Abstract
We previously have shown that the zinc finger transcription factor Egr-1 blocked granulocytic differentiation of HL-60 cells, restricting differentiation along the monocytic lineage. Egr-1 also was observed to block granulocyte colony-stimulating factor (G-CSF)–induced differentiation of interleukin-3 (IL-3)–dependent 32Dcl3 hematopoietic precursor cells, endowing the cells with the ability to be induced by granulocyte-macrophage colony-stimulating factor (GM-CSF) for terminal differentiation along the macrophage lineage. To better understand the function of Egr-1 as a positive modulator of monocytic differentiation, in this work we have studied the effect of ectopic expression of Egr-1 on the murine myeloblastic leukemic cell line M1, which is induced for differentiation by the physiological inducer IL-6. It is shown that, unlike in HL-60 and 32Dcl3 cells, ectopic expression of Egr-1 in M1 cells resulted in activation of the macrophage differentiation program in the absence of differentiation inducer. This included the appearance of morphologically differentiated cells, decreased growth rate in mass culture, and cloning efficiency in soft agar, and expression of endogenous c-myb and c-myc mRNAs was markedly downregulated. Untreated M1Egr-1 cells also exhibited cell adherence, expression of Fc and C3 receptors, and upregulation of the myeloid differentiation primary response genes c-Jun, junD, andjunB and the late genetic markers ferritin light-chainand lysozyme. Ectopic expression of Egr-1 in M1 cells also dramatically increased the sensitivity of the cells for IL-6–induced differentiation, allowed a higher proportion of M1 cells to become terminally differentiated under conditions of optimal stimulation for differentiation, and decreased M1 leukemogenicity in vivo. These findings demonstrate that the functions of Egr-1 as a positive modulator of macrophage differentiation vary, depending on the state of lineage commitment for differentiation of the hematopoietic cell type.
© 1998 by The American Society of Hematology.
THE COMPLEX PROCESS of blood cell formation provides a profound example of cell homeostasis that is regulated throughout life, whereby a hierarchy of hematopoietic progenitor cells in the bone marrow proliferate and terminally differentiate along multiple, distinct cell lineages, including the proliferation and differentiation of myeloid precursor cells into mature granulocytes and macrophages.1-5 The murine M1 myeloid leukemic cell line proliferates autonomously and can be induced with the physiological inducers interleukin-6 (IL-6), leukemia inhibitory factor (LIF),6 or conditioned media of mouse lungs (LUCM), containing both IL-6 and LIF,7 to undergo terminal differentiation and growth arrest, which culminates in programmed cell death.8
To identify genes that may play a role in the regulation of hematopoietic cell differentiation, we have isolated cDNA clones of myeloid differentiation primary response (MyD) genes, activated in the absence of de novo protein synthesis, in HL-60 and M1 cells after induction for macrophage or granulocyte differentiation.7,9,10 In the course of this work, the gene encoding the zinc finger transcription factor Egr-1 (Krox24, NGIF-A, or Zif268 Tis8) has been identified as a myeloid differentiation primary response gene, specifically induced upon HL-60 macrophage differentiation.10
Egr-1 was initially identified as an early growth response gene in cultured fibroblasts11,12 and was subsequently shown to be induced in response to B-cell maturation as well as during differentiation of nerve, bone, myeloid, and erythroleukemic cells.10,13-17 Recently, Egr-1 also has been shown to be involved in cell proliferation,18-20 negative regulation of cell growth,21,22 and apoptosis.23 The Egr-1 protein has been localized to the nucleus and shown to bind specifically to the consensus sequence 5′-GCGGGGGCG-3′ as well as to transactivate a promoter containing this sequence.24-29
Egr-1 was found by us to be a macrophage differentiation primary response gene that restricts the differentiation of human HL-60 cells along the monocytic lineage10 and potentiates macrophage differentiation of the hematopoietic precursor cell line 32Dcl3.30 Egr-1 is induced in M1 cells immediately after stimulation with physiological inducers IL-6, LIF, or LUCM, and blocking Egr-1 expression by antisense oligonucleotides results in repression of monocytic differentiation.10 These findings raised the possibility that deregulated expression of Egr-1 in M1 cells may predispose the cells for terminal differentiation. To test this hypothesis, we studied the effect of deregulated expression of Egr-1 on macrophage differentiation of the murine myeloblastic leukemic cell line M1. It was shown that ectopic expression of Egr-1 in M1 cells activated the macrophage differentiation program and included the appearance of morphologically differentiated cells. In addition, the cells were sensitized for further induction of terminal differentiation by IL-6, and a higher proportion of M1 cells became terminally differentiated under conditions of optimal stimulation for differentiation. Ectopic expression of Egr-1 in M1 cells decreased the leukemogenicity in vivo.
MATERIALS AND METHODS
Cells, mice, and cytokines.
The differentiation competent murine myeloid leukemic cell line M1 has been described previously.31 The cells were cultured in Dulbecco’s modified Eagle’s medium (DMEM; Cellgro, Mediatech Inc, Heyndon, VA) supplemented with 10% heat-inactivated horse serum (HS; GIBCO-BRL, Grand Island, NY) plus 1% penicillin and streptomycin (Cellgrow) in a humidified atmosphere with 10% CO2 at 37°C. Conditions to stimulate the cells for terminal differentiation were described in detail previously.31 Briefly, the cells were seeded at 0.15 × 106 cells/mL with or without IL-6 at 1 or 50 ng/mL, as indicated. For RNA extractions, cell concentrations were adjusted to give a final density of greater than 0.25 × 106cells/mL at the time of extraction. Viable cell numbers were determined by trypan blue dye exclusion. Experiments were repeated at least three times. PA317 (American Type Culture Collection, Rockville, MD), a retrovirus packaging cell line,32 was cultured in DMEM supplemented with 10% heat-inactivated newborn calf serum (GIBCO-BRL) plus 1% penicillin and streptomycin in a humidified atmosphere with 10% CO2 at 37°C. PA317 cells were periodically selected in HAT (hypoxanthine, aminopterin, and thymidine) medium to maintain their packaging function. For M1 leukemogenicity assays, 4- to 6-week-old CD-1 homozygous nude mice were obtained from Charles River Laboratories (Wilmington, MA). In the leukemogenicity assay, nude mice were intravenously injected (tail vein) with 104 or 105 cells prepared in 200 μL of 1× phosphate-buffered saline (PBS) for each cell type. Control animals were injected with the same volume of 1× PBS. Recombinant human IL-6 was a generous gift from Amgen Inc (Thousand Oaks, CA).
