Abstract
Karyotypes in multiple myeloma (MM) are complex and exhibit numerous structural and numerical aberrations. The largest subset of structural chromosome anomalies in clinical specimens and cell lines involves aberrations of chromosome 1. Unbalanced translocations and duplications involving all or part of the whole long arm of chromosome 1 presumably occur as secondary aberrations and are associated with tumor progression and advanced disease. Unfortunately, cytogenetic evidence is scarce as to how these unstable whole-arm rearrangements may take place. We report nonrandom, unbalanced whole-arm translocations of 1q in the cytogenetic evolution of patients with aggressive MM. Whole-arm or “jumping translocations” of 1q were found in 36 of 158 successive patients with abnormal karyotypes. Recurring whole-arm translocations of 1q involved chromosomes 5,8,12,14,15,16,17,19,21, and 22. A newly delineated breakpoint present in three patients involved a whole-arm translocation of 1q to band 5q15. Three recurrent translocations of 1q10 to the short arms of different acrocentric chromosomes have also been identified, including three patients with der(15)t(1;15)(q10;p10) and two patients each with der(21)t(1;21)(q10;p13) and der(22)t(1;22) (q10;p10). Whole-arm translocations of 1q10 to telomeric regions of nonacrocentric chromosomes included der(12)t(1;12) (q10;q24.3) and der(19)t(1;19)(q10;q13.4) in three and two patients, respectively. Recurrent whole-arm translocations of 1q to centromeric regions included der(16)t(1;16)(q10;q10) and der(19)t(1;19)(q10;p10). The mechanisms involved in the 1q instability in MM may be associated with highly decondensed pericentromeric heterochromatin, which may permit recombination and formation of unstable translocations of chromosome 1q. The clonal evolution of cells with extra copies of 1q suggests that this aberration directly or indirectly provides a proliferative advantage.
CHROMOSOME 1 aberrations are very common in most hematologic malignancies and constitute the most common structural aberration in multiple myeloma (MM). Up to 40% of patients with abnormal cytogenetics show chromosome 1 rearrangements,1 which are the most common secondary findings in the complex karyotypes of MM.1-5 To date no distinct clinical and prognostic features have been associated with extra copies of 1q, whereas aberrations involving chromosomes 13 and 11q are associated with a poor prognosis in MM.6,7 Little is known about the progression of nonrandom secondary chromosome events involving chromosome 1. Duplications of all or part of 1q and whole-arm translocations of 1q are widely reported in neoplasia, but the origin of these major genomic rearrangements remains obscure. Extra copies of 1q can occur as translocated unbalanced derivative chromosomes, isochromosomes, or “jumping translocations”; however, the essential genetic characteristic is the same, resulting in partial trisomies for the 1q segment.8-12
Whole-arm translocations of 1q become jumping translocations when the 1q segment moves (jumps) around the karyotype to more than one nonhomologous chromosome.
The cytogenetic changes associated with extra copies of 1q have been attributed in part to cytotoxic treatments and in part to the natural evolution of disease progression. Aberrations in the centromeric regions of chromosomes can result in chromosome instability, which can lead to a generalized breakdown in normal chromosome segregation, resulting in nondisjunction or unbalanced translocations during mitosis. The extra copies of 1q present in B-cell acute lymphoblastic leukemia and many advanced neoplasias may confer a proliferative advantage.13 Although present in a wide variety of tumors, the movement of chromosome 1q to one or more nonhomologous chromosomes and the resulting increase in copy number appear to be a special type of chromosome instability, because it has been reported only in a small fraction of patients with any given malignancy as jumping translocations. Unfortunately, the exact mechanisms by which whole chromosome arms separate and rejoin with other centromeres, telomeres, or interstitial sites is unknown.
We have analyzed chromosome 1 aberrations in 158 patients with abnormal karyotypes and have found a subset of patients with evidence of nonrandom whole-arm 1q aberrations. The observation that extra copies of 1q occurred in patients with the decondensation of centromeric heterochromatin prompted an expanded study of this group. The decondensation of the centromeric heterochromatin of 1q suggests that hypomethylation of this region may play a role in the somatic pairing, fragility, and formation of triradial configurations involving the long arm of chromosome 1. These events may be the precursors to the subsequent jumping translocations found in some patients. The striking similarity between chromosome 1q aberrations in MM patients and those with high-grade lymphomas suggests the possibility of a common mechanism in a number of malignancies.
MATERIALS AND METHODS
Bone marrow of MM patients was processed for chromosome studies as previously described.4 Twenty cells were studied in each case for routine analysis. An abnormal clone was identified as two or more metaphases displaying either the same structural abnormality or the same extra chromosome or at least three cells with the same missing chromosome. Aberrations were designated according to ISCN.14
RESULTS
Complete karyotype designations are provided in Table 1. These data represent a subset of cytogenetic findings in a group of 427 MM patients previously reported.5 To briefly summarize this patient population, 187 patients (44%) had normal, 158 had abnormal (37%), and 82 had inevaluable karyotypes (19%). Within the subset of 158 patients with abnormal karyotypes, 50 patients (32%) showed aberrations of all or part of 1q in the myeloma clone. These aberrations thus constituted the most common recurring secondary abnormalities in our MM patients. Unbalanced whole-arm translocations were found in 26 patients, whereas jumping translocations where the 1q was observed on more than one nonhomologous chromosome were found in 10 patients (Nos. 1,2,3,4,8,9,14,17,22,36).
