Abstract
The differentiation inhibitory factor nm23 can inhibit the differentiation of murine and human myeloid leukemia cells. We recently reported that nm23 genes were overexpressed in acute myelogenous leukemia (AML), and a higher level of nm23-H1expression was correlated with a poor prognosis in AML, especially in AML-M5 (acute monocytic leukemia). To evaluate the importance ofnm23 expression as a prognostic factor in AML, we compared it with other putative prognostic factors in AML. An analysis of the correlation between nm23 expression and the clinical parameters of 110 patients with AML demonstrated that increased nm23-H1mRNA levels were associated with resistance to initial chemotherapy and with reduced overall survival. Multivariate analysis using Cox's proportional hazard model also showed that elevated nm23-H1mRNA levels significantly contributed to the prognosis of patients with AML. Especially in AML-M5, nm23-H1 status was the most important prognostic factor. Furthermore, to determine whether we can apply the results observed in AML to other hematologic malignancies, we investigated the relative levels of nm23-H1 and nm23-H2transcripts in 149 patients with hematologic neoplasms, including 110 with de novo AML, 9 with de novo acute lymphoblastic leukemia, 14 with myelodysplastic syndrome, 16 with chronic myelogenous leukemia (CML), and 5 normal subjects by the reverse transcriptase-polymerase chain reaction. Expression of nm23-H1 was significantly higher in all the hematologic neoplasms, except CML in chronic phase, than in normal blood cells. nm23 may have a prognostic effect in these hematologic malignancies as well as in AML.
ALTHOUGH NORMAL hematopoiesis can be controlled by various positive and negative regulatory molecules, leukemic cells are arrested in less-differentiated stages of development. This suggests that both positive and negative regulators are important for the differentiation of leukemic cells. We previously reported that a nondifferentiating mouse myeloid leukemia cell line produced differentiation inhibiting factors (I-factors). Suppression of the production of these I-factors resulted in nondifferentiating leukemic cells becoming sensitive to differentiation inducers. One of these I-factors was purified as a homologue of nm23.1-3
The differentiation inhibitory factor nm23 can inhibit the differentiation of murine and human myeloid leukemia cells, andnm23 expression is greatly increased during blast formation in normal lymphocytes.1-4 These findings suggest thatnm23 genes play a role in the growth and differentiation of normal and malignant hematopoietic cells. Few studies have focused on the role of nm23 in human hematopoietic malignancies.5 We recently reported that nm23 genes were overexpressed in acute myelogenous leukemia (AML) and thatnm23-H1 expression was significantly correlated with a poor prognosis in AML, especially in AML-M5.6 It has been reported that high-grade non-Hodgkin's lymphoma (NHL) and Hodgkin's lymphoma exhibited significantly higher levels of nm23-H1expression than low-grade NHL.7 These studies suggest thatnm23-H1 expression in human hematopoietic malignancies is associated with the aggressiveness of the disease.
In the present study, to evaluate the importance of nm23expression as a prognostic factor in AML, we compared the levels ofnm23 expression with other putative prognostic factors in AML by a multivariate analysis. Furthermore, to determine whether we can apply the results observed in AML to other hematologic malignancies, we examined the expression of nm23-H1 and -H2 genes in various human hematologic malignancies other than AML.
MATERIALS AND METHODS
Patients' samples.