General recombinant DNA techniques, expression vectors, and DNA probes.
Plasmid preparations, restriction enzyme digestions, DNA fragment preparations, and agarose gel electrophoresis were performed as previously described.31,33,34 The retroviral plasmid expression vector, MSCV EB neo, used in this study was a gift from Dr Robert G. Hawley (University of Toronto, Toronto, Ontario, Canada).35 The 2.3-kb BamHI and Sal I fragment of the full-length murine Egr-1 cDNA10 was cloned into the Xho I site of the MSCV EB neo retroviral vector by blunt end ligation. DNA probes for murine c-Jun, junD,junB, MyD88, c-fos, Icam-1,c-myc, c-myb, b-actin, ferritin, andlysozyme have been previously described.30,31,33,34stat3 cDNA was excised from pRcCMV-stat3 plasmid by ApaI/NotI digestion (James E. Darnell Jr, Invitrogen Inc, Carlsbad, CA). The probes were labeled by random priming (GIBCO-BRL; RadPrime DNA labeling, catalogue no. 18428-011) to a specific activity equal to or greater than 109 cpm/μg. Genomic DNA extraction and Southern blot analysis were performed as described previously.10 30
Establishment of M1 cells that ectopically express the Egr-1 transgene.
Virus was generated from the plasmid forms of retroviral vectors, MSCV EB neo (as a control) and MSCV EB neo Egr-1, by transfection of the packaging cell line PA317 using calcium phosphate-DNA precipitation.36 Transfected PA317 cells were selected using 800 μg/mL G418 (Geneticin; 400 μg/mL; GIBCO-BRL) in growth medium. Several clones were expanded and the viral titer of the supernatants (viral conditioned medium [VCM]) was determined to be 0.8 × 105/mL by infecting NIH 3T3 cells.37 The VCM was passed through a 0.4-μm filter and immediately used to infect M1 cells. Infection of M1 cells was accomplished by resuspending pellet of cells (0.5 × 106 cells) in 4 mL of VCM in the presence of 8 mg/mL polybrene for 4 hours. After infection, the cells were washed once with growth medium and incubated for 48 hours. For neomycin-resistant colony selection, infected cells were seeded at 100 cells/mL in growth media containing G418, and 1-mL aliquots were dispensed into 24-well trays. After 10 to 15 days, cultures from wells containing surviving cells were expanded. The infectants were maintained in growth media containing 200 μg/mL of G418. Four independent clones with different integration sites were detected using Southern blot analysis as described previously10 34 and used throughout the study. All experiments in this study were initiated with nonadherent M1Egr-1 cells, and the results of all experiments represent the mean of at least three independent determinations, with standard deviations indicated in the appropriate figure legend.
Assays for differentiation-associated properties.
Morphological differentiation was determined by counting at least 300 cells on May-Grunwald-Giemsa–stained cytospin smears and scoring the proportion of immature blast cells, cells at intermediate stages of differentiation, and mature macrophages.7,9 Immature blast cells are characterized by scant cytoplasm and round or oval nuclei; cells at intermediate monocyte stages of differentiation are flattened, with a larger cytoplasm to nucleus ratio, and contain irregularly shaped nuclei and few interspersed or no vacuoles; mature macrophage-like cells are flattened; and spread out cells are interspersed with numerous vacuoles in a greatly enlarged cytoplasm. Fc and C3 receptor assays7 and cell adherence were determined as previously described.9 Agar colony assays were performed as previously described.33 Colonies in soft agar were examined after 7 days and scored after 14 days.
RNA extraction, Northern blotting, and hybridization.
RNA was extracted using Trizol (GIBCO-BRL) reagent according to the manufacturer’s specifications. Total RNA (10 μg/lane, equal amounts of RNA in each lane were confirmed by equal intensity of ethidium bromide staining of ribosomal RNA bands) was electrophoresed on 1% agarose formaldehyde gels. Northern blots, using Duralon-UV membranes (Stratagene, La Jolla, CA), were prepared and UV cross-linked (Stratalinker; Stratagene) before baking at 60°C under vacuum for 2 hours. Blots were hybridized in 50% deionized formamide, 10% dextran sulfate, 1 mol/L NaCl, 1% sodium dodecyl sulfate (SDS), and 100 μg/mL sheared salmon sperm DNA at 42°C with 106 cpm/mL of probe for 12 to 16 hours. Blots were washed at room temperature twice for 5 minutes in 2× SSC, 0.1% SDS and at 60°C twice for 30 minutes in 0.1× SSC, 1% SDS and exposed to x-ray film at −80°C for 48 to 72 hours. Stripping blots of probe to rehybridize was performed as described previously.9,30 34
Reverse transcriptase-polymerase chain reaction (RT-PCR)8.
Primers for murine IL-6 and gp130 genes were selected with the aid of the program PCRPLAN (PCGENE; Intelligenetics Inc, Mountain View, CA). The primers corresponded to bases 1946 to 1966 (5′-3′ sequence of primer) and to 2964 to 2985 of the murine gp130gene and bases 27 to 48 (5′-3′ sequence of primer) and 656 to 675 of the murine IL-6 gene. To detect the transcripts encoding for gp130 and IL-6, RT-PCR was performed on aliquots of RNA, essentially as described previously.34 38 Briefly, 3 μg of total RNA, extracted using Trizol reagent, was reverse transcribed (RT) with the GIBCO-BRL Superscript preamplification system (catalogue no. 180-89-011), used according to the manufacturer’s instructions, in a final volume of 21 μL, using oligo dT as primer. For PCR, 2 μL of cDNA was taken from each RT reaction volume and samples were diluted to 50 μL with buffer (Boehringer Mannheim Biochemicals [BMB], Indianapolis, IN; 10×), yielding 0.1 mmol/L dNTPs, 0.5 mmol/L MgCl2, 10 mmol/L Tris (pH 8.3), 50 mmol/L KCl; each primer, at a final concentration of 0.1 μmol/L, and 5 U Taq DNA polymerase (BMB) were added. Samples were covered with 50 μL mineral oil, heated at 94°C for 5 minutes, and subjected to 15 cycles of PCR in a Perkin-Elmer thermal cycler, using 1 minute of denaturation at 94°C, 1 minute of annealing at 62°C, and 2 minutes of polymerization at 72°C; finally, 5 minutes of polymerization was performed at 72°C. To monitor for efficiency and reproducibility of PCR amplification, β-actin transcripts were amplified using murine β-actin amplimers (Clontech Laboratories Inc, Palo Alto, CA). After extraction with CHCl3, 40 μL of PCR products was electrophoresed on 1% agarose gel, blotted, and hybridized with gp130 and IL-6 probe or β-actin probe (Clontech; catalogue no. 9800-1; within the amplified PCR region). Control samples not reverse transcribed were used to monitor for possible contamination with genomic DNA.