Summary of Cytogenetic Findings
| Patient No. . | Karyotype . |
|---|---|
| Translocations of 1q10 to 5q15 | |
| 1 | 52∼59,XY,−Y,+1,add(1)(q32),del(1)(p11),+dic(1;20)(p22;p13), inv(3)(p26q23),t(4;19)(q21;q13.3),+5,add(5)(q22),+der(5)t(1;5) (q10;p15),der(5)t(1;5)(q10;q15),del(5)(p13),+6,add(6)(q11),+7,+7,der(8)t(8;11)(q24;q13)×2,+der(8)t(8;11)(q24;q13), t(1;8)(q10;p23),+15,der(16)t(1;16)(q10;p13.3),del(17)(p11),+19,+20,add(20)(p13),+21,add(22)(p11),add(22)(q13)[cp20] |
| 2 | 57∼59,X,−X,der(1)t(1;11)(q10;q10),t(1;8)(q42;p23),+der(5)t(1;5)(q10;q15)×2,del(6)(q14q15),+7,+8,+8,+11,del(13)(q12q14),+15, +15,+17,+der(19)t(1;19)(q21;q13.3),+20,+21,?inv(22)(q12q13)[cp20]. |
| 3 | 46,XX[4] 54∼55,X,−X,add(2)(q21),+3,+5,+der(5)t(1;5)(q10;q15)+7,der(7)t(1;7)(q21;q36),i(8)(q10)add(8)(q24.1),+der(8)t(8;15)(q10;q10) add(8)(q24.1),+9,der(11;22)(q10;q10),13,der(16)t(2;16)(q23;p13),der(17)t(1;17)(q10;q25),add(19)(p13),+21,+21, add(22)(p11), −22[cp16] |
| Translocations of 1q10 to 8p23 | |
| 4 | 44∼45,XY,?add(5)(q34),t(5;19)(q31;p11),i(6)(p10),der(8)t(1;8)(q10;p23),t(12;18;20)(q13;q21;p13),del(13)(q13q22),−14,der(1;16) (q10;p10)[cp4]5=63∼92,XXYY,t(1;2)(q25;q37)×2,t(5;19)×2,i(6)(p10)×2,der(8)t(8;11)(p11.2;q13)×2,der(11)t(1;8;11) (q11;p23;q13)×2,del(13)(q12q23)×2,−14,−14,der(1;16)(q10;p10)×2[cp5]. 46,XY[13] 45,X,−Y[6] |
| 5 | 43,XY,i(6)(p10),der(8)t(1;8)(q10;p23),−13,−14,−22[4] 46,XY[16] |
| See patient No. 1 | |
| Translocations of 1q10 to 12q24 | |
| 6 | 46,XX[18] 44,XX,tas(2;9)(q37;p24),add(8)(p11),der(12)t(1;12)(q10;q24.3),−13, −21[2] |
| 7 | 51,XY,+3,+7,−8,+9,der(12)t(1;12)(q10;q24.3),−13,14q+, +der(14)t(11;14)(q11;p11),+15,+19,+21[15] 46,XY[5] |
| 8 | 46,X[6] 43,XY,−1,trp(1)(q12q25),der(1)t(1;8)(q10;q10), der(12)t(1;12)(q10;q24),−13,−14,add(14)(q32)[14] |
| 9 | 44∼46,XX,t(6;7)(q23;q22),−8,der(12)t(1;12)(q10;q24.3),del(13)(q12q21),der(13)t(13;17)(q10;q10),der(14)t(14;17)(q10;q10),+17, +19,der(19)t(1;19)(q10;q11)[19] 46,XX[1] |
| Translocations of 1q10 to 14p11 | |
| 10 | 47,XY,+9,der(14)t(1;14)(q10;p11.2)[18] 46,XY[2] |
| 11 | 44∼46,XY,del(1)(p31p35),t(8;13)(q24.3;q13),del(12)(p11),del(13)(q14q22),−14,der(14)t(1;14)(q10;p11),−20[cp6] 46,XY[16] |
| Translocations of 1q10 to 15p10 | |
| 12 | 74∼76,XX,+X,+del(1)(p21),add(2)(q27),+add(2)(q27),+der(3)t(3;21)(p11;?q11)×2,+4,+5,+5,+6,+6,del(7)(p15),+del(7)(p15), t(8;21)(q22;q22),+9,+9,+12,+12,+der(15)t(1;15)(q10;p10)×2,+16,+16,+17,+17,+18,+18,+19,+20,+20,+21,+21, +der(21)t(8;21)(q22;q22),+22[cp16] 46,XX[4] |
| 13 | 46,XY[12] 43,X,−Y,der(4)t(1;4)(q11;q35),−13,−14,−15,+der(15)t(1;15)(q10;p10),−21,+der(21)t(1;21)(q11;p11),−22[8] |
| 14 | 46,XY[18] 47,X,−Y,+5,t(8;14)(q24;q32),+9,−13,der(15)t(1;15)(q10;p22),der(16)t(1;16)(q10;p10),+19,add(19)(q13)[cp2]. |
| Translocations of 1q to 16p10 | |
| See patient No. 4 | |
| See patient No. 14 | |
| 15 | 41∼44,XY,add(1)(q21)×2,der(2)add(2)(p24)t(2;3)(q31;p14),add(3)(p21),add(3)(q29),t(5;19)(p15;q13),add(7)(p13),−10,−13,−14, add(14)(q32),der(16)t(1;16)(q10;p10),del(17)(p11),−18,add(21)(p13),add(22)(p11),del(22)(q13)[cp20] |
| 16 | 46,XY,t(8;14)(q24;q32),del(11)(q12),?der(14)t(11;14)(q13;q11),der(16)t(1;16)(p10;q10)[cp4]. 46,XY[13] |
| (Continued on following page) | |
| 17 | 45∼46,XY,−1,add(3)(q26.2),t(4;9)(q35;p11),add(8)(q22),t(11;14)(q13;q32),add(12)(q22),der(16)t(1;16)(q10;p10), der(19)t(1;19)(q10;p10)[cp3] 74∼88,idem,×2[cp8] 46,XY[6] |
| Translocations of 1q10 to 16p13 | |
| See patient No. 1 | |
| 18 | 52,XY,+7,+9,+9,+11,t(14;17)(q32;q?11),+15,der(16)t(1;16)(q10;p13),+17,add(18)(p11)[24] 53,idem,+8[5] |
| 19 | 46,XX[13] 44,XX,−13,−14,add(14)(q32),−14,der(16)t(1;16)(q10;p13)[7] |
| Translocations of 1q10 to 17q25 | |
| 20 | 45,X,−X,−1,+5,+6,del(6)(q15),+7,add(8)(p11.1),−12,−13,−14,+15,add(15)(p11),−16,add(17)(p11),der(17)t(1;17)(q10;q25), add(18)(p11.1),add(19)(q13.4)[cp3] 73∼77,XX,−X,−1,+2,+3,+5,+6,+7,+7,+7,−8,add(8)(p11;1),+11,+11,−12,−13,−14,+15,add(15)(p11)×2,−16,del(16)(q22),+17, der(17)t(1;17)(q10;q25)×2,add(18)(p11;1)×2,−19,+20,+22[cp7] 46,XX[10] |