Bone marrow (BM) or peripheral blood (PB) samples were obtained from 149 patients with newly diagnosed hematologic neoplasms, consisting of 110 with de novo AML, including 42 previously reported cases,6 9 with de novo acute lymphoblastic leukemia (ALL), 9 with myelodysplastic syndrome (MDS; 1 with chronic myelomonocytic leukemia [CMMoL], 4 with refractory anemia with excess of blasts [RAEB], and 4 with RAEB-T), 5 with MDS overt leukemia, 9 with chronic myelogenous leukemia in the chronic phase (CML-CP), and 7 with CML in blast crisis (CML-BC) with their informed consent at onset, before chemotherapy. De novo AML, ALL, and MDS were classified according to the criteria devised by the French-American-British (FAB) Committee. AML patients were treated with cytosine arabinoside (or behenoyl cytosine arabinoside), daunorubicin, and with or without prednisolone and/or 6-mercaptopurine, and AML-M3 patients were consecutively treated with all-transretinoic acid for remission induction therapy.8,9 Treated patients were judged to have achieved complete remission (CR) when BM aspirates showed trilineage regeneration with less than 5% blasts by morphology and immunocytochemistry in the presence of a normal blood count that persisted for at least 1 month. Patients who died of toxic complication (infection or bleeding) before the time of expected marrow recovery were not evaluated. All other patients were considered nonresponsive (NR). To purify leukemic cells, heparinized PB cells or BM aspirates were mixed with an equal volume of RPMI-1640 medium and centrifuged on Ficoll-Hypaque (Pharmacia, Uppsala, Sweden). Normal BM and PB cells were obtained from healthy volunteers after obtaining their informed consent. Mononuclear cells from normal BM and PB cells were separated over Ficoll-Hypaque. Total RNA was extracted as described by Chomczynski and Sacchi, using guanidium thiocyanate.10
Reverse transcriptase-polymerase chain reaction (RT-PCR).
Quantitative RT-PCR was performed using a GeneAmp RNA PCR kit (Takara, Tokyo, Japan), as reported previously.6 The oligonucleotides used in PCR amplification were as follows: sense strand, 5′-ATGGCCAACTGTGAGCGTACC-3′; antisense strand, 5′-CATG TATTTCACCAGGCCGGC-3′ for nm23-H1; sense strand, 5′-ATGGCCAACCTGGAGCGCACC-3′; antisense strand, 5′-TCCC CACGAATGGTGCCTGGC-3′ for nm23-H2; and sense strand, 5′-ACATCGCTCAGACACCATGG-3′; antisense strand, 5′-GTAGTTGAGGTCAATGAAGGG-3′ for glyceraldehyde-3-phosphate dehydrogenase (gapdh). PCR consisted of 35 cycles fornm23-H1 and 25 for nm23-H2 and gapdh, with denaturing at 95°C for 1 minute, annealing at 60°C for 1 minute, and extension at 72°C for 0.5 minutes. The reaction was performed in a GeneAmp PCR system 9600 (Perkin Elmer, Norwalk, CT). The PCR products were then subjected to 6% polyacrylamide gel electrophoresis, and the dried gel was exposed to imaging plates (Fuji Film Co, Ltd, Tokyo, Japan) at room temperature for 15 to 20 minutes. The results of autoradiography were quantified using a Fuji Bio-Image Analyzer BAS2000 (Fuji Film Co).
Statistical analysis.
Differences between groups were evaluated by Mann-Whitney's U-test (nonparametric analysis), and P < 0.05 indicated a significant difference. Pearson's correlation coefficient was used to evaluate the correlation between paired values. Survival curves of patients were prepared by the Kaplan-Meier method, and differences between the survival curves were evaluated using the log-rank and generalized Wilcoxon's tests. A multivariate analysis of the prognosis was performed using Cox's proportional hazard model.
RESULTS
Relationship between nm23 expression and clinical data in AML.
We examined the levels of nm23 expression in BM and PB samples from 110 patients with newly diagnosed de novo AML, including 3 M0, 20 M1, 29 M2, 19 M3, 19 M4, 18 M5 (7 M5a and 11 M5b), and 2 M6. To normalize the differences in RNA loading for RT-PCR and in RNA degradation in individual samples, the values of nm23-H1and -H2 gene expression were divided by that of thegapdh gene for comparison with the values in erythroleukemia HEL cells, which were defined as 100 (the expression “Index”). Both nm23-H1 and -H2 genes were overexpressed in these 110 AML samples (Table 1). To evaluate the relative importance of nm23 expression as a prognostic factor in AML, we determined the correlation of nm23-H1 or-H2 expression with age, sex, white blood cell (WBC) count, lactate dehydrogenase (LDH) level, surface marker CD7, chromosomal aberration, and the response to initial chemotherapy (Table 2). Increased LDH was correlated with nm23-H1 (P = .006) and nm23-H2expression (P = .038). CD7+ AML showed highernm23-H1 expression than CD7− AML (P = .019). A good response to initial chemotherapy was inversely correlated with nm23-H1 (P = .020). A total of 100 samples were evaluable in terms of the response to treatment. Thirty-two patients failed to achieve remission after the initial chemotherapy. The drug-resistant AML samples expressed a significantly highernm23-H1 level (nm23-H1 index = 144 ± 212) than those from the 68 AML patients who achieved CR (nm23-H1index = 97 ± 115). The expression levels of nm23-H1 and-H2 in AML-M3 leukemia with t(15;17) were lower than those in the other AMLs without chromosomal aberrations (H1, P= .010; H2, P = .045; Table 2).