RESULTS
Establishment of M1Egr-1 cells ectopically expressing the Egr-1transgene.
M1Egr-1 and M1neo cell lines were established via infection of M1 cells with the retrovirus derived expression vectors MSCV EB neoEgr-1 and MSCV EB neo, as described in Materials and Methods. As shown in Fig 1, the M1Egr-1 clones expressed exogenous Egr-1 transcripts, whereas parental M1 cells or M1 cells infected with the MSCV EB neo vector carrying the selectable neo marker did not. Southern blot analysis of M1Egr-1 cell lines confirmed that each clone was an independent isolate, as evident by the distinct integration sites of the Egr-1 transgene in different M1Egr-1 clones (Fig 1B). Four distinct clones of M1Egr-1 and four clones of M1neo have been established.
Establishment of cell lines that ectopically express Egr-1. (A) Northern blot analysis of Egr-1 expression in M1, M1neo, and M1Egr-1 clones. Ten micrograms of total RNA was analyzed by Northern blots as described in Materials and Methods. (B) Southern blot analysis of genomic DNA from parental M1, M1neo, and M1Egr-1 clones. Genomic DNA (10 μg) was digested with EcoRI, resolved on a 1% agarose gel, transferred to Gene Screen Plus membranes (NEN, Boston, MA), and hybridized to a murine Egr-1 cDNA probe.
Establishment of cell lines that ectopically express Egr-1. (A) Northern blot analysis of Egr-1 expression in M1, M1neo, and M1Egr-1 clones. Ten micrograms of total RNA was analyzed by Northern blots as described in Materials and Methods. (B) Southern blot analysis of genomic DNA from parental M1, M1neo, and M1Egr-1 clones. Genomic DNA (10 μg) was digested with EcoRI, resolved on a 1% agarose gel, transferred to Gene Screen Plus membranes (NEN, Boston, MA), and hybridized to a murine Egr-1 cDNA probe.
The effect of constitutive expression of the Egr-1 transgene on the growth and differentiation characteristics of the M1 myeloblastic leukemia cell line was studied both in untreated cells as well as in cells treated with IL-6. Results are shown for representative M1Egr-1 and M1neo clones and are similar to what was observed with the other clones tested. The number of clones used in each experiment is indicated in the appropriate figure legend.
Ectopic expression of Egr-1 in M1 leukemic myeloblasts activates the macrophage differentiation program.
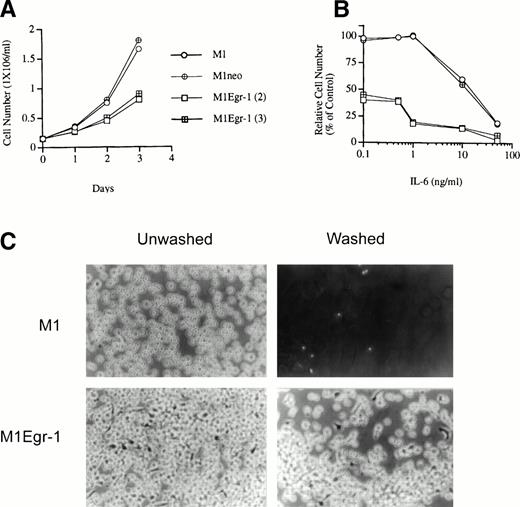
Ectopic expression of Egr-1 in untreated M1 cells was observed to markedly inhibit the growth of the cells. Figure 2A depicts the growth kinetics of untreated M1, M1neo, and M1Egr-1 cells analyzed in mass culture. M1 cells expressing an exogenous Egr-1 transgene exhibited a significantly slower growth rate than the control parental M1 and M1neo cells (Fig 2A). Starting with a culture of nonadherent M1Egr-1 cells, the cells grew in aggregates and after 3 days about 50% of the cells adhered to the culture dish and exhibited an elongated morphology, characteristic of monocytic differentiation (Table 1 and Fig 2C). Analysis of morphology by May Grunwald-Giemsa–stained cytospin smears showed that greater than 90% of the untreated M1Egr-1 cells differentiated into either intermediate or mature macrophages, with fewer than 10% retaining blast morphology (Table 1). In contrast, none of the uninduced M1 or M1neo clones grew in aggregates, adhered to the culture plate, or exhibited the morphology of differentiated cell types (Table1 and Fig 2C). It should be noted that, while establishing the M1Egr-1 cell lines, some of the clones could not be expanded in culture, because most of the cell population underwent differentiation.
Growth characteristics of M1, M1neo, and M1Egr-1 cells in mass culture. (A) Growth kinetics in culture medium in the absence of IL-6. (B) Growth kinetics in culture medium in the presence of different concentrations of IL-6. Cells were cultured in the presence of varying concentrations of IL-6 for 3 days. The results are presented as the percentage of untreated M1 cells (% control). For (A) and (B), data presented are the mean of three independent determinations, with standard deviations up to 13%. Cells were seeded as indicated in Materials and Methods and viable cell numbers were determined by trypan blue dye exclusion, with counting in a hemocytometer. All experiments were initiated with nonadherent cells. In each experiment, four M1Egr-1 clones were used; all gave similar results, and data are shown only for two. Similarly, in addition to parental M1 cells, 2 M1neo clones were used. All control cell lines gave similar results. (C) Representative photo-micrographs (original magnification × 100) of M1 and M1Egr-1 cells in mass culture. Cells were seeded as indicated in Materials and Methods and cultured in the absence of IL-6 for 4 days. Photomicrographs were taken before and after washing the plates with DMEM; thus, after washing, only the cells that remained attached to the surface of the tissue culture plate are shown.