| See patient No. 3 | |
| Translocations of 1q10 to 19p13 | |
| 21 | 68∼71,X,−X,−Y,+4,−5,add(5)(q31),del(6)(q21),+7,+7,del(8)(p11)×2,−13,+16,+der(16)t(3;16)(q10;q10),+17,−18,+19, +der(19)t(1;19)(q10;p13.3),+20,−21,−21,−22,+mar[cp17] 46,XY[3] |
| 22 | 44∼47,X,−X,+1,del(1)(p13p31),der(7)t(1;7)(q10;p22),add(8)(p23),−13, −14,+15,+15,der(19)t(1;19)(q10;p13.3)[cp8] 46,XX[12] |
| Translocations of 1q10 to 19q13 | |
| 23 | 77,XY,+Y,+1,+1,del(1)(p13),+2,+2,+3,+4,+6,+7,+7,+8,+9,+9,+11,+11,+12,+14,add(14)(q32)×2,+15,+16,+16, add(17)(p13)×2,+add(18)(q23)×2,+19,+der(19)t(1;19)(q10;q13.3),+20,+20,+21,+21,+22,+22,+mar[cp9] 46,XY[8] |
| 24 | 47∼48,X,−X,der(2)t(2;3)(p21;p21),add(3)(p21),+del(3)(p23),+6,add(8)(q24),del(8)(p11),der(9)t(6;9)(q13;p13),del(10)(q25), del(13)(q13q22),−14,+15,−16,add(16)(p13),add(17)(p11),del(18)(q12q21),der(19)t(1;19)(q10;q13),add(20)(p13),add(21)(q22), +22,+mar1,+mar2[cp4] 46,XX[16] |
| Translocations of 1q10 to 19p10 | |
| 25 | 46,XY,t(9;12)(q11;q22),t(11;14)(q13;q32),der(19)t(1;19)(q10;p10)7=90∼92,idem×2[cp7] 46,XY[6] |
| 26 | 50∼52,X,−X,+3,+5,+6,+7,+8,i(8)(q10)×2,+9,−13,der(13)t(9;13)(q11;q11.2),+15,+19,der(19)t(1;19)(q10;p10),+20[cp20] |
| 27 | 53∼54,XY,+3,+5,+6,add(6)(q23),+7,t(7;17)(p11.1;p12),−8,+9,+15,+19,+21,+mar[cp11] 53∼54,idem,der(19)t(1;19)(q10;p10)[cp4] 46,XY[5] |
| 28 | 57∼59,XX,+del(1)(p13),+2,+del(3)(p13),+der(3)add(3)(q29)t(3;13)(q26;?q12),+i(6)(p10),+7,+7,+9,der(9)add(9)(p24), +der(10)t(10;?22)(p11.2;?q11;2),i(12)(p10),−13,+15,+der(15)t(1;15)(q21;p11.1),−16,+18,+der(19)t(1;19)(q10;p10), del(20)(q11.1),del(22)(q12),+add(22)(q13)[cp20] |
| See patient No. 17 | |
| 29 | 53,X,−X,del(1)(p13p22),+der(1)t(1;19)(q10;p10)×2,+3,+6,del(6)(q21),+9,+15,+18,+21[2] 106,idem×2[1] 46,XX[17] |
| Translocations of 1q10 to 21q22 | |
| 30 | 46,XX[14] 47,XX,−2,add(2)(q31),+19,−21,+der(21)t(1;21)(q10;q22)[cp6] |
| 31 | 46,XX[17] 45∼48,X,−X,+7,+9,−13,+der(21)t(1;21)(q10;q22)[cp4] |
| Translocations of 1q10 to 21p11 | |
| 32 | 44,X,−X,der(6)add(6)(p25)t(1;6)(q12;q12),?inv(7)(p14p22),del(8)(p11),−13,del(20)(q12),der(21)t(1;21)(q10;p11)[cp9] 46,XX[11] |
| 33 | 143∼46,XY,t(1;21)(q21;p11),−2,add(6)(q?21),add(7)(p?22),−8,+9,−12,add(13)(p11),add(17)(p?11),+19[cp18] 46,XY[2] |
| Translocations of 1q10 to 22p11 | |
| 34 | 52,X,−X,+3,+5,+6,+7,der(8)t(?X;8)(?q13;p21),+11,+15,der(22)t(1;22)(q10;p11.2),+mar[2] 46,XX[13] |
| 35 | 46,XY[16] 40∼43,X,−Y,−13,add(14)(q32),−18,der(22)t(1;22)(q10;p11)[cp3] |
| 36 | 46,XY,+1,del(1)(p11),t(2;19)(q37;p13;1),inv(3)(p14q29),del(6)(p22),der(6)t(1;6)(q21;q23),add(7)(p22),−13,der(16)t(1;16)(q10;q22), +19,der(22)t(1;22)(q10;p11.2)[3] |
| Patient No. . | Karyotype . |
|---|---|
| Translocations of 1q10 to 5q15 | |
| 1 | 52∼59,XY,−Y,+1,add(1)(q32),del(1)(p11),+dic(1;20)(p22;p13), inv(3)(p26q23),t(4;19)(q21;q13.3),+5,add(5)(q22),+der(5)t(1;5) (q10;p15),der(5)t(1;5)(q10;q15),del(5)(p13),+6,add(6)(q11),+7,+7,der(8)t(8;11)(q24;q13)×2,+der(8)t(8;11)(q24;q13), t(1;8)(q10;p23),+15,der(16)t(1;16)(q10;p13.3),del(17)(p11),+19,+20,add(20)(p13),+21,add(22)(p11),add(22)(q13)[cp20] |
| 2 | 57∼59,X,−X,der(1)t(1;11)(q10;q10),t(1;8)(q42;p23),+der(5)t(1;5)(q10;q15)×2,del(6)(q14q15),+7,+8,+8,+11,del(13)(q12q14),+15, +15,+17,+der(19)t(1;19)(q21;q13.3),+20,+21,?inv(22)(q12q13)[cp20]. |
| 3 | 46,XX[4] 54∼55,X,−X,add(2)(q21),+3,+5,+der(5)t(1;5)(q10;q15)+7,der(7)t(1;7)(q21;q36),i(8)(q10)add(8)(q24.1),+der(8)t(8;15)(q10;q10) add(8)(q24.1),+9,der(11;22)(q10;q10),13,der(16)t(2;16)(q23;p13),der(17)t(1;17)(q10;q25),add(19)(p13),+21,+21, add(22)(p11), −22[cp16] |
| Translocations of 1q10 to 8p23 | |
| 4 | 44∼45,XY,?add(5)(q34),t(5;19)(q31;p11),i(6)(p10),der(8)t(1;8)(q10;p23),t(12;18;20)(q13;q21;p13),del(13)(q13q22),−14,der(1;16) (q10;p10)[cp4]5=63∼92,XXYY,t(1;2)(q25;q37)×2,t(5;19)×2,i(6)(p10)×2,der(8)t(8;11)(p11.2;q13)×2,der(11)t(1;8;11) (q11;p23;q13)×2,del(13)(q12q23)×2,−14,−14,der(1;16)(q10;p10)×2[cp5]. 46,XY[13] 45,X,−Y[6] |