Levels of nm23-H1 and -H2 Index in Normal and Hematologic Neoplasms
| Diagnosis (no.) . | nm23-H1 Index . | P . | nm23-H2 Index . | P . |
|---|---|---|---|---|
| Normal (5) | 26 ± 26 | 39 ± 24 | ||
| AML (110) | 107 ± 147 | .003-150 | 95 ± 110 | .023-150 |
| ALL (9) | 87 ± 41 | .007-150 | 54 ± 19 | .223 |
| MDS (9) | 91 ± 81 | .011-150 | 58 ± 30 | .184 |
| MDS overt leukemia (5) | 147 ± 141 | .012-150 | 52 ± 34 | .569 |
| CML-CP (9) | 35 ± 20 | .204 | 42 ± 10 | .633 |
| CML-BC (7) | 102 ± 34 | .006-150 | 83 ± 39 | .013-150 |
| Diagnosis (no.) . | nm23-H1 Index . | P . | nm23-H2 Index . | P . |
|---|---|---|---|---|
| Normal (5) | 26 ± 26 | 39 ± 24 | ||
| AML (110) | 107 ± 147 | .003-150 | 95 ± 110 | .023-150 |
| ALL (9) | 87 ± 41 | .007-150 | 54 ± 19 | .223 |
| MDS (9) | 91 ± 81 | .011-150 | 58 ± 30 | .184 |
| MDS overt leukemia (5) | 147 ± 141 | .012-150 | 52 ± 34 | .569 |
| CML-CP (9) | 35 ± 20 | .204 | 42 ± 10 | .633 |
| CML-BC (7) | 102 ± 34 | .006-150 | 83 ± 39 | .013-150 |
The mRNA levels were normalized for gapdh mRNA. The positive control (the Index = 100) is represented by RNA extracted from the HEL cell line. Values are means ± SD. Analyzed by Mann-Whitney's U-test (v normal).
P < .05.
Relationship Between nm23 Expression and Clinical Factors in AML
| Clinical Factor (no.) . | nm23-H1 Index . | P . | nm23-H2 Index . | P . |
|---|---|---|---|---|
| Age (yr) | ||||
| ≤50 (53) | 101 ± 171 | 95 ± 119 | ||
| >50 (57) | 112 ± 123 | .275 | 95 ± 101 | .862 |
| Sex | ||||
| M (68) | 102 ± 87 | 89 ± 58 | ||
| F (42) | 115 ± 212 | .107 | 104 ± 162 | .216 |
| WBC (×109/L) | ||||
| ≤50 (79) | 103 ± 153 | 85 ± 102 | ||
| >50 (29) | 121 ± 136 | .266 | 119 ± 130 | .079 |
| LDH | ||||
| ≥N, ≤5 × N (92) | 96 ± 142 | 84 ± 96 | ||
| >5 × N (14) | 188 ± 179 | .006* | 159 ± 175 | .038* |
| CD7 | ||||
| Negative (70) | 105 ± 115 | 87 ± 94 | ||
| Positive (17) | 188 ± 279 | .019* | 141 ± 193 | .121 |
| Chromosomal aberration | ||||
| − (42) | 97 ± 80 | 85 ± 53 | ||
| + (65) | 116 ± 180 | .398 | 100 ± 136 | .438 |
| − (42) | 97 ± 80 | 85 ± 53 | ||
| t(8;21) (13) | 113 ± 108 | .898 | 79 ± 60 | .677 |
| t(15;17) (17) | 51 ± 29 | .010* | 63 ± 52 | .045* |
| inv(16) (3) | 78 ± 21 | .891 | 89 ± 43 | .733 |
| 11q23 (3) | 81 ± 66 | >.999 | 77 ± 44 | >.999 |
| MAKA (6) | 300 ± 471 | .554 | 213 ± 317 | .513 |
| others (25) | 122 ± 148 | .866 | 112 ± 28 | .856 |
| Response to initial chemotherapy | ||||
| CR (68) | 97 ± 115 | 91 ± 96 | ||
| NR (32) | 144 ± 212 | .020* | 110 ± 147 | .597 |