Growth characteristics of M1, M1neo, and M1Egr-1 cells in mass culture. (A) Growth kinetics in culture medium in the absence of IL-6. (B) Growth kinetics in culture medium in the presence of different concentrations of IL-6. Cells were cultured in the presence of varying concentrations of IL-6 for 3 days. The results are presented as the percentage of untreated M1 cells (% control). For (A) and (B), data presented are the mean of three independent determinations, with standard deviations up to 13%. Cells were seeded as indicated in Materials and Methods and viable cell numbers were determined by trypan blue dye exclusion, with counting in a hemocytometer. All experiments were initiated with nonadherent cells. In each experiment, four M1Egr-1 clones were used; all gave similar results, and data are shown only for two. Similarly, in addition to parental M1 cells, 2 M1neo clones were used. All control cell lines gave similar results. (C) Representative photo-micrographs (original magnification × 100) of M1 and M1Egr-1 cells in mass culture. Cells were seeded as indicated in Materials and Methods and cultured in the absence of IL-6 for 4 days. Photomicrographs were taken before and after washing the plates with DMEM; thus, after washing, only the cells that remained attached to the surface of the tissue culture plate are shown.
Differentiation-Associated Properties of M1, M1neo, and M1Egr-1 Cells
| Cell Lines . | IL-6 (ng/mL) . | Blast . | Cells Types (%)* . | % Adhered Cells* . | Receptors (%)* . | ||
|---|---|---|---|---|---|---|---|
| Inter. . | Mature . | Fc . | C3 . | ||||
| M1 | 0 | 99 | <1 | 0 | 0 | <2 | <1.5 |
| 1 | 95 | 5 | 0 | 10 | 12 | 8 | |
| 50 | 0 | 32 | 68 | 62 | 52 | 74 | |
| M1neo.11 | 0 | 98 | <2 | 0 | 0 | <2 | 1 |
| 1 | 94 | 6 | 0 | 9 | 13 | 9 | |
| 50 | 0 | 29 | 71 | 61 | 49 | 70 | |
| M1neo.12 | 0 | 98 | 2 | 0 | 0 | 2 | <2 |
| 1 | 96 | 4 | 0 | 8 | 11 | 7 | |
| 50 | 0 | 33 | 67 | 60 | 48 | 69 | |
| M1Egr.1.2 | 0 | 7 | 54 | 39 | 57 | 35 | 39 |
| 1 | 4 | 27 | 69 | 65 | 58 | 64 | |
| 50 | 0 | 18 | 82 | 90 | 68 | 78 | |
| M1Egr.1.3 | 0 | 9 | 58 | 33 | 52 | 28 | 32 |
| 1 | 5 | 30 | 65 | 60 | 56 | 63 | |
| 50 | 0 | 22 | 78 | 82 | 66 | 77 | |
| Cell Lines . | IL-6 (ng/mL) . | Blast . | Cells Types (%)* . | % Adhered Cells* . | Receptors (%)* . | ||
|---|---|---|---|---|---|---|---|
| Inter. . | Mature . | Fc . | C3 . | ||||
| M1 | 0 | 99 | <1 | 0 | 0 | <2 | <1.5 |
| 1 | 95 | 5 | 0 | 10 | 12 | 8 | |
| 50 | 0 | 32 | 68 | 62 | 52 | 74 | |
| M1neo.11 | 0 | 98 | <2 | 0 | 0 | <2 | 1 |
| 1 | 94 | 6 | 0 | 9 | 13 | 9 | |
| 50 | 0 | 29 | 71 | 61 | 49 | 70 | |
| M1neo.12 | 0 | 98 | 2 | 0 | 0 | 2 | <2 |
| 1 | 96 | 4 | 0 | 8 | 11 | 7 | |
| 50 | 0 | 33 | 67 | 60 | 48 | 69 | |
| M1Egr.1.2 | 0 | 7 | 54 | 39 | 57 | 35 | 39 |
| 1 | 4 | 27 | 69 | 65 | 58 | 64 | |
| 50 | 0 | 18 | 82 | 90 | 68 | 78 | |
| M1Egr.1.3 | 0 | 9 | 58 | 33 | 52 | 28 | 32 |
| 1 | 5 | 30 | 65 | 60 | 56 | 63 | |
| 50 | 0 | 22 | 78 | 82 | 66 | 77 | |
*Determined 3 days after the cells were seeded at a density of 0.15 × 106 cells/mL. For details of analysis of differentiated phenotypes, receptor assays and determination of percentage of cell adherence, see Materials and Methods. All experiments were initiated with nonadherent cells. Four M1Egr1 clones and four M1neo clones were used in these experiments. All clones of the same type gave similar results and data are shown for two clones. All values represent the mean of three independent determinations with standard deviations up to 12% (ie, 10% ± 1.2%).
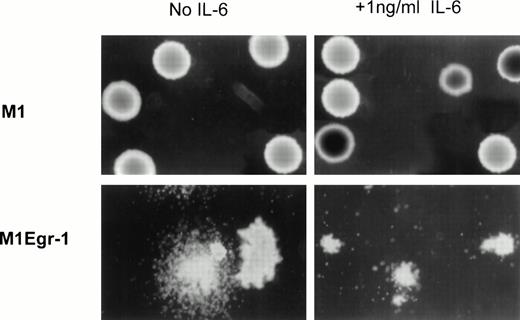
Consistent with the proliferation and differentiation characteristics of the M1Egr-1 cells in mass culture, the average cloning efficiency in soft agar of uninduced M1Egr-1 cells was 37% of that obtained for control M1 and M1neo cell lines (Table 2). Furthermore, about 30% of the colonies formed by the M1Egr-1 cells displayed a diffuse morphology at 7 days, characteristic of colonies with differentiated cell types. In contrast, no diffuse colonies were observed with the M1 and M1neo cells (Fig 3and Table 2).