| 5 | 43,XY,i(6)(p10),der(8)t(1;8)(q10;p23),−13,−14,−22[4] 46,XY[16] |
| See patient No. 1 | |
| Translocations of 1q10 to 12q24 | |
| 6 | 46,XX[18] 44,XX,tas(2;9)(q37;p24),add(8)(p11),der(12)t(1;12)(q10;q24.3),−13, −21[2] |
| 7 | 51,XY,+3,+7,−8,+9,der(12)t(1;12)(q10;q24.3),−13,14q+, +der(14)t(11;14)(q11;p11),+15,+19,+21[15] 46,XY[5] |
| 8 | 46,X[6] 43,XY,−1,trp(1)(q12q25),der(1)t(1;8)(q10;q10), der(12)t(1;12)(q10;q24),−13,−14,add(14)(q32)[14] |
| 9 | 44∼46,XX,t(6;7)(q23;q22),−8,der(12)t(1;12)(q10;q24.3),del(13)(q12q21),der(13)t(13;17)(q10;q10),der(14)t(14;17)(q10;q10),+17, +19,der(19)t(1;19)(q10;q11)[19] 46,XX[1] |
| Translocations of 1q10 to 14p11 | |
| 10 | 47,XY,+9,der(14)t(1;14)(q10;p11.2)[18] 46,XY[2] |
| 11 | 44∼46,XY,del(1)(p31p35),t(8;13)(q24.3;q13),del(12)(p11),del(13)(q14q22),−14,der(14)t(1;14)(q10;p11),−20[cp6] 46,XY[16] |
| Translocations of 1q10 to 15p10 | |
| 12 | 74∼76,XX,+X,+del(1)(p21),add(2)(q27),+add(2)(q27),+der(3)t(3;21)(p11;?q11)×2,+4,+5,+5,+6,+6,del(7)(p15),+del(7)(p15), t(8;21)(q22;q22),+9,+9,+12,+12,+der(15)t(1;15)(q10;p10)×2,+16,+16,+17,+17,+18,+18,+19,+20,+20,+21,+21, +der(21)t(8;21)(q22;q22),+22[cp16] 46,XX[4] |
| 13 | 46,XY[12] 43,X,−Y,der(4)t(1;4)(q11;q35),−13,−14,−15,+der(15)t(1;15)(q10;p10),−21,+der(21)t(1;21)(q11;p11),−22[8] |
| 14 | 46,XY[18] 47,X,−Y,+5,t(8;14)(q24;q32),+9,−13,der(15)t(1;15)(q10;p22),der(16)t(1;16)(q10;p10),+19,add(19)(q13)[cp2]. |
| Translocations of 1q to 16p10 | |
| See patient No. 4 | |
| See patient No. 14 | |
| 15 | 41∼44,XY,add(1)(q21)×2,der(2)add(2)(p24)t(2;3)(q31;p14),add(3)(p21),add(3)(q29),t(5;19)(p15;q13),add(7)(p13),−10,−13,−14, add(14)(q32),der(16)t(1;16)(q10;p10),del(17)(p11),−18,add(21)(p13),add(22)(p11),del(22)(q13)[cp20] |
| 16 | 46,XY,t(8;14)(q24;q32),del(11)(q12),?der(14)t(11;14)(q13;q11),der(16)t(1;16)(p10;q10)[cp4]. 46,XY[13] |
| (Continued on following page) | |
| 17 | 45∼46,XY,−1,add(3)(q26.2),t(4;9)(q35;p11),add(8)(q22),t(11;14)(q13;q32),add(12)(q22),der(16)t(1;16)(q10;p10), der(19)t(1;19)(q10;p10)[cp3] 74∼88,idem,×2[cp8] 46,XY[6] |
| Translocations of 1q10 to 16p13 | |
| See patient No. 1 | |
| 18 | 52,XY,+7,+9,+9,+11,t(14;17)(q32;q?11),+15,der(16)t(1;16)(q10;p13),+17,add(18)(p11)[24] 53,idem,+8[5] |
| 19 | 46,XX[13] 44,XX,−13,−14,add(14)(q32),−14,der(16)t(1;16)(q10;p13)[7] |
| Translocations of 1q10 to 17q25 | |
| 20 | 45,X,−X,−1,+5,+6,del(6)(q15),+7,add(8)(p11.1),−12,−13,−14,+15,add(15)(p11),−16,add(17)(p11),der(17)t(1;17)(q10;q25), add(18)(p11.1),add(19)(q13.4)[cp3] 73∼77,XX,−X,−1,+2,+3,+5,+6,+7,+7,+7,−8,add(8)(p11;1),+11,+11,−12,−13,−14,+15,add(15)(p11)×2,−16,del(16)(q22),+17, der(17)t(1;17)(q10;q25)×2,add(18)(p11;1)×2,−19,+20,+22[cp7] 46,XX[10] |
| See patient No. 3 | |
| Translocations of 1q10 to 19p13 | |
| 21 | 68∼71,X,−X,−Y,+4,−5,add(5)(q31),del(6)(q21),+7,+7,del(8)(p11)×2,−13,+16,+der(16)t(3;16)(q10;q10),+17,−18,+19, +der(19)t(1;19)(q10;p13.3),+20,−21,−21,−22,+mar[cp17] 46,XY[3] |
| 22 | 44∼47,X,−X,+1,del(1)(p13p31),der(7)t(1;7)(q10;p22),add(8)(p23),−13, −14,+15,+15,der(19)t(1;19)(q10;p13.3)[cp8] 46,XX[12] |
| Translocations of 1q10 to 19q13 | |
| 23 | 77,XY,+Y,+1,+1,del(1)(p13),+2,+2,+3,+4,+6,+7,+7,+8,+9,+9,+11,+11,+12,+14,add(14)(q32)×2,+15,+16,+16, add(17)(p13)×2,+add(18)(q23)×2,+19,+der(19)t(1;19)(q10;q13.3),+20,+20,+21,+21,+22,+22,+mar[cp9] 46,XY[8] |
| 24 | 47∼48,X,−X,der(2)t(2;3)(p21;p21),add(3)(p21),+del(3)(p23),+6,add(8)(q24),del(8)(p11),der(9)t(6;9)(q13;p13),del(10)(q25), del(13)(q13q22),−14,+15,−16,add(16)(p13),add(17)(p11),del(18)(q12q21),der(19)t(1;19)(q10;q13),add(20)(p13),add(21)(q22), +22,+mar1,+mar2[cp4] 46,XX[16] |
| Translocations of 1q10 to 19p10 | |
| 25 | 46,XY,t(9;12)(q11;q22),t(11;14)(q13;q32),der(19)t(1;19)(q10;p10)7=90∼92,idem×2[cp7] 46,XY[6] |