| Clinical Factor (no.) . | nm23-H1 Index . | P . | nm23-H2 Index . | P . |
|---|---|---|---|---|
| Age (yr) | ||||
| ≤50 (53) | 101 ± 171 | 95 ± 119 | ||
| >50 (57) | 112 ± 123 | .275 | 95 ± 101 | .862 |
| Sex | ||||
| M (68) | 102 ± 87 | 89 ± 58 | ||
| F (42) | 115 ± 212 | .107 | 104 ± 162 | .216 |
| WBC (×109/L) | ||||
| ≤50 (79) | 103 ± 153 | 85 ± 102 | ||
| >50 (29) | 121 ± 136 | .266 | 119 ± 130 | .079 |
| LDH | ||||
| ≥N, ≤5 × N (92) | 96 ± 142 | 84 ± 96 | ||
| >5 × N (14) | 188 ± 179 | .006* | 159 ± 175 | .038* |
| CD7 | ||||
| Negative (70) | 105 ± 115 | 87 ± 94 | ||
| Positive (17) | 188 ± 279 | .019* | 141 ± 193 | .121 |
| Chromosomal aberration | ||||
| − (42) | 97 ± 80 | 85 ± 53 | ||
| + (65) | 116 ± 180 | .398 | 100 ± 136 | .438 |
| − (42) | 97 ± 80 | 85 ± 53 | ||
| t(8;21) (13) | 113 ± 108 | .898 | 79 ± 60 | .677 |
| t(15;17) (17) | 51 ± 29 | .010* | 63 ± 52 | .045* |
| inv(16) (3) | 78 ± 21 | .891 | 89 ± 43 | .733 |
| 11q23 (3) | 81 ± 66 | >.999 | 77 ± 44 | >.999 |
| MAKA (6) | 300 ± 471 | .554 | 213 ± 317 | .513 |
| others (25) | 122 ± 148 | .866 | 112 ± 28 | .856 |
| Response to initial chemotherapy | ||||
| CR (68) | 97 ± 115 | 91 ± 96 | ||
| NR (32) | 144 ± 212 | .020* | 110 ± 147 | .597 |
Values are means ± SD. Analyzed by Mann-Whitney's U-test.
Abbreviation: MAKA, major karyotypic abnormalities.
P < .05.
Overall survival in AML.
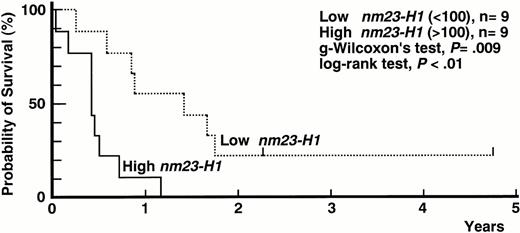
One hundred three evaluable AML patients were classified into groups based on age, sex, WBC count, LDH, CD7, chromosomal aberration, andnm23-H1 and -H2 expression. Overall survival in each group of patients is shown in Table 3. There were significant differences in the survival time between the patients classified by age, WBC count, LDH level, presence or absence of chromosomal aberration on t(8;21) and t(15;17), and nm23-H1and -H2 expression levels. Cox's proportional hazard model was used to evaluate the relative importance of the putative prognostic factors in Table 3. Of these factors, chromosomal aberration on t(15;17) and t(8;21) had a good prognostic effect. However, if these were excluded from a multivariate analysis of the overall survival of AML patients, nm23-H1 was the most important factor (P= .034, Table 4). Especially in AML-M5, nm23-H1 mRNA expression significantly contributed to the prognosis, and nm23-H1 status was the most important prognostic factor (P = .003, Table 4 and Fig 1). These results showed that the levels of nm23 gene expression represent a new prognostic factor for AML.