Cloning Efficiency of M1, M1neo, and M1Egr-1 Cells
| Cell Type . | No. of Colonies . | ||
|---|---|---|---|
| Untreated . | IL-6 (ng/mL) . | ||
| 1 . | 5 . | ||
| M1 | 197 | 120 | 16 |
| M1neo (11) | 199 | 116 | 28 |
| M1neo (12) | 202 | 124 | 22 |
| M1Egr-1 (2) | 72 | 36 | 2 |
| M1Egr-1 (3) | 64 | 30 | 7 |
| M1Egr-1 (4) | 90 | 60 | 5 |
| M1Egr-1 (5) | 69 | 34 | 6 |
| Cell Type . | No. of Colonies . | ||
|---|---|---|---|
| Untreated . | IL-6 (ng/mL) . | ||
| 1 . | 5 . | ||
| M1 | 197 | 120 | 16 |
| M1neo (11) | 199 | 116 | 28 |
| M1neo (12) | 202 | 124 | 22 |
| M1Egr-1 (2) | 72 | 36 | 2 |
| M1Egr-1 (3) | 64 | 30 | 7 |
| M1Egr-1 (4) | 90 | 60 | 5 |
| M1Egr-1 (5) | 69 | 34 | 6 |
Colonies in soft agar were counted 14 days after seeding 300 cells per 60-mm dish. All experiments were initiated with nonadherent cells. Numbers represent the mean of three independent experiments with standard deviations up to ±15%.
Photomicrographs of M1 and M1Egr-1 colonies in soft agar. Representative photomicrographs (original magnification × 100) of M1 and M1Egr-1 colonies 7 days after the cells were seeded in soft agar, in the absence or presence of IL-6. The number of colonies for the different cell types is presented in Table 2.
Photomicrographs of M1 and M1Egr-1 colonies in soft agar. Representative photomicrographs (original magnification × 100) of M1 and M1Egr-1 colonies 7 days after the cells were seeded in soft agar, in the absence or presence of IL-6. The number of colonies for the different cell types is presented in Table 2.
The proto-oncogenes c-myc and c-myb are expressed in proliferating M1 and other hematopoietic cells and are downregulated during terminal differentiation.31,39 40 Analysis ofc-myc and c-myb transcripts showed that the expression of both of these proto-oncogenes was appreciably reduced in untreated M1Egr-1 compared with parental M1 cells (Fig 4A), consistent with the reduced proliferation and the differentiated state of the M1Egr-1 cell population.
Analysis of expression of the proto-oncogenes c-myb and c-myb (A), MyD genes (B), and late genetic markers (C) before and after stimulation of M1 and M1Egr-1 cells with 1 and 50 ng/mL of IL-6. RNA was extracted from the cells at the indicated times after stimulation with IL-6. The RNA was resolved on formaldehyde-agarose gels and transferred to Durolon nylon membranes for Northern blot analysis. These data were generated using two RNA blots prepared at the same time from common RNA samples. Blots were stripped and reprobed as described in Materials and Methods.
Analysis of expression of the proto-oncogenes c-myb and c-myb (A), MyD genes (B), and late genetic markers (C) before and after stimulation of M1 and M1Egr-1 cells with 1 and 50 ng/mL of IL-6. RNA was extracted from the cells at the indicated times after stimulation with IL-6. The RNA was resolved on formaldehyde-agarose gels and transferred to Durolon nylon membranes for Northern blot analysis. These data were generated using two RNA blots prepared at the same time from common RNA samples. Blots were stripped and reprobed as described in Materials and Methods.
The effect of ectopic expression of Egr-1 on activation of the macrophage differentiation program was further investigated by analysis of the expression of Fc and C3 receptors, early markers of M1 myeloid differentiation.33 As shown in Table 1, 28% to 39% of M1Egr-1 cells displayed Fc and C3 receptors, compared with only 2% of the M1 and M1neo control cell lines.
Further characterization of the effects of constitutive expression of the Egr-1 transgene on the myeloid differentiation program was achieved by examining expression of the myeloid differentiation primary response (MyD) genes c-Jun, junD, junB, MyD88,Icam-1,6 and stat3,41 which are induced in the absence of de novo protein expression after stimulation of M1 cells for terminal differentiation. As shown in Fig 4B, untreated M1 cells expressed low basal levels of junD, whereas expression of MyD genes cJun and junB was undetectable. In sharp contrast, all of these MyD genes were highly expressed in M1Egr-1 cells, at levels comparable to their expression in M1 cells induced for differentiation by IL-6. However, expression of MyD88,Icam-1, and stat3 remained undetectable in untreated M1Egr-1, similar to M1 and M1neo cell lines.
The expression of late genetic markers associated with M1 myeloid terminal differentiation, namely ferritin light chain andlysozyme,31 42 was examined as well. As shown in Fig 4C, expression of ferritin light-chain and lysozymemRNAs was undetectable in unstimulated M1 cells. In contrast, unstimulated M1Egr-1 cells expressed high basal levels of bothferritin light-chain and lysozyme mRNAs, similiar to what was observed in M1 stimulated with optimal concentration (50 ng) of IL-6.
Taken together, these observations indicate that ectopic expression of Egr-1 in M1 cells was sufficient for activation of a subset of early to late cellular, biochemical, and genetic markers of the IL-6-induced macrophage differentiation program.
Ectopic expression of Egr-1 increases the propensity of M1 cells to be induced for macrophage differentiation by IL-6.
The growth response of M1, M1neo, and M1Egr-1 cells after treatment with varying concentrations of IL-6 for 3 days is depicted in Fig 2B. It can be seen that low concentrations of IL-6 (up to 1 ng/mL) did not inhibit and even somewhat stimulated the proliferation of control M1 and M1neo cell lines; in contrast, proliferation of M1Egr-1 cell lines, which was markedly inhibited in the absence of IL-6, was further suppressed (compare Fig 2A and B). Consistent with the effect of low concentrations of IL-6 on growth inhibition of M1Egr-1, low concentrations of IL-6 (1 ng/mL) further increased the percentage of M1Egr-1 cells that adhered to the culture dish compared with untreated cells (60% to 65% compared with 52% to 57%), whereas at most 10% of M1 and M1neo cell lines were adherent (Table 1). Analysis of cell morphology showed that, at low concentrations of IL-6 (1 ng/mL), control M1 and M1neo cells retained predominantly blast-like morphology (94% to 96%). In sharp contrast, low concentrations of IL-6 further increased the percentage of M1Egr-1 cells that differentiated into mature cell types compared with untreated cells (65% to 69% compared with 33% to 39%) and decreased the percentage of cells at intermediate stages of differentiation, with not more than 5% of the cells retaining the blast morphology (Table 1). These data are consistent with the notion that ectopic expression of Egr-1, in addition to allowing M1 cells to undergo differentiation in the absence of any exogenous stimuli, also renders M1 cells responsive to low levels of IL-6 that have no or a minimal effect on parental M1 or M1neo cells.