| 26 | 50∼52,X,−X,+3,+5,+6,+7,+8,i(8)(q10)×2,+9,−13,der(13)t(9;13)(q11;q11.2),+15,+19,der(19)t(1;19)(q10;p10),+20[cp20] |
| 27 | 53∼54,XY,+3,+5,+6,add(6)(q23),+7,t(7;17)(p11.1;p12),−8,+9,+15,+19,+21,+mar[cp11] 53∼54,idem,der(19)t(1;19)(q10;p10)[cp4] 46,XY[5] |
| 28 | 57∼59,XX,+del(1)(p13),+2,+del(3)(p13),+der(3)add(3)(q29)t(3;13)(q26;?q12),+i(6)(p10),+7,+7,+9,der(9)add(9)(p24), +der(10)t(10;?22)(p11.2;?q11;2),i(12)(p10),−13,+15,+der(15)t(1;15)(q21;p11.1),−16,+18,+der(19)t(1;19)(q10;p10), del(20)(q11.1),del(22)(q12),+add(22)(q13)[cp20] |
| See patient No. 17 | |
| 29 | 53,X,−X,del(1)(p13p22),+der(1)t(1;19)(q10;p10)×2,+3,+6,del(6)(q21),+9,+15,+18,+21[2] 106,idem×2[1] 46,XX[17] |
| Translocations of 1q10 to 21q22 | |
| 30 | 46,XX[14] 47,XX,−2,add(2)(q31),+19,−21,+der(21)t(1;21)(q10;q22)[cp6] |
| 31 | 46,XX[17] 45∼48,X,−X,+7,+9,−13,+der(21)t(1;21)(q10;q22)[cp4] |
| Translocations of 1q10 to 21p11 | |
| 32 | 44,X,−X,der(6)add(6)(p25)t(1;6)(q12;q12),?inv(7)(p14p22),del(8)(p11),−13,del(20)(q12),der(21)t(1;21)(q10;p11)[cp9] 46,XX[11] |
| 33 | 143∼46,XY,t(1;21)(q21;p11),−2,add(6)(q?21),add(7)(p?22),−8,+9,−12,add(13)(p11),add(17)(p?11),+19[cp18] 46,XY[2] |
| Translocations of 1q10 to 22p11 | |
| 34 | 52,X,−X,+3,+5,+6,+7,der(8)t(?X;8)(?q13;p21),+11,+15,der(22)t(1;22)(q10;p11.2),+mar[2] 46,XX[13] |
| 35 | 46,XY[16] 40∼43,X,−Y,−13,add(14)(q32),−18,der(22)t(1;22)(q10;p11)[cp3] |
| 36 | 46,XY,+1,del(1)(p11),t(2;19)(q37;p13;1),inv(3)(p14q29),del(6)(p22),der(6)t(1;6)(q21;q23),add(7)(p22),−13,der(16)t(1;16)(q10;q22), +19,der(22)t(1;22)(q10;p11.2)[3] |
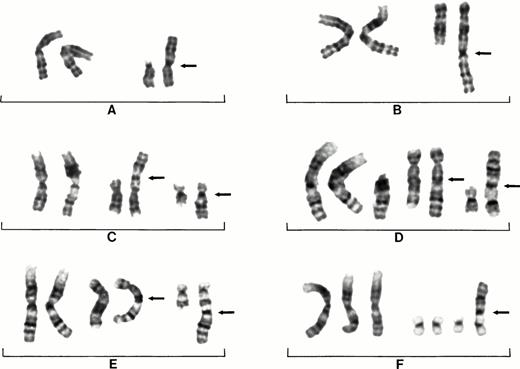
In decreasing order of frequency, 1q was translocated to 15pter in 10 patients (Fig 1A), 22pter in 6 patients, and to 13pter and 21pter in 3 patients each. One 1q was translocated to 21qter in 3 patients. Translocations to nonacrocentric chromosomes included 1q to 19qter in 5 patients, to 19pter in 2 patients, to 12qter in 3 patients (Fig 1B), to 8pter in 3 patients (Fig 1C), to 9pter in 2 patients, and to 17qter in 2 patients (Fig 1E). Whole-arm centromere to centromere translocations occurred most frequently between 16p and 1q in 9 patients.
Partial karyotypes from six different patients showing examples of recurring 1q aberrations seen in MM. Patient No.14 showing normal chromosomes 1 on left and der(15)t(1;15)(q10;q10) on right (A). Patient No. 7 showing normal chromosomes 1 on left and der(12)t(1;12)(q10;q24) on right (B). Patient No. 4 showing normal chromosomes 1 on left, der(8)t(1;8)(q10;p23) in middle, and der(16)t(1;16)(q10;p10) on right (C). Patient No. 1 showing two normal chromosomes 1 and an extra copy of 1q on left, a der(5)t(1;5)(q10;q15) in the middle, and der(16)t(1;16)(q10;p13) on right (D). Patient No. 2 showing der(5)t(1;5)(q10;q15) in middle and der(17)t(1;17)(q10;q25) on right (E). Patient No. 21 showing three chromosomes 1 on left with three normal copies of chromosome 19 and a der(19)t(1;19)(q10;p13) on right (F). Arrows indicate chromosome fusion points.
Partial karyotypes from six different patients showing examples of recurring 1q aberrations seen in MM. Patient No.14 showing normal chromosomes 1 on left and der(15)t(1;15)(q10;q10) on right (A). Patient No. 7 showing normal chromosomes 1 on left and der(12)t(1;12)(q10;q24) on right (B). Patient No. 4 showing normal chromosomes 1 on left, der(8)t(1;8)(q10;p23) in middle, and der(16)t(1;16)(q10;p10) on right (C). Patient No. 1 showing two normal chromosomes 1 and an extra copy of 1q on left, a der(5)t(1;5)(q10;q15) in the middle, and der(16)t(1;16)(q10;p13) on right (D). Patient No. 2 showing der(5)t(1;5)(q10;q15) in middle and der(17)t(1;17)(q10;q25) on right (E). Patient No. 21 showing three chromosomes 1 on left with three normal copies of chromosome 19 and a der(19)t(1;19)(q10;p13) on right (F). Arrows indicate chromosome fusion points.