Correlation of Overall Survival With Clinical and Biologic Features of 103 Patients With AML
| Clinical Factor (no.) . | Log-Rank (P) . | g-Wilcoxon (P) . |
|---|---|---|
| Age (yr) | ||
| ≤50 (50) | ||
| >50 (53) | .005 | <.001 |
| Sex | ||
| M (63) | ||
| F (40) | NS | NS |
| WBC (×109/L) | ||
| ≤50 (74) | ||
| >50 (28) | .004 | <.001 |
| LDH | ||
| ≥N, ≤5 × N (87) | ||
| >5 × N (14) | .003 | <.001 |
| CD7 | ||
| Negative (67) | ||
| Positive (17) | NS | NS |
| Chromosomal aberration | ||
| − (40) | ||
| + (61) | NS | NS |
| − (40) | ||
| t(8;21) (13) | .041 | NS |
| t(15;17) (14) | <.005 | <.005 |
| inv(16) (3) | NS | NS |
| 11q23 (3) | NS | NS |
| MAKA (5) | NS | NS |
| Others (24) | NS | NS |
| nm23-H1 Index | ||
| ≤100 (71) | ||
| >100 (32) | <.001 | <.005 |
| nm23-H2 Index | ||
| ≤110 (77) | ||
| >110 (26) | NS | <.05 |
| Clinical Factor (no.) . | Log-Rank (P) . | g-Wilcoxon (P) . |
|---|---|---|
| Age (yr) | ||
| ≤50 (50) | ||
| >50 (53) | .005 | <.001 |
| Sex | ||
| M (63) | ||
| F (40) | NS | NS |
| WBC (×109/L) | ||
| ≤50 (74) | ||
| >50 (28) | .004 | <.001 |
| LDH | ||
| ≥N, ≤5 × N (87) | ||
| >5 × N (14) | .003 | <.001 |
| CD7 | ||
| Negative (67) | ||
| Positive (17) | NS | NS |
| Chromosomal aberration | ||
| − (40) | ||
| + (61) | NS | NS |
| − (40) | ||
| t(8;21) (13) | .041 | NS |
| t(15;17) (14) | <.005 | <.005 |
| inv(16) (3) | NS | NS |
| 11q23 (3) | NS | NS |
| MAKA (5) | NS | NS |
| Others (24) | NS | NS |
| nm23-H1 Index | ||
| ≤100 (71) | ||
| >100 (32) | <.001 | <.005 |
| nm23-H2 Index | ||
| ≤110 (77) | ||
| >110 (26) | NS | <.05 |
Abbreviation: NS, not significant.
Multivariate Analysis of Clinical Factors on Overall AML and AML-M5 Patient Survival
| Variable* . | All AML (P) . | AML-M5 (P) . |
|---|---|---|
| Age (≤50/>50 yr) | .037 | .218 |
| Sex (M/F) | .145 | .926 |
| WBC (+Lz50/>50 × 109/L) | .036 | .516 |
| LDH (≥N, ≤5 × N/>5 × N) | .113 | .722 |
| nm23-H1(≤100/>100) | .034 | .003 |
| nm23-H2 (≤110/>110) | .691 | .396 |
| Variable* . | All AML (P) . | AML-M5 (P) . |
|---|---|---|
| Age (≤50/>50 yr) | .037 | .218 |
| Sex (M/F) | .145 | .926 |
| WBC (+Lz50/>50 × 109/L) | .036 | .516 |
| LDH (≥N, ≤5 × N/>5 × N) | .113 | .722 |
| nm23-H1(≤100/>100) | .034 | .003 |
| nm23-H2 (≤110/>110) | .691 | .396 |
*Except the factors of chromosomal aberration, t(15;17) and t(8;21).
Survival curves of AML-M5 patients. High nm23-H1(>100) patients (n = 9, solid line) had a worse prognosis than lownm23-H1 (100) patients (n = 9, broken line) (generalized Wilcoxon's test, P = .009; log-rank test,P < .01).
Survival curves of AML-M5 patients. High nm23-H1(>100) patients (n = 9, solid line) had a worse prognosis than lownm23-H1 (100) patients (n = 9, broken line) (generalized Wilcoxon's test, P = .009; log-rank test,P < .01).
Application of the expression of the nm23-H1 and -H2 genes to other hematologic neoplasms.
