Chromosomal translocations leading to deregulation of specific oncogenes characterize approximately 50% of cases of diffuse large B-cell lymphomas (DLBL). To characterize additional genetic features that may be of value in delineating the clinical characteristics of DLBL, we studied a panel of 96 cases at diagnosis consecutively ascertained at the Memorial Sloan-Kettering Cancer Center (MSKCC) for incidence of gene amplification, a genetic abnormality previously shown to be associated with tumor progression and clinical outcome. A subset of 20 cases was subjected to comparative genomic hybridization (CGH) analysis, which identified nine sites of chromosomal amplification (1q21-23, 2p12-16, 8q24, 9q34, 12q12-14, 13q32, 16p12, 18q21-22, and 22q12). Candidate amplified genes mapped to these sites were selected for further analysis based on their known roles in lymphoid cell and lymphoma development, and/or history of amplification in tumors. Probes for six genes, which fulfilled these criteria,REL (2p12-16), MYC (8q24), BCL2 (18q21),GLI, CDK4, and MDM2 (12q13-14), were used in a quantitative Southern blotting analysis of the 96 DLBL DNAs. Each of these genes was amplified (four or more copies) with incidence ranging from 11% to 23%. This analysis is consistent with our previous finding that REL amplification is associated with extranodal presentation. In addition, BCL2 rearrangement and/or REL, MYC, BCL2, GLI,CDK4, and MDM2 amplification was associated with advanced stage disease. These data show, for the first time, that amplification of chromosomal regions and genes is a frequent phenomenon in DLBL and demonstrates their potential significance in lymphomagenesis.
B-CELL DIFFUSE LARGE cell lymphomas (DLBLs) are clinically and genetically heterogeneous. Approximately 50% of patients relapse after treatment and succumb to recurrent lymphoma.1,2 The frequently recurring chromosomal translocations, t(3;14)(q27;q32), t(8;14)(q24;q32), and t(14;18)(q32;q21), have been shown to characterize genetic subsets, which together make up approximately 50% of DLBLs.3 In these translocations, the expression of BCL6 (3q27),MYC (8q24), and BCL2 (18q21) genes is deregulated as a result of their juxtaposition to the IG genes.3However, the genetic basis of the clinical heterogeneity of DLBL is poorly understood. One genetic lesion frequently associated with progression and clinical outcome of diverse tumor types is gene amplification.4 Although sporadic cases of DLBL with gene amplification detected by cytogenetic and/or molecular genetic methods have been reported,5-7 the incidence or biologic significance of gene amplification in DLBL has not been studied in detail. Recently, by comparative genomic hybridization (CGH) analysis of a case of DLBL with double minute chromosomes (dmins), we identified high level amplification of the chromosomal region 2p13-15.6 Following a Southern blotting-based candidate gene approach analysis, we showed that the REL proto-oncogene, mapped to this chromosomal region, is amplified in this single tumor, as well as in 23% of cases of DLBL.6 To compare gains and losses of DNA in DLBLs with a t(14;18)(q32;q21) translocation to those without an IG gene site-associated translocation, we undertook detailed CGH and follow-up candidate gene analysis of a cohort of previously untreated B-cell non-Hodgkin's lymphoma (NHL) specimens diagnosed as DLBL by the International Lymphoma Study Group (ILSG) classification.8 We identified nine sites of chromosomal amplification and frequent amplification ofREL, MYC, BCL2, GLI, CDK4, andMDM2, and found that gene amplification was associated with advanced stage disease at diagnosis.
MATERIALS AND METHODS
Tumor specimens.
A subset of 96 at diagnosis B-cell NHL biopsy specimens histologically classified as DLBL according to ILSG criteria (diffuse large cell, diffuse mixed small, and large cell, immunoblastic),8 were selected for this study from our ongoing prospective ascertainment of consecutive NHL cases for genetic and clinical analyses, initiated in 1984 and previously described in detail.9,10 None of the cases belonged to the subset primary mediastinal (thymic) large B-cell lymphoma (PMLBL) or showed evidence of follicularity. Of the 96 cases, 78 were previously studied for REL amplification.6
Cytogenetic analysis and CGH.