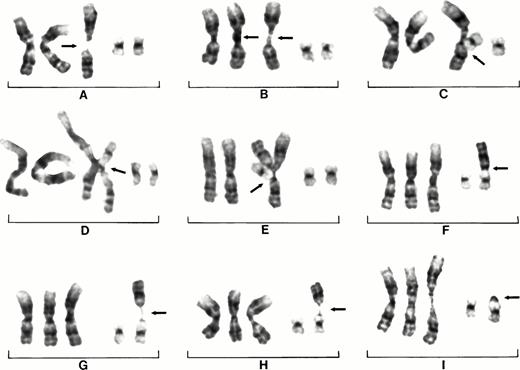
The association of centromeric decondensation, separation, and subsequent jumping 1q is illustrated in detail by partial karyotypes of nine cells each from three different patients. Patient No. 3 shows the jumping of 1q to 17q and subsequently to 7q (Fig 2A to I). The instability of chromosome 1 is associated with partial duplications but also with decondensed chromosomes 1 crossed at the centromere (Fig 2A to C). The chromosome crossovers in the decondensed centromeric regions suggest somatic association or pairing of centromeric sequences. Even the der(17)t(1;17)(q10;q25) fusion chromosome is involved in crossovers with the chromosomes 1 at the centromere, which also suggests somatic pairing of the centromeric 1q sequences between the chromosomes (Fig 2D and E). In some cells extreme decondensation of both chromosomes 1 and crossing over shows the fragility of these configurations (Fig 2F).
Partial karyotypes of nine different cells from patient No. 3 showing centromeric heterochromatin decondensation of chromosome 1 and association of 1q heterochromatin with 17q25. Duplications of 1q (open arrow) are found in addition to extra copies of translocated 1q (closed arrow) (A). Subtle decondensation of chromosomes 1 and crossing over of chromosomes 1 (B and C). Crossing over of der(17)t(1;17)(q10;q24) with chromosome 1 (arrows) (D and E). An extra free copy of 1q (G). Decondensation of chromosome 1 and der(17) (H). Chromosome 1q has jumped from der(17) to 7q leaving heterochromatin on 17q24 (arrow) (I).
Partial karyotypes of nine different cells from patient No. 3 showing centromeric heterochromatin decondensation of chromosome 1 and association of 1q heterochromatin with 17q25. Duplications of 1q (open arrow) are found in addition to extra copies of translocated 1q (closed arrow) (A). Subtle decondensation of chromosomes 1 and crossing over of chromosomes 1 (B and C). Crossing over of der(17)t(1;17)(q10;q24) with chromosome 1 (arrows) (D and E). An extra free copy of 1q (G). Decondensation of chromosome 1 and der(17) (H). Chromosome 1q has jumped from der(17) to 7q leaving heterochromatin on 17q24 (arrow) (I).
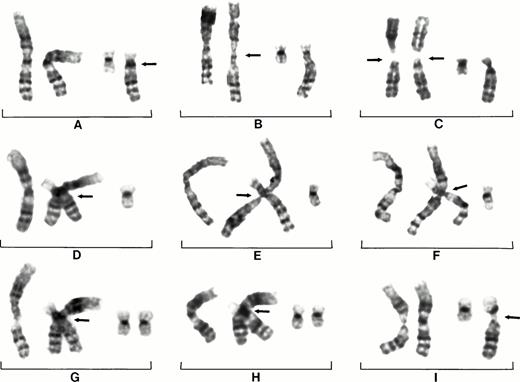
Patient No. 22 (Fig 3A to I) shows the sequence of events leading to the der(19)t(1;19)(q10;p13) with the 1q jumping to the telomere of the short arm of chromosome 19. First there is the decondensation of 1qh and apparent separation of 1q in some cells (Fig 3A), whereas other cells show decondensation of two copies of chromosome 1 (Fig 3B). The association of the short arm of 19 in the decondensed region of 1q can be clearly seen (Fig 3C), as can the association of 19p with an extra copy of 1q while still in the decondensed regions of a triradial configuration of 1q (Fig 3D). The best illustration of a triradial 1 configuration and the association of 19p is shown in Fig 3E. This cell appears to show the formation of an extra copy of 1q from the triradial and the initial fusion of 19p with the extra copy of 1q. This type of triradial configuration, which shows an apparent endoreduplication of 1q and association of the short arm of chromosome 19 with 1q, indicates the likely origin of der(19).
Partial karyotypes of nine different cells from patient No. 22 showing centromeric heterochromatin decondensation of chromosome 1, formation of triradial configurations, and movement of 1q. Decondensation of centromeric heterochromatin and apparent separation of 1p and 1q of one chromosome 1 (arrow), normal chromosomes 19 on right (A). Decondensation of two chromosomes 1 (B). Decondensation of chromosomes 1 with chromosome 19p associating in region of decondensation (arrow) (C). Decondensation of 1qh and assocation of 19p13 with decondensed heterochromatin (arrow); note there are now four copies of 1q (D). Decondensation of 1qh and association of 19p13 with 1qh (arrow) and an extra copy of 1q. Note clear triradial of chromosome 1 (E). The translocation of 1q to 19p13 as it is seen in the vast majority of cells (F). The continuing instability of 1q is illustrated by the apparent decondensation of 1q sequences as it is lost from 19p (arrows); note thread-like chromatin. (G and H). The loss of 1q from 19p is shown by only heterochromatin remaining on 19p (arrow) (I). Note small segments of heterochromtin left on the short arm of 19 in cells (G and H).
Partial karyotypes of nine different cells from patient No. 22 showing centromeric heterochromatin decondensation of chromosome 1, formation of triradial configurations, and movement of 1q. Decondensation of centromeric heterochromatin and apparent separation of 1p and 1q of one chromosome 1 (arrow), normal chromosomes 19 on right (A). Decondensation of two chromosomes 1 (B). Decondensation of chromosomes 1 with chromosome 19p associating in region of decondensation (arrow) (C). Decondensation of 1qh and assocation of 19p13 with decondensed heterochromatin (arrow); note there are now four copies of 1q (D). Decondensation of 1qh and association of 19p13 with 1qh (arrow) and an extra copy of 1q. Note clear triradial of chromosome 1 (E). The translocation of 1q to 19p13 as it is seen in the vast majority of cells (F). The continuing instability of 1q is illustrated by the apparent decondensation of 1q sequences as it is lost from 19p (arrows); note thread-like chromatin. (G and H). The loss of 1q from 19p is shown by only heterochromatin remaining on 19p (arrow) (I). Note small segments of heterochromtin left on the short arm of 19 in cells (G and H).
Patient No. 26 illustrates centromeric instability not only in chromosome 1 but also in chromosome 19. In this patient, der (19) is created by the joining of centromeric sequences rather than the centromeric telomeric fusions described above. This patient showed centromeric decondensation and fragility (Fig 4A to C), and many cells showed the crossing over of the der(19) with a decondensed chromosome 1, again suggesting somatic pairing of centromeric 1q sequences (Fig 4D to H).
Partial karyotypes of nine different cells from patient No. 26 showing centromeric heterochromatin decondensation of chromosomes 1, formation of triradial configurations, and movement of 1q. Cell showing normal 1s and der(19)t(1;19)(q10;p10) (arrow) (A). Chromosome 1 on right showing decondensation (arrow) (B). Both chromosomes 1 showing separations of short and long arms (arrows) (C). Somatic pairings of der (19)t(1;19)(q10;p10) and chromosomes 1 (arrows) (D through H). Subsequent instability of 19p10 and 1q10 (arrow) (I).
Partial karyotypes of nine different cells from patient No. 26 showing centromeric heterochromatin decondensation of chromosomes 1, formation of triradial configurations, and movement of 1q. Cell showing normal 1s and der(19)t(1;19)(q10;p10) (arrow) (A). Chromosome 1 on right showing decondensation (arrow) (B). Both chromosomes 1 showing separations of short and long arms (arrows) (C). Somatic pairings of der (19)t(1;19)(q10;p10) and chromosomes 1 (arrows) (D through H). Subsequent instability of 19p10 and 1q10 (arrow) (I).
DISCUSSION
The primary numerical chromosome aberrations seen in MM karyotypes apparently evolve over an extended period of time as a subclinical phenomenon. In later stages of progressive MM, cytogenetic evolution takes place, resulting in secondary chromosomal aberrations commonly involving chromosome 1. Structural aberrations of both arms involving reciprocal translocations are the most common findings. However, a special type of whole-arm or jumping translocation somehow including an extra copy of 1q and its subsequent movement to another chromosome creates a partial trisomy for the whole long arm. Whole-arm translocations of 1q are different from jumping translocations because in jumping translocations the 1q segment becomes unstable and moves (jumps) around the karyotype to more than one nonhomologous chromosome. Trisomy for the long arm of chromosome 1 is common in many types of cancer15-18 and has been reported previously in leukemias and lymphomas showing multiple telomeric associations with different chromosomes.13,19-30 Experimental evidence has shown that dup 1q might be a secondary aberration associated with disease progression26; however, they may also be primary aberrations in some cases.27 The correlation of trisomy for 1q with the progression of malignancy has been correlated with the metastatic potential in colon and renal cell carcinomas, including the involvement of the SKI oncogene located at 1q21.31 32
The derivative (der) chromosomes we report have been reported previously, with the exception of the der(5)t(1;5) (q10;q15) in the present study. The der(5) was found only in conjunction with other 1q aberrations and thus may constitute a further unique step in the secondary evolution of the MM karyotype. The recurring der(15)t(1;15)(q10;q10) in this report is a rare but nonrandom change also associated with myelodysplastic syndrome and myeloproliferative disorders. This aberration has been reported as the sole aberration in most patients.33,34 The der(16)t(1;16)(q10;p10) has been reported in a wide variety of malignancies, including breast cancer, Ewing's sarcomas, and Wilms' tumors. This aberration has also been reported as the sole aberration in some cases, but as a secondary aberration in most patients.35-38 This whole-arm translocation has been confirmed by fluorescence in situ hybridization using probes reacting with alphoid and classic satellite DNA.39 It may be that the probability of recombination of these centromeric repeats is favored by the sequence homology shared in the regions corresponding to the t(1;16) exchange points. The centromeric regions of chromosomes 1,9,16 and Y contain satellite III DNA consensus sequences largely consisting of (GGAAT)nrepeats and small clusters of satellite III DNA interspersed among the alpha-satellite DNA.40 Guanine-rich motifs, such as telomere sequences (TTAGGG)n, adopt highly stable intra-strand and inter-strand duplexes and possibly tetraplex structures that may favor recombination in this region.41,42 It has further been suggested that tetra-strand DNA has a function related to nonhomologous recombination, telomere-telomere recombination, and immunoglobulin switch recombination.42
Jumping translocations involving multiple chromosomes are a rare phenomenon, the mechanisms of which remain obscure. However, the types of chromosome 1 centromeric decondensation observed in our patients appear to be similar and reminiscent of changes observed in cells treated with the hypomethylating agent 5-azacytidine.43 44This suggests that undermethylation is associated with the decondensation of the heterochromatic regions. Hypomethylation could be induced as a side effect of cytotoxic therapy, have a viral association, or be part of an unknown process associated with tumor progression.
A viral origin for jumping translocations and juxta-centromeric fragility in neoplasia has been suggested.19 It is known that gene products of certain DNA cancer viruses (SV-40, human papilloma virus, and adenovirus) can alter cellular proteins and affect cell-cycle checkpoints, thereby inducing karyotype instability.45 A variety of chromosome aberrations, including telomeric associations, dicentric chromosomes, and aneuploidy, have been induced in human fibroblasts by the SV-40 virus,46-48 as have jumping translocations.48,49 An alternative explanation to viral induction could be that, following DNA duplication, the hypomethylated decondensed state of the paracentromeric heterochromatic regions of homologous chromosomes preserves the interphase somatic pairing and accounts for the multiradial associations observed at metaphase.50 The persistent somatic pairing could result in multibranched chromosomes of varying sizes from duplications of 1q occurring in these cells. In fact, azacytidine-treated cells show uncoiling and somatic associations and indicate molecular exchanges between classical satellite-containing regions in homologous and nonhomologous chromosomes.51 52
As there are several possible mechanisms involved in jumping translocations, our findings suggest that a number of chromosomal landmarks may be associated with the process of “jumping copies of 1q.” In our patients we found recurrent centromeric decondensations and centromeric separations as signs of hypomethylation of the centromeric heterochromatin. The duplication of part of 1q is often seen in the same patients who subsequently show duplications of the entire 1q. Decondensed chromosomes 1 frequently cross over apparently as a result of sequence homology (somatic pairing) in the stretched regions (Figs 2-4). Triradials as seen in patient No. 22 (Fig 3E) are rare events and are believed to arise from the partial endoreduplication of a chromosome arm.53 Interestingly, the combination of hypomethylation and the appearance of triradial chromosome configurations as observed here have been reported elsewhere in both neoplastic and non-neoplastic disorders. A rare pediatric immunodeficiency syndrome (ICF syndrome) shows the most striking array of triradial and multiradial chromosomes.54 These patients show an embryonic-like methylation pattern of classical satellite DNA and multibranched 1q in peripheral blood lymphocytes.55 In neoplastic disorders, decondensation of 1q and jumping translocations have been reported in an HIV-related non-Hodgkin's lymphoma.30 Although the factors involved in the induction of the centromeric decondensation may be different, and the resulting clonal expansion is different, the striking similarity of chromosome triradials is intriguing.