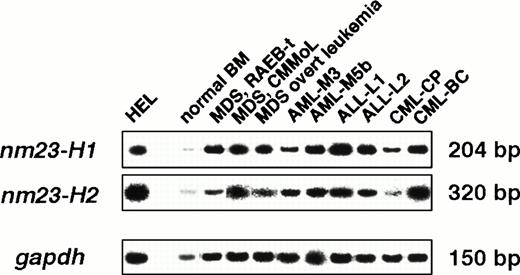
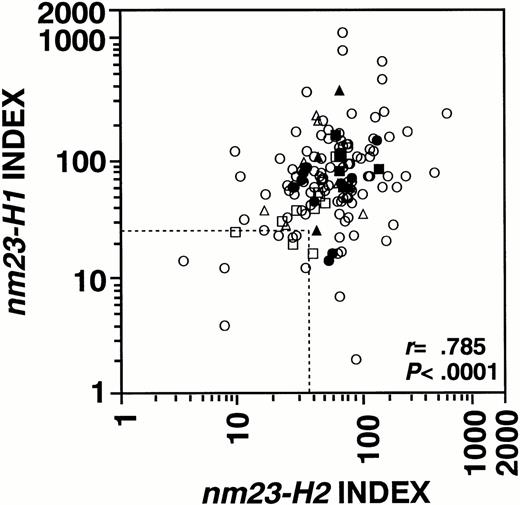
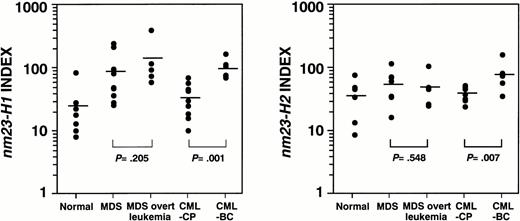
To apply nm23 expression to hematologic neoplasms other than AML, we examined the mRNA levels of the nm23-H1 and -H2genes in hematologic neoplasms, as shown in Table 1 and Fig 2. The expression levels of thenm23-H1 gene were significantly higher in MDS (P = .011), MDS overt leukemia (P = .012), AML (P = .003), ALL (P = .007), and CML-BC (P = .006) than in normal blood cells. Figure 3 shows thenm23-H1 and -H2 expression levels in all of the cases. Although a statistically significant correlation was observed between the expression levels of nm23-H1 and -H2 (r= .785, P < .0001), the expression levels of thenm23-H2 gene were significantly higher in only AML (P = .023) and CML-BC (P = .013) compared with those in normal blood cells. The levels of nm23-H1 and -H2 mRNA in CML-CP cells were similar to the average levels in normal blood cells. With respect to the clinical stage of hematologic neoplasms, the expression of both nm23-H1 and -H2 was significantly higher in CML-BC than in CML-CP (P = .001 and P = .007, respectively), as shown in Fig 4. In contrast, the expression of both genes in MDS overt leukemia was similar to the average levels in MDS (P = .205 and P = .548, respectively; Fig 4). These results indicate that (1)nm23-H1 is overexpressed in hematologic neoplasms, except CML-CP; (2) this increase in nm23-H1 is observed in the stages of MDS; and (3) the progression of CML is accompanied by the overexpression of nm23-H1 and -H2 mRNA.
Quantitative RT-PCR analysis of nm23-H1 andnm23-H2 mRNA in human normal BM cells, MDS, MDS overt leukemia, AML, ALL, CML-CP, and CML-BC samples. Normal BM cells were from normal volunteers. Neoplasm cells were identified as the mononuclear cell fraction.
Quantitative RT-PCR analysis of nm23-H1 andnm23-H2 mRNA in human normal BM cells, MDS, MDS overt leukemia, AML, ALL, CML-CP, and CML-BC samples. Normal BM cells were from normal volunteers. Neoplasm cells were identified as the mononuclear cell fraction.
Relationship between the levels of nm23-H1 andnm23-H2 in human hematologic neo plasms. (---) Average levels of normal BM cells; (○) AML; (•) ALL; (□) CML-CP; (▪) CML-BC; (▵) MDS; (▴) MDS overt leukemias
Relationship between the levels of nm23-H1 andnm23-H2 in human hematologic neo plasms. (---) Average levels of normal BM cells; (○) AML; (•) ALL; (□) CML-CP; (▪) CML-BC; (▵) MDS; (▴) MDS overt leukemias
nm23-H1 and nm23-H2 expression in MDS in comparison with MDS overt leukemia and in CML-CP in comparison with CML-BC. Analyzed by Mann-Whitney's U-test.
nm23-H1 and nm23-H2 expression in MDS in comparison with MDS overt leukemia and in CML-CP in comparison with CML-BC. Analyzed by Mann-Whitney's U-test.