At 50 ng/mL of IL-6, the optimal concentration of IL-6 for M1 differentiation,33 M1Egr-1 cells exhibited more pronounced inhibition of growth compared with M1 and M1neo cells. In addition, 80% to 90% of M1Egr-1 cells adhered to the culture dish, whereas only 60% to 62% of M1 and M1neo cell lines were adherent (Table 1). At the optimal concentration of IL-6, 78% to 82% of the M1Egr-1 cells terminally differentiated into mature macrophages, whereas only 67% to 71% of the control cells displayed a mature morphology7,9 34 (Table 1). Thus, ectopic Egr-1 expression allows a higher proportion of M1 cells to become terminally differentiated under conditions of optimal stimulation for M1 differentiation.
Consistent with the results in mass culture, all of the colonies formed in soft agar by M1Egr-1 cells in the presence of 1 ng/mL IL-6 displayed a diffuse morphology at 7 days, characteristic of differentiated cells, compared with 30% for untreated M1Egr-1 cells. In contrast to M1Egr-1, no diffuse morphology was observed with similarly treated M1 and M1neo cells (Fig 3). For all cell types, colony formation was inhibited with increasing concentrations of IL-6 (Table 2).
To corroborate and extend our findings, further analysis of markers associated with the differentiation phenotype was performed. These markers include the proto-oncogenes c-myc and c-myb, Fc and C3 receptors, myeloid differentiation (MyD) genes, and the late markers ferritin and lysozyme.
The proto-oncogenes c-myb and c-myc were completely suppressed in M1Egr-1 cells after treatment with 1 ng/mL of IL-6, yet this concentration of IL-6 had little, if any, effect on c-myb and c-myc expression in M1 (Fig 4A) or M1neo cells (data not shown). These data are consistent with the observed growth suppression of M1Egr-1 cells.
Analysis of the expression of Fc and C3 receptors showed that, at low IL-6 concentrations (1 ng/mL), the percentage of M1Egr-1 cells expressing Fc (56% to 58%) and C3 (63% to 64%) receptors increased compared with untreated M1Egr-1 cells, as well as compared with similarly treated M1 or M1neo cells. After treatment with the optimum concentration of IL-6 (50 ng/mL; Table 1), the percentage of cells expressing Fc and C3 receptors was similar for all cell types.
Analysis of MyD gene expression has shown that the low concentration (1 ng/mL) of IL-6 was sufficient to induce cJun, junD, junB, MyD88, Icam-1, and stat3 in both parental M1 and M1Egr-1 cells (in which expression of c-Jun, junD, and junB was already increased in untreated M1Egr-1; Fig 4B). However, the levels of MyD88, Icam-1, and stat3 decrease at later times in M1 cells, but not in M1Egr-1 cells. Egr-1 was only transiently induced to barely detectable levels by 1 ng/mL IL-6 in M1 cells. At the optimum concentration of IL-6 (50 ng/mL), all seven MyD genes assessed are stably induced in both M1 and M1Egr-1 cells (Fig 4B). Thus, it appears that induction of MyD genes in itself is not an indicator of induction of the differentiated state of the cell.
Finally, analysis of the expression of the late genetic markers, ie, the ferritin light-chain and lysozyme, has shown that 1 ng/mL of IL-6 further elevated the high basal expression levels offerritin light-chain and lysozyme in the M1Egr-1 cells (Fig 4C). In contrast, in M1 cells, 1 ng/mL of IL-6 induced very low expression of these genes (Fig 4C), and it should be noted that, even with the optimal concentration (50 ng/mL) of IL-6, expression offerritin light-chain and lysozyme was lower in M1 compared with M1Egr-1 cells, either untreated or treated with the low (1 ng/mL) concentration of IL-6. Expression of these late markers reflects the differentiated state of each cell population.
Ectopic expression of Egr-1 decreases the leukemogenicity of M1 cells.
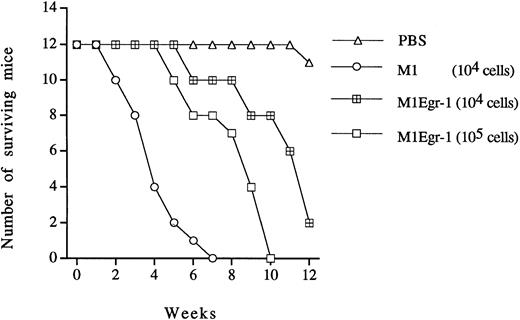
M1 cells are leukemogenic when injected into syngeneic or nude mice, and their leukemogenicity is lost after induction of differentiation in vitro or in vivo.33,43 It has been shown that constitutive expression of Egr-1 in M1 cells activated the monocytic differentiation program, in which a substantial portion of the cells underwent differentiation in the absence of external stimuli, reduced the growth rate, and allowed the cells to be highly responsive to low levels of the differentiation inducer IL-6. Therefore, to better understand the relationship between these acquired traits in vitro and leukemogenicity in vivo, the leukemogenicity of the M1Egr-1 cells was compared with the parental M1 cells. As shown in Fig 5, 12 control nude mice injected with PBS survived for the observed 12 weeks, whereas all nude mice that were injected with 104 M1 cells died within 7 weeks. In contrast, only 2 of 12 nude mice injected with the same number of M1Egr-1 cells died within this time period. Even when the nude mice were injected with 105 M1Egr-1 cells, only 4 of 12 animals died during the 7-week time period, in which the surviving nude mice showed no sign of leukemogenicity. Also, 5 weeks after injection, no myeloid leukemic cells were recovered from bone marrow of animals injected with M1Egr-1 cells, as determined by growth autonomy and differentiation characteristics.8 In contrast, during the same period of time, leukemic myeloid cells were recovered from the bone marrow of nude mice injected with M1 cells. Thus, constitutive expression of Egr-1 decreased the leukemogenicity of the cells in vivo.
Leukemogenicity of M1 and M1Egr-1 cells in nude mice. For each cell type, 12 nude mice were intravenously injected (tail vein) with 104 or 105 cells prepared in 200 μL of 1× PBS. Control animals were injected with same volume of 1× PBS. The experiment was terminated after 12 weeks. All surviving mice were asymptomatic. (▵) PBS; (○) M1 (104 cells); (⊞) M1Egr-1 (104 cells); (□) M1Egr-1 (105cells).
Leukemogenicity of M1 and M1Egr-1 cells in nude mice. For each cell type, 12 nude mice were intravenously injected (tail vein) with 104 or 105 cells prepared in 200 μL of 1× PBS. Control animals were injected with same volume of 1× PBS. The experiment was terminated after 12 weeks. All surviving mice were asymptomatic. (▵) PBS; (○) M1 (104 cells); (⊞) M1Egr-1 (104 cells); (□) M1Egr-1 (105cells).
Effect of deregulated Egr-1 on the expression of IL-6 and its receptor subunit gp130.
Multiple cytokine- and second messenger-responsive elements have been located within the 5′ regulatory region of the IL-6 gene, including AP-1 binding sites that are recognized by the AP-1 transcription factor complexes encoded by proto-oncogenes of the fos/jun family.44,45 We have reported that constitutive expression of a c-fos transgene in M1 cells, which increased the sensitivity of M1fos cells for differentiation by IL-6, resulted in stimulation of endogenous synthesis of IL-6 and higher inducibility of the IL-6 gene.33 These observations led us to explore whether Egr-1 mediated the activation of endogenous IL-6 and/or upregulation of expression of its receptor gp130, thereby contributing to the predisposition of M1Egr-1 cells for terminal differentiation.
To explore this possibility, we have used quantitative PCR to determine the mRNA levels of IL-6 and its receptor subunit gp130 in M1Egr-1 cells compared with M1 cells, before and after stimulation for differentiation. No difference in the levels of IL-6 and gp130 mRNAs was observed between M1neo and M1Egr-1 cells (data not shown). Taken together, these data indicate that the Egr-1-mediated predisposition of M1 cells for terminal differentiation is not due to either induction of IL-6 or upregulation of the IL-6 receptor signal transducer gp130 subunit.
DISCUSSION
Egr-1 was found by us to be a macrophage differentiation primary response gene that is essential for and restricts differentiation along the macrophage lineage.10 More recently, it has been shown that Egr-1 potentiates macrophage differentiation of the hematopoietic precursor cell line 32Dcl3.30 In the present work, it is shown that ectopic expression of Egr-1 in the myeloblastic leukemic cell line M1 resulted in activation of the macrophage differentiation program, rendered M1 cells responsive to low levels of IL-6, allowed a higher proportion of M1 cells to become terminally differentiated under conditions of optimal stimulation for differentiation, and decreased the leukemogenicity of M1 cells.
Constitutive expression of many differentiation associated characteristics was observed in the absence of IL-6 treatment, including the appearance of morphologically differentiated cells. Ectopic expression of Egr-1 in M1 cells decreased the growth rate in mass culture, the cloning efficiency in soft agar, and leukemogenicity in vivo and, consistent with these observations, expression of endogenous c-myb and c-myc mRNAs was markedly downregulated. Untreated M1Egr-1 cells also exhibited cell adherence, expression of Fc and C3 receptors, and upregulation of the myeloid differentiation primary response genes c-Jun, junD, andjunB and the late genetic markers ferritin light-chainand lysozyme. Whether any of the observed changes in gene expression is regulated directly by Egr-1 is under investigation. Interestingly, most untreated M1Egr-1 cells exhibit either intermediate or mature monocyte morphologies, yet all the MyD genes normally associated with differentiation of both normal myeloid cells and M1 cells are not expressed. Therefore, these monocytes represent cells that have not undergone complete macrophage differentiation despite the mature morphological phenotype.
The ability of Egr-1 to activate the macrophage differentiation program in M1 cells is contrary to the situation in HL-60Egr-1 and 32DEgr-1 cells, in which ectopic expression of Egr-1 did not result in the onset of spontaneous differentiation along the monocytic lineage. Ectopic Egr-1 expression blocked the dimethyl sulfoxide (DMSO)-induced granulocytic differentiation of HL-60 cells10 and granulocyte colony-stimulating factor (G-CSF)–induced differentiation of IL-3–dependent 32Dcl3 hematopoietic precursor cells, endowing the cells with the new ability to be induced by granulocyte-macrophage colony-stimulating factor (GM-CSF) for terminal differentiation exclusively along the macrophage lineage.30 Interestingly, in 32Dcl3 cells, expression of Egr-1 increased expression of NSE and ferritin in unstimulated cells, whereas no evidence for Egr-1–induced activation of macrophage markers was observed in unstimulated HL60 cells.10 The difference in the reponses of unstimulated M1, 32Dcl3, and HL-60 cells to expression of an Egr-1 transgene may be attributed to their state of lineage commitment for differentiation. Unlike M1 cells, which are predetermined for terminal monocytic differentiation,7,9,31 and 32D cl3 cells, which are predetermined for granulocytic differentiation,46 HL-60 cells are bipotential and capable of undergoing differentiation into macrophages in response to phorbol 12-myristate 13-acetate (PMA)47 and into granulocytes in response to DMSO.48 Also, HL-60 cells respond more strongly to nonphysiological inducers (PMA and DMSO) than to physiological inducers of differentiation.49 The results of the present work, thus, extend our previous studies, demonstrating that the functions of Egr-1 as a positive modulator of macrophge differentiation vary, depending on the hematopoietic cell type.
In M1 and HL-60 cells, the proto oncogenes c-myb andc-myc, which are expressed at high levels in proliferating cells, are downregulated upon induction of differentiation subsequent to the induction of Egr-1 expression.10,31,50 In this study, we have shown that enforced expression of Egr-1 downregulatesc-myb and c-myc mRNA. In view of the reported findings that EGR-1 protein interacts with the promoter regions ofc-myb51 and c-myc genes,52 it is possible that Egr-1 may be directly involved in downregulating the expression of c-myb and c-myc genes during M1 myeloid cell differentiation. In contrast, in HL-60Egr-1 cells, in which ectopic Egr-1 expression was observed to block DMSO-induced granulocytic differentiation, c-myb expression was markedly upregulated compared with its expression in the parental cells.10 Thus, it will be interesting to study the relationship between Egr-1, c-myb, and c-myc in M1 compared with HL-60 cells to understand the mechanism of interaction of these genes during myeloid cell differentiation.
Ectopic expression of Egr-1, in addition to allowing M1 cells to undergo differentiation in the absence of any exogenous stimuli, also renders M1 cells responsive to low levels of IL-6 that have no effect or a minimal effect on parental M1 or M1neo cells. Even with an IL-6 concentration as low as 1 ng/mL, M1Egr-1 cells displayed all the morphological characteristics and stably expressed early and late genetic markers associated with terminal differentiation. The increased propensity of M1Egr-1 cells for differentiation was not due to the upregulation of IL-6 or its receptor signal transducer gp130 by Egr-1, because there is no detectable induction of transcripts for these genes. However, ectopic expression of Egr-1 has a profound effect on M1 cells, activating the macrophage differentiation program, and any one or combination of the Egr-1–mediated changes may account for the increased sensitivity to respond to IL-6.
It should be noted that all the MyD genes that were assessed, with the exception of Egr-1, were induced in both parental and M1neo cells using low levels of IL-6. This suggests that expression of several MyD genes is not sufficient to activate the differentiation program. These data further implicate Egr-1 as a major player in regulating myeloid differentiation.
Previously, we have observed that, whereas c-fos andJuns (cJun, junB, and junD) are stably induced during normal macrophage and/or granulocyte differentiation, only the Juns are induced upon macrophage differentiation of the M1 cells.33,53,54 Because it has been demonstrated that M1 cells ectopically expressing c-fosexhibit some differentiation associated markers and respond to low levels of IL-6 in a manner similar to M1Egr-1 cells,33 it was of interest to ascertain if c-fos is expressed in M1Egr-1. No c-fos expression was detected (data not shown). It is possible that the high levels of constitutive expression ofcJun, junD, and junB in M1Egr-1, which is not observed in M1fos,33 may substitute for c-fosand contribute to the increased susceptibility for induction of terminal differentiation by IL-6. The marked reduction in the expression of c- myc and c-myb in M1Egr-1 cells, compared with parental cells, may also play a role in increasing the propensity of M1Egr-1 for induction of terminal differentiation by IL-6. Clearly, it is of future interest to determine whether any one or a combination of these molecular events may be responsible for the Egr-1–mediated increase in the sensitivity of M1 cells to be induced for terminal differentiation.
Recently, it has been reported that stat3 is essential for and induces M1 myeloid cell macrophage differentiation in the absence of Egr-1 induction.41 On the other hand, our results demonstrate that enforced expression of Egr-1 predisposes M1 cells and activates the macrophage differentiation program in the absence ofstat3 expression. These observations raise the possibility that Egr-1 and stat3 may use distinct pathways to induce differentiation.
The Icam-1 gene, which was recently identified as an Egr-1 target gene in primary B lymphocytes and B-cell lines,27was not among the genes induced in response to ectopic expression of Egr-1 in unstimulated M1Egr-1 cells. Thus, Icam-1 appears not to be an Egr-1 target gene in M1 myeloid cells. However, our findings do not rule out the possibility that Egr-1 may play a role in maintaining the stable expression of Icam-1 during myeloid differentiation. Additional experiments with M1 cells, as well as other differentiation-inducible myeloid precursor cell lines, are needed to determine whether Icam-1 may be a primary target for Egr-1 in myeloid cells and/or whether Egr-1 may contribute to the regulation of Icam-1 expression during myeloid differentiation.
Our results demonstrate that ectopic expression of Egr-1 impairs the leukemogenicity of M1 myeloblastic leukemia cells in vivo. Consistent with these findings is the report that deletion of the Egr-1 gene on human chromosome 5 results in acute myeloid leukemia.55Furthermore, many tumor cell lines express little or no Egr-1, and enforced expression of Egr-1 suppresses the growth of different types of tumor cell lines in vivo.21 22
Recently, it was reported that macrophage differentiation was not affected in mice lacking the Egr-1 gene, suggesting that Egr-1 may not be required for macrophage differentiation in vivo.56However, it has been reported that the Egr family members, Egr-2, Egr-3, and Egr-4,12 share a high degree of structural and functional homology with Egr-1, including the DNA binding domains. This suggests that proteins of the Egr family may be capable of substituting for Egr-1 function in the control of macrophage differentiation; this possibility currently is being investigated.
In conclusion, the findings reported here increase our understanding of how cell differentiation and its associated growth control is regulated by Egr-1 and how these phenomena influence the leukemogenicity of cells. This work extends our previous work demonstrating that the function of Egr-1 as a positive modulator of macrophage differentiation varies, depending on the state of lineage commitment for differentiation of the hematopoietic cell type.
ACKNOWLEDGMENT
The authors thank Dr Arthur G. Balliet for his critical evaluation and comments.
Supported by National Institutes of Health Grants No. 1R01CA59774 (D.A.L.) and 1R01CA51162 (B.H.), by the core program on carcinogenesis (5P3CA12227), and by Amgen, Inc. (Thousand Oaks, CA) (B.H. and D.A.L.).
Address reprint requests to Dan A. Liebermann, PhD, or Barbara Hoffman, PhD, Fels Institute for Cancer Research and Molecular Biology and Department of Biochemistry, Temple University School of Medicine, 3307 N Broad St, Philadelphia, PA 19140; e-mail: dan@sgil.fels.temple.edu.
The publication costs of this article were defrayed in part by page charge payment. This article must therefore be hereby marked "advertisement" is accordance with 18 U.S.C. section 1734 solely to indicate this fact.



This feature is available to Subscribers Only
Sign In or Create an Account Close Modal