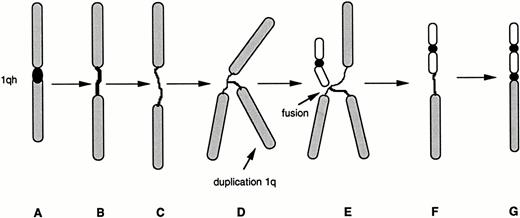
The hypothesized model of the clonal evolution of tumor-cell populations suggests that during the cytogenetic progression of malignancy acquired genetic lability permits the stepwise selection of variant subclones.56 During this evolution tumor-cell populations emerge that may or may not be viable. Nearly all variants are eliminated, but occasionally one has a selective advantage and becomes the predominant subpopulation. It is likely that hypomethylation is induced by a variety of mechanisms. However, hypomethylation appears to be the critical event associated with the decondensation and subsequent instability of the classical satellite sequences associated with the pericentromeric heterochromatin of chromosome 1 (Fig 2). This decondensation in some patients is apparently followed by duplication of 1q regions adjacent to the heterochromatin of chromosome 1 resulting in what presents as triradial chromosomes 1q (Fig 3). These configurations may result from somatic pairings of chromosome 1 with the resulting loss of 1p and the subsequent translocation or jumping of the 1q to other chromosomes. The finding of triradial chromosomes in patients is extremely rare because these configurations are unstable and probably lost as micronuclei. Apparently, in some patients, these configurations do not evolve, whereas in other patients the entanglement of other chromosomes in the decondensed heterochromatic regions adjacent to an extra copy of 1q may cause chromosome arm exchanges (Fig 4). The highly decondensed heterochromatin may provide an opportunity for the fusion of this chromosome segment to other chromosomes because the hypomethylated segments may favor recombination. Once the 1q has translocated to another chromosome it is likely the only stable chromosome change to survive from the transitional (unstable) triradial. Our data suggest a speculative model for heterochromatin decondensation in the dynamics of 1q translocations (Fig 5).
Possible model for the decondensation of juxtacentromeric heterochromatin in jumping translocations of 1q. A spectrum of 1qh decondensations occur in MM cells, ranging from an apparently normal 1qh region (A), to slightly elongated 1qh (B), or highly decondensed and thread-like 1qh (C). Partial endoreduplication of 1q apparently occurs while heterochromatin is decondensed (D). Fusions with telomeres of nonhomologous chromosomes may be facilitated by the highly decondensed heterochromatin (E). The origin of a new derivative chromosome with 1q fused to telomere (F). Condensation of heterochromatin on derivative chromosome (G) creates the appearance of a typical whole-arm jumping translocation.
Possible model for the decondensation of juxtacentromeric heterochromatin in jumping translocations of 1q. A spectrum of 1qh decondensations occur in MM cells, ranging from an apparently normal 1qh region (A), to slightly elongated 1qh (B), or highly decondensed and thread-like 1qh (C). Partial endoreduplication of 1q apparently occurs while heterochromatin is decondensed (D). Fusions with telomeres of nonhomologous chromosomes may be facilitated by the highly decondensed heterochromatin (E). The origin of a new derivative chromosome with 1q fused to telomere (F). Condensation of heterochromatin on derivative chromosome (G) creates the appearance of a typical whole-arm jumping translocation.
The equilibrium between proliferation and programmed cell death in MM cells is believed to be controlled in part by cytokines. In this respect, growth control of MM cells may be affected by increased gene dosage related to duplications of part or all of the long arm. The observation of extra copies of 1q suggests several possibilities for low-level gene amplification indicated by the presence of genes related to MM biology. The interleukin-6 (IL-6) signaling pathway may possibly be affected by the amplification of the 1q21 region, which is the site of IL-6RA.57 Other genes of interest in this region include C-reactive protein (CRP) and amyloid P component (APCS), both localized to 1q21-23,58 and pre–B-cell leukemia transcription factor 1 (PBX1) at 1q23.59
Chromosome aberrations often have diagnostic and prognostic significance. The roles played by cytotoxic drugs, ionizing radiation, or oncogenic viruses in the evolution of secondary chromosomal aberrations in MM are still far from clear. It seems likely that these factors interact with the cell genome in a variety of ways to bring about at least a gene dosage effect caused by the extra copies of 1q. The evolution of centromeric instability appears to be the precursor for subsequent telomeric fusions and jumping translocations in some patients. Decondensation and stretching of centromeric heterochromatin is associated with the persistence of somatic pairing, multibranched chromosome arms, whole-arm deletion, duplication, isochromosomes, and centromeric fragility.52,53 60 The progression of centromeric destabilization in these patients, from simple heterochromatic decondensation to subsequent multibranching and jumping translocations, shows a sequence of events in its progression. We speculate that hypomethylation-induced pericentromeric heterochromatin decondensation is an initiating event.
ACKNOWLEDGMENT
We gratefully acknowledge the expert technical assistance of cytogenetic technologists Eddie Thomas, Charles Swanson, Linda Goosen, Mamie Crowson, Gael Sammartino, Emmett Jones, and Janet Lukacs.
Supported in part by Grant No. CA55819 from the National Cancer Institute, National Institutes of Health, Bethesda, MD.
Address reprint requests to Jeffrey R. Sawyer, PhD, Cytogenetics Laboratory, Arkansas Children's Hospital, 800 Marshall St, Little Rock, AR 72202.
The publication costs of this article were defrayed in part by page charge payment. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. section 1734 solely to indicate this fact.


This feature is available to Subscribers Only
Sign In or Create an Account Close Modal