DISCUSSION
The nm23 gene was originally identified by the differential hybridization of a cDNA library with total RNA extracted from mildly and highly metastatic melanoma cell lines.11 Its expression was inversely correlated with the tumor's metastatic potential in experimental rodent cells and in some human tumors.12Transfection with nm23 cDNA reduced the metastatic potential in vivo and the ability of cells to migrate in response to cytokines in vitro.13-15 Reduced nm23 expression levels are correlated with increased metastatic potential and an aggressive disease in human breast,16 hepatocellular,17ovarian18 and gastric carcinoma,19 and in melanoma.20 However, an opposite trend is observed in neuroblastoma and pancreatic carcinoma.21-23 In other tumor types, including colorectal, thyroid, and lung carcinomas, nm23expression does not correlate with disease progression.21These results suggest a functional difference in nm23 gene expression in several types of human tumor.21 In leukemia, as shown in this report, nm23 gene expression correlated with a poor prognosis in AML and with disease progression of CML. Especially in AML-M5, nm23-H1 mRNA overexpression was the most important poor prognostic factor (Table 4). These results suggest that there is a connection between nm23 function and malignant phenotypes in leukemia, such as malignant growth, differentiation resistance, and chemotherapy resistance, although it remains unclear why AML-M5 cells exhibit a higher expression of nm23 nmRNA levels.
We preliminally tried to evaluate the effect of nm23-H1 mRNA level on prognosis for each of the FAB subtypes, although each group is a small cohorts of patients. For FAB-M5, there were 9 nm23 low versus 9 nm23 high; as shown in Fig 1 and Table 4, the elevatednm23-H1 expression in FAB-M5 significantly contributed to the prognosis, and nm23-H1 status is the most important prognostic factor. For FAB-M4, there were 12 nm23 low versus 4nm23 high; the elevated nm23-H1 expression in FAB-M4 significantly predicts a poor response to initial therapy (CR ratios ofnm23 low expression group and nm23 high group were 83.3% and 25.0%, respectively; P = .029, χ2test for independence). The correlation between elevatednm23-H1 expression and poor survival was observed in M4 cases, but the statistical significance has not been observed yet. For FAB-M3, there were 15 nm23 low versus 0 nm23 high; theses cases showed the lowest expression levels of nm23-H1 among FAB subtypes. Because all cases showed nm23-H1 ≤100 (low expression), we could not analyse the effect of the overexpression of nm23-H1 (>100) on overall survival. The low expression levels of nm23-H1 in FAB-M3 may be associated with the good prognosis in M3 patients. For FAB-M2, there were 18nm23 low versus 9 nm23 high; the correlation between elevated nm23-H1 expression and a poor response to initial therapy and poor survival was observed, but the statistical significance was not shown. For FAB-M1, there were 12 nm23 low versus 8 nm23 high; the elevated nm23-H1 expression in these cases did not contribute to the response to initial therapy and the prognosis, although FAB-M1 cases had significantly higher expression levels of nm23-H1 and nm23-H2 than did normal subjects. Therefore, further analysis of nm23-H1 andnm23-H2 mRNA levels on prognosis for each of FAB subtypes in lager cohorts is required.
nm23-H1 and -H2 show 88% amino acid sequence homology24,25 and are located on the same region of chromosome 17q21 in tandem.26-28 Based on an analysis of the promoter regions of the nm23-H1 and -H2 genes, it has been suggested that these nm23 genes are independently and differentially regulated.29 However, a statistically significant correlation between the expression levels of nm23-H1and -H2 was observed in AML (Fig 3). Elevated expressions of both nm23 genes were observed in AML, whereas a poor prognosis and a low percentage of CR in AML were associated only with the nm23-H1 expression level. Postel et al30reported that nm23-H2 was a transcription factor (PuF) and that one of its targets was the c-myc gene. Overexpression of the c-myc gene has also been reported to inhibit the differentiation of human and mouse leukemia cells.31-33 However, in our previous experiment, we did not find a correlation between c-myc expression and nm23-H2 expression in AML.6 Thus, c-myc expression in AML does not appear to be influenced by the overexpression of nm23-H2 (PuF).