Karyotypic analysis using G-banding after conventional methods was attempted on each biopsy ascertained. CGH analysis of a subset of 20 tumors with G-banding karyotype data was performed on DNA extracted from frozen tumor tissue as described6 using the Quantitative Image Processing System (QUIPS, Vysis, IL). For each hybridization, a minimum of five metaphases was analyzed and green to red fluorescence ratio profiles for eight to 10 chromosomes exhibiting comparable levels of fluorescence intensity were normalized to standard length and combined statistically to display mean and 95% confidence intervals of the ratio. Chromosomal imbalances were detected on the basis of the ratio profiles deviating from the balance value of green to red ratio of 1.0. Ratio values of 1.20 and 0.80 were used as upper and lower thresholds to define gains and losses, respectively.6 Regions near the centromeres of chromosomes 1, 9, 13-16, 21, and 22 were not scored in the CGH analysis because of the highly repeated nature of DNA in these sites. High level amplification was defined as occurrence of fluorescence intensity values in excess of 2.0 and strong localized fluorescein isothiocyanate (FITC) signal at the chromosomal site. For chromosomal definition of such regions of high level amplification, the peaks of the ratio profiles were compared with the corresponding 4′,6-diamidino-2-phenylindole (DAPI) banding of individual chromosomes. Chromosomal or subchromosomal gain or loss was considered recurrent if detected in two or more tumors by G-banding or CGH.
Southern blot analysis and determination of gene copy number.
Southern blotting using probes for the REL,MYC, BCL2, GLI, CDK4, and MDM2 genes and the restriction fragment length polymorphism (RFLP) probe D2S48 (control of copy number) was performed on the DNA isolated from snap-frozen biopsies of the entire panel of 96 tumors, as described recently by us, to determine their gene copy number.6 In addition, BCL2 gene rearrangements were ascertained from the same hybridization filters using appropriate probes as described by us.11 All the probes used were as described previously.5,6,11 12 A probe for CDK4 was generated by polymerase chain reaction (PCR).
Clinical correlation analysis.
To analyze the effect of genetic alterations taken in aggregate, a score was computed for each case with one point given for each of the following alterations: BCL2 rearrangement, or ≥4 copies ofREL, MYC, BCL2, GLI, CDK4, orMDM2 determined as described above. The mean score for these genetic variables was compared in groups with different clinical features (eg, stage at presentation, extranodal involvement at presentation). In addition, each genetic variable was analyzed independently. Aggregate comparisons were performed using two-samplet-test, and proportions of subsets with individual genetic variables were compared using a two-sided Fischer's exact test.
RESULTS
Clonal chromosomal abnormalities detected by G-banding.
Clonal chromosome abnormalities were documented in 72 of the 96 cases. Recurring gains and losses detected in the entire group are summarized in Fig 1A. Homogeneously staining regions (HSRs) or HSR-like marker chromosomes were noted in two tumors (218 and 806) and dmins in one other tumor (1533). Recurring translocations affecting bands 3q27, 8q24, and 18q21 were represented by 8, 3, and 13 cases, respectively (one case had translocations affecting both the 3q27 and 18q21 bands). The remaining 49 clonally abnormal cases did not display a 14q32-associated translocation.
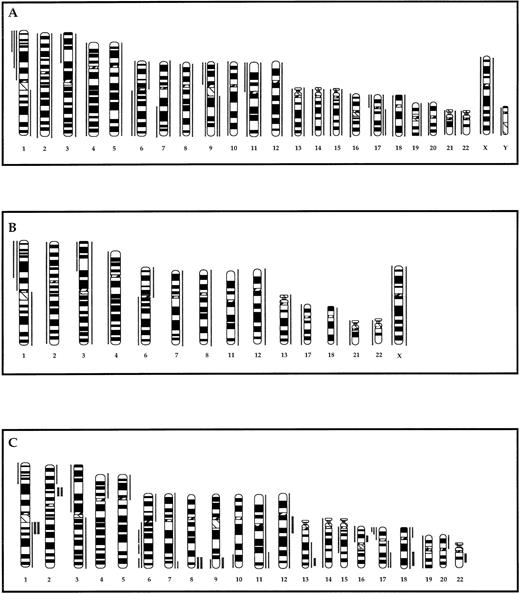
Partial ideograms comparing the recurring chromosomal gains and losses detected by G-banding in 72 of the 96 tumors assayed for gene amplification (A), the subset of 20 tumors assayed by CGH (B), and gains and loses detected by CGH in the same subset of 20 tumors (C). Thin vertical lines on either side of the chromosome ideogram indicate only recurrent gains (right) or losses (left) of a chromosome or a chromosomal region. Gains/losses were considered recurrent if observed in two or more tumors. Thick lines in (C) indicate regions of high copy number (amplification) as defined in the text.
Partial ideograms comparing the recurring chromosomal gains and losses detected by G-banding in 72 of the 96 tumors assayed for gene amplification (A), the subset of 20 tumors assayed by CGH (B), and gains and loses detected by CGH in the same subset of 20 tumors (C). Thin vertical lines on either side of the chromosome ideogram indicate only recurrent gains (right) or losses (left) of a chromosome or a chromosomal region. Gains/losses were considered recurrent if observed in two or more tumors. Thick lines in (C) indicate regions of high copy number (amplification) as defined in the text.
CGH analysis.
A subset of 20 specimens from the 72 with documented clonal chromosome abnormalities were subjected to CGH analysis to identify gains and losses of DNA in tumors with a t(14;18)(q32;q21) translocation, as well as those without an IG gene site-associated translocation (Fig1B and C). Seven cases (102, 251, 486, 721, 885, 956, and 1659) had the t(14;18)(q32;q21) translocation, while the remaining 13 (23, 150, 178, 216, 248, 293, 331, 375, 970, 1368, 1533, 1593, and 1664) did not exhibit an IG gene site-associated translocation. Comparison of gains and losses of chromosomes and chromosomal regions detected by CGH with those detected by G-banding showed an overall concordance in the subset of 20 tumors with CGH detecting a higher number of gains and losses compared with G-banding (Fig 1B and C). Discordances may be due to previously detailed reasons.6
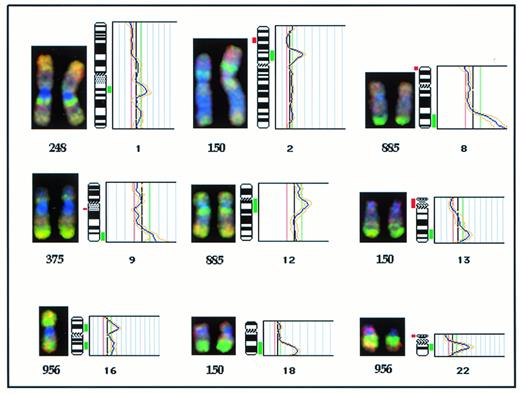
High-level amplification was detected at nine sites in six tumors: 1q21-23, 2p12-16, 8q24, 9q34, 12q12-14, 13q32, 16p12, 18q21-22, and 22q12 (Figs 1C and 2). Recurring sites of amplification were noted in five tumors and comprised two cases with amplification of 1q21-23 (248 and 1659), two cases with amplification of 2p12-16 (150 and 248), and two cases with amplification of 8q24 (375 and 885). Among these sites, amplification of 1q21-23, 2p12-16, and 8q24 was noted in tumors with, as well as without, the t(14;18)(q32;q21) translocation, while amplification of 12q12-14, 16p12, and 22q12 sites was noted only in tumors with the t(14;18)(q32;q21) translocation. Of the three tumors, which showed cytogenetic evidence of gene amplification, one with dmins (1533) was studied by CGH and did not identify an amplified region. The G-banded karyotype of this tumor was 46,XX, +3dmin[20] suggesting that the amplification itself was of low level.
Partial CGH karyotypes (left) and corresponding ratio profiles (right) illustrating high level amplification of chromosomal regions in DLBL tumors studied. Hybridized tumor DNA was visualized via FITC (green) and control DNA via Texas Red (red). The averaged green to red fluorescent signal ratio along the length of the chromosome is shown. The blue line in the ratio profile represents the mean of eight to 10 chromosomes and the yellow line represents the standard deviation. The vertical red and green bars on the right of the ideogram indicate threshold values of 0.80 and 1.20 for loss and gain, respectively.
Partial CGH karyotypes (left) and corresponding ratio profiles (right) illustrating high level amplification of chromosomal regions in DLBL tumors studied. Hybridized tumor DNA was visualized via FITC (green) and control DNA via Texas Red (red). The averaged green to red fluorescent signal ratio along the length of the chromosome is shown. The blue line in the ratio profile represents the mean of eight to 10 chromosomes and the yellow line represents the standard deviation. The vertical red and green bars on the right of the ideogram indicate threshold values of 0.80 and 1.20 for loss and gain, respectively.
Southern blot analysis for copy numbers of candidate amplified genes.
Genes mapped to the chromosomal regions of amplification were considered as candidate amplified genes if they previously had been shown to be amplified in human tumors and/or if their cellular functions included regulation of signal transduction events. Such genes, identified by searching the Genome Database (GDB) are listed in Table 1.6,13-35Among these genes, we decided to determine the incidence of amplification of two groups of genes. One comprised REL,MYC, and BCL2 genes. These were selected because of their previously established roles in lymphoid lineage development and deregulated expression in lymphomagenesis.3,16,17,36REL and the related NFKB2 gene were previously shown to be deregulated by rare IG gene site-associated translocations,37,38 and we recently showed REL to be frequently amplified in DLBL.6MYC andBCL2 are frequently deregulated by IG gene site-associated translocations in various lymphoma subsets,39 while anecdotal instances of amplification of these genes have previously been reported in CGH assays.7,20 40-42 In this study of DLBLs, four or more copies indicative of amplification were noted in 23% of cases forREL, 16% of cases for MYC, and 11% of cases forBCL2 (Table 2 and Fig 3).
Candidate Genes Mapped to Regions of Amplification Identified by CGH
| Chromosomal Region . | Candidate Genes . | Normal Function . | Amplification in Tumorigenesis . |
|---|---|---|---|
| 1q21-23 | IL6R | Signal transduction (receptor)13 | |
| MCL1 | Apoptosis (BCL2 family member)14 | ||
| SKI | Transcription factor15 | ||
| 2p12-16 | REL | Transcription factor16 | DLBL6 |
| 8q24 | MYC | Transcription factor17 | Breast carcinoma,18 Ovarian carcinoma,19FCL20 |
| 9q34 | ABL | Signal transduction (kinase)21 | |
| TAN1 | Signal transduction (receptor)22 | ||
| 12q12-14 | CDK2 | Cell cycle kinase23 | Colorectal carcinoma24 |
| GLI | Transcription factor25 | Gliomas,26Sarcomas27 | |
| SAS | Transmembrane protein28 | Gliomas,29 Sarcomas28 | |
| CDK4 | Cell cycle kinase23 | Gliomas,29 Sarcomas30 | |
| MDM2 | p53 inactivation31 | Gliomas,29Sarcomas30 | |
| 13q32 | — | — | — |
| 16p12 | IL4R | Signal transduction (receptor)32 | |
| 18q21-22 | BCL2 | Apoptosis14 | NHL,33 DLBL7 |
| 22q12 | SIS/YES2 | Growth factor34 | Atypical NHL35 |
| Chromosomal Region . | Candidate Genes . | Normal Function . | Amplification in Tumorigenesis . |
|---|---|---|---|
| 1q21-23 | IL6R | Signal transduction (receptor)13 | |
| MCL1 | Apoptosis (BCL2 family member)14 | ||
| SKI | Transcription factor15 | ||
| 2p12-16 | REL | Transcription factor16 | DLBL6 |
| 8q24 | MYC | Transcription factor17 | Breast carcinoma,18 Ovarian carcinoma,19FCL20 |
| 9q34 | ABL | Signal transduction (kinase)21 | |
| TAN1 | Signal transduction (receptor)22 | ||
| 12q12-14 | CDK2 | Cell cycle kinase23 | Colorectal carcinoma24 |
| GLI | Transcription factor25 | Gliomas,26Sarcomas27 | |
| SAS | Transmembrane protein28 | Gliomas,29 Sarcomas28 | |
| CDK4 | Cell cycle kinase23 | Gliomas,29 Sarcomas30 | |
| MDM2 | p53 inactivation31 | Gliomas,29Sarcomas30 | |
| 13q32 | — | — | — |
| 16p12 | IL4R | Signal transduction (receptor)32 | |
| 18q21-22 | BCL2 | Apoptosis14 | NHL,33 DLBL7 |
| 22q12 | SIS/YES2 | Growth factor34 | Atypical NHL35 |
Copy Number of REL, MYC, BCL2, GLI, CDK4, andMDM2 Genes in DLBL Determined by Southern Blotting
| Copy No. . | No. and Percent (parentheses) of Tumors . | |||||
|---|---|---|---|---|---|---|
| REL . | MYC . | BCL2 . | GLI . | CDK4 . | MDM2 . | |
| <4 | 70 (77) | 73 (84) | 82 (89) | 78 (87) | 82 (89) | 79 (86) |
| 4-10 | 19 (21) | 13 (15) | 10 (11) | 12 (13) | 10 (11) | 13 (14) |
| >10 | 2 (2) | 1 (1) | 0 | 0 | 0 | 0 |
| Total | 91 (100) | 87 (100) | 92 (100) | 90 (100) | 92 (100) | 92 (100) |
| Copy No. . | No. and Percent (parentheses) of Tumors . | |||||
|---|---|---|---|---|---|---|
| REL . | MYC . | BCL2 . | GLI . | CDK4 . | MDM2 . | |
| <4 | 70 (77) | 73 (84) | 82 (89) | 78 (87) | 82 (89) | 79 (86) |
| 4-10 | 19 (21) | 13 (15) | 10 (11) | 12 (13) | 10 (11) | 13 (14) |
| >10 | 2 (2) | 1 (1) | 0 | 0 | 0 | 0 |
| Total | 91 (100) | 87 (100) | 92 (100) | 90 (100) | 92 (100) | 92 (100) |
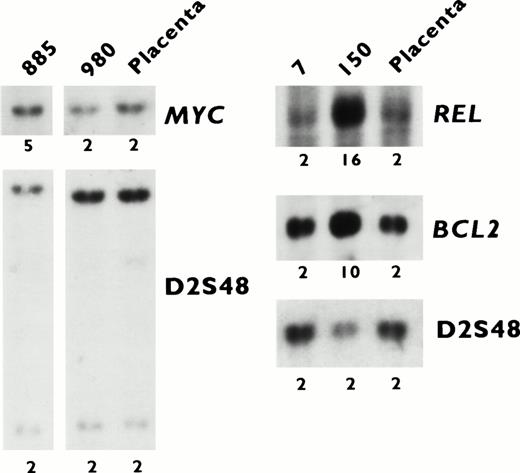
Southern blot analysis of DNA extracted from tumors (7, 150, 885, and 980) and normal placenta with MYC, REL,BCL2, and D2S48 probes. The calculated copy numbers are listed. Tumor 150 exhibited amplification of 2p12-16 (REL) and 18q21-22 (BCL2), and tumor 885 exhibited amplification of 8q24 (MYC) by CGH.
Southern blot analysis of DNA extracted from tumors (7, 150, 885, and 980) and normal placenta with MYC, REL,BCL2, and D2S48 probes. The calculated copy numbers are listed. Tumor 150 exhibited amplification of 2p12-16 (REL) and 18q21-22 (BCL2), and tumor 885 exhibited amplification of 8q24 (MYC) by CGH.
In a subset of 78 tumors from this cohort, we previously identified 20 with REL amplification; we noted one additional tumor in this study (Table 2). No REL rearrangements suggesting 2p13-16 translocation were encountered. None of the 14 cases, which showed ≥4 copies of MYC, exhibited the t(8;14)(q24;q32) translocation. Similarly, a t(14;18)(q32;q21) translocation or BCL2 gene rearrangement was not noted in the 10 cases, which showed ≥4 copies ofBCL2. A BCL2 gene rearrangement was identified in 15% of all the cases studied.
The second group of genes screened were GLI, CDK4, andMDM2, which have previously been mapped to amplicons in band 12q13, a chromosomal region that has frequently been found to be amplified in a variety of tumors, particularly gliomas and sarcomas.26,27,29,30 None of the DLBLs assayed exhibited more than 10 copies of any of the three genes (Table 2), and in seven cases (150, 251, 252, 1404, 1584, 1593, and 1712), all three genes were found to be in higher copy number. In two of these cases (251 and 1593), CGH indicated a gain of chromosome 12. In four other cases (311, 1319, 1602, and 1693), two of the genes were in >4 copies (311:GLI and MDM2; 1319 and 1602: CDK4 andMDM2; 1693: GLI and CDK4). Discontinuous amplification of genes mapped to this region, such as that exhibited in case 311, has been reported in other tumors.29 In case 885, which showed amplification of 12q12-14 by CGH, none of the three genes tested exhibited higher than normal copy numbers suggesting possible amplification of other genes.
Clinical correlation analysis.
The median follow-up for survivors in the cohort was 27 months. As expected, median survival was greater among those with limited stage (I-II) disease (P = .08, log rank; P = .03, Breslow). A correlation was noted between genetic alterations and stage of disease at presentation; thus, cases presenting with stage I-III disease had a lower frequency of BCL2 rearrangement and/or ≥4 copies of REL, MYC, BCL2, GLI, CDK4, or MDM2 compared with cases presenting with stage IV disease (P = .03). With the exception of the cohort of cases withREL amplification, there were no associations between the genetic alterations and extranodal involvement, when analyzed in aggregate or separately. Of the 21 cases with RELamplification, 18 presented with extranodal involvement, compared with 47 of 70 cases without REL amplification (P = .1), indicating a trend towards REL amplification in extranodal lesions. In 13 of the 18 cases with REL amplification and extranodal involvement, the gastrointestinal system was involved.
DISCUSSION
In this study, we found a good concordance in gains and losses detected by G-banding and CGH, although the latter analysis identified many more changes in addition to those detected by the former. In addition, G-banding did not identify any evidence of high copy number of chromosomal regions detected by CGH. This higher efficiency of CGH to detect copy number changes is not unexpected because this method scans the DNA of the entire genome taking into account subpopulations of tumor cells, which may not proliferate adequately in short-term culture, to be detected in conventional cytogenetic analysis. In addition, increased copy numbers of chromosomal regions hidden in marker chromosomes are usually undetectable by G-banding.
Gene amplification, commonly associated with tumor progression and clinical outcome in solid tumors, has not been widely identified in hematopoietic tumors. Recent CGH analysis of small series of NHL samples comprising multiple histologic subsets has identified several sites of amplification, including 8q24 and 18q21.7,20,40-42The only large series representing a specific histologic subgroup assayed for gene amplification was DLBL, in which REL was shown to be amplified in 23% of cases.6 To determine the incidence and possible role of gene amplification in an at diagnosis, histologically-defined, and clinically significant subset of NHL, we undertook a CGH analysis, followed by identification of candidate genes, in a cohort of DLBL.
The genetic and clinical heterogeneity of DLBL presents a biologic, as well as a clinical challenge. The commonly recurring IG gene site-associated translocations leading to deregulation of specific genes occurs only in a proportion of cases, as shown in this study.BCL6, one of the three genes deregulated by such translocations, plays an important role in the genesis of DLBL.3 In addition, BCL6 undergoes mutations (hypermutation) in the 5′ regulatory region in more than 70% of cases of DLBL, providing another possible mechanism for BCL6deregulation.3 Thus, nearly all DLBLs contain deregulatedBCL6 alleles. Deregulation of BCL2, an antiapoptosis gene, and MYC, a DNA-binding transcription factor, have been known to be the primary events in the genesis of follicular center cell (FCL) and Burkitt's lymphoma (BL), respectively.39 The BL phenotype is invariably associated with MYC deregulation; its role in the genesis of DLBL is unknown. However, MYCderegulation has been suggested to comprise a second and contributing genetic event in the progression of BCL2-deregulated FCL to DLBL.43-45
The data presented here suggest that REL, MYC, andBCL2 genes may be more frequently involved in DLBL than is indicated by the frequency with which they are deregulated by chromosomal rearrangement. In the tumor panel assayed, there was no overlap between tumors with amplification and translocation affectingREL, MYC, and BCL2, suggesting that amplification and rearrangement are independent pathways to deregulation or overexpression of these genes. A similar conclusion was recently reached in a study of BCL2 overexpression in DLBLs with rearrangement versus amplification of the gene.41 They also indicate that the role of aberrant expression of these genes in lymphomagenesis is more prevalent than hitherto realized based on the incidence of cytogenetic translocations or DNA rearrangements. The relationship between rearrangement, amplification, and expression remains to be characterized. The site of BCL6 was not seen to be amplified in the CGH analysis, which is consistent with its frequent deregulation by translocation, as well as mutation in DLBLs.46
We also identified in this panel of at diagnosis DLBL, a higher copy number of three genes mapped to 12q13, (GLI, CDK4, andMDM2) with approximately the same frequencies as were noted forMYC and BCL2. In addition, all three genes were found to be in high copy number in seven cases, indicating a large amplified region. The role of amplification of these three genes or other candidate genes mapping to 12q13-14 region in lymphomagenesis remains to be determined.
In this study, we have identified the spectrum of chromosomal sites amplified in at diagnosis DLBL shown by CGH analysis and, using a candidate gene approach, show frequent amplification of REL,MYC, BCL2, GLI, CDK4, and MDM2 genes. Whether these amplifications comprise primary events leading to transformation or secondary events that underlie progression and clinical outcome, remains to be determined. However, our clinical correlation studies have indicated that BCL2 rearrangement and/or amplification of REL, MYC, BCL2,GLI, CDK4, and MDM2 are associated with advanced stage disease at presentation. As noted above, MYCderegulation has been associated with progression ofBCL2-deregulated FCL, and BCL2 overexpression itself has been associated with adverse clinical outcome in FCL, as well as DLBL, in some studies.36 Our previous data of a pretreatment and posttreatment cohort of DLBL suggested thatREL amplification may be associated with extranodal presentation.6 The results of our present study of at diagnosis DLBL are consistent with this observation. Further candidate gene and/or positional cloning approaches can be expected to identify additional amplified genes mapping in the amplified chromosomal regions identified, thereby providing new clues to the genetic basis of progression and clinical behavior of DLBL.
Supported by Grant No. CA-66999 from the National Institutes of Health/National Cancer Institute, Bethesda, MD, and the Lymphoma Foundation.
Address reprint requests to R.S.K. Chaganti, PhD, Memorial Sloan-Kettering Cancer Center, 1275 York Ave, New York, NY 10021.
The publication costs of this article were defrayed in part by page charge payment. This article must therefore be hereby marked "advertisement" is accordance with 18 U.S.C. section 1734 solely to indicate this fact.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal