosis. Thus, the introduction of RARβ might induce growth arrest and differentiation preceding apoptosis in these cells. We are presently testing this hypothesis by expressing RARβ in transfected PLB-985 cells. This model would indicate that specific RARs (and perhaps RXRs as well) are responsible for different pathways (apoptosis v differentiation) and that RARβ serves as an RA-inducible signal for differentiation in these cells.
Ultimately, we need to determine which genes are directly affected by ligand-bound RARs and RXRs in PLB-985 cells. First, on one end of the cascade, we have shown that both RAR and RXR need to be stimulated by retinoic acid to trigger apoptosis in this cell line. These ligand-bound receptors initiate a gene response, of which bcl-2 and CPP32β/Yama are downstream effectors. However, we have not yet identified the genes that are immediate responders to the activated receptors. Discovery of these genes will provide a clearer flowchart of the events responsible for RA-induced apoptosis. This, in turn, will help identify specific ligands targeting very selective events and cell types in the treatment of human cancer.
ACKNOWLEDGMENT
We thank Dr Richard Heyman from Ligand Pharmaceuticals, Inc, for the selective ligands; Dr Henry Sucov for kindly providing the βRE-tk-CAT response element; Dr Pierre Laneuville from the Royal Victoria Hospital, Montréal, Canada, for cell-cycle analysis; Franca Sicilia from the Division of Hematology, SMBD Jewish General Hospital, for excellent technical assistance in cell-surface marker analysis and cytospin preparation; and Angelika Rosenauer for technical help and discussion.
Supported by a grant from the Medical Research Council of Canada. Y.M. is a Robert Stewart Fellow in Hematology Research, SMBD Jewish General Hospital. W.H.M. Jr is a Scholar of the Medical Research Council of Canada.
Address reprint requests to Wilson H. Miller, Jr, MD, PhD, 3755 Chemin de la Côte-Ste-Catherine, Montréal, Québec, Canada H3T 1E2.






This feature is available to Subscribers Only
Sign In or Create an Account Close Modal