Drug resistance, either inherent or acquired, is an important cause of treatment failure in hematologic neoplasms. Ferguson et al34 reported a functional link between nm23expression and cancer cell sensitivity to the alkylating agent cisplatin. We investigated, mdr1, mrp,gst-π,topo I, topo IIα, and topo IIβ expression in 20 AML patients by RT-PCR, but none of these drug resistance-related genes were associated with nm23-H1 expression (data not shown). Other mechanisms may be involved in drug resistance. nm23 genes can modulate differentiation, proliferation (cell cycle), and drug resistance in AML, and these three events are closely linked. Inducing differentiation in leukemic cells is associated with the concomitant block of the cell cycle at the G0/G1 phase. Transforming growth factor-β (TGF-β) is a negative regulator of proliferation, induced growth arrest in the G1 phase of the cell cycle in many cell types, and the loss of the cellular response to this ligand that occurs during oncogenesis in some systems. Transfection of melanoma K-1735 TK and breast carcinoma MDA-MB-435 cells with the nm23 gene decreases the response to TGF-β.13,14 We previously isolated a differentiation-resistant mouse myeloid leukemia M1 cells from the parent differentiation-sensitive M1 cells. We then characterized the differentiation-resistant cells and showed that the differentiation-resistant cells had higher leukemogenicity and produced I-factors/nm23 more than the parent differentiation-sensitive M1 cells. The differentiation-resistant M1 cells showed a more progressive potential, which was resistant to not only differentiation-induction but also to growth-suppressive cytokines such as TGF-β.33 35-37 The role of nm23 expression in the malignant growth of leukemia cells remains to be clarified, butnm23 may play key roles in various aspects of hematopoietic cell biology, including differentiation, proliferation (cell cycle), and drug resistance.
MDS expressed higher nm23-H1 mRNA levels, and this level was similar to that in AML (Table 1 and Fig 4). In this study, MDS included CMMoL, RAEB, and RAEB in transformation (RAEB-T), but excluded RA and RA with ringed sideroblasts (RARS). Although it is necessary to study RA and RARS (low-risk MDS), this study indicated that, if there were at least 11% blasts in marrow, nm23-H1 expression could be detected at the leukemic level by RT-PCR.
The median survival time in CML is 3 to 4 years. Acute transformation may occur rapidly over weeks or even days. These patients are usually refractory to conventional chemotherapy. These clinical features seem to be associated with the elevation of nm23 expression (Table 1and Fig 4). Recently, Venturelli et al38 isolated a novel cDNA DR-nm23 that was differentially expressed in a CML blast crisis cDNA library. The sequence of DR-nm23 is highly similar to that of nm23, and DR-nm23 inhibits granulocyte differentiation and induces apoptosis in 32Dc13 myeloid cells. These results suggest that the nm23 family, which includesDR-nm23, plays an important role in CML blastic crisis as well as in AML.
Cox's proportional hazard model indicated that the chromosomal aberrations t(15;17) and t(8;21) had good prognostic value, but they were limited to just AML-M3 and AML-M2. Because the prognostic evaluation of nm23 is applicable to all stages of AML,nm23-H1 status will be the most important prognostic factor. Furthermore, because the overexpression of nm23-H1 is also observed in other hematologic malignancies, such as ALL and MDS,nm23 expression could also have prognostic value in these diseases.
ACKNOWLEDGMENT
The authors gratefully acknowledge Dr Yoshiharu Hoshiyama (Showa University School of Medicine, Tokyo, Japan) for helpful discussions in statistical analysis.
Supported in part by a grant from the Ministry of Health and Welfare, Grants-in-Aid for Scientific Research (C) and Cancer Research; a grant from the Ministry of Education, Science, Sports and Culture, Japan; and a grant from the Kawano Memorial Foundation for the Promotion of Pediatrics.
Address reprint requests to Junko Okabe-Kado, PhD, Department of Chemotherapy, Saitama Cancer Center Research Institute, 818 Komuro, Ina, Saitama 362-0806, Japan.
The publication costs of this article were defrayed in part by page charge payment. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. section 1734 solely to indicate this fact.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal