Abstract
Whether Kaposi’s sarcoma herpesvirus (KSHV) is associated with multiple myeloma (MM) remains controversial. We assayed for KSHV DNA sequences in long-term bone marrow stromal cells (BMSCs) from 26 patients with MM and 4 normal donors. Polymerase chain reaction (PCR) using primers which amplify a KSHV gene sequence to yield a 233-bp fragment (KS330233 within open reading frame 26) was negative in all cases. Aliquots of these PCR products were used as templates in subsequent nested PCR, with primers that amplify a 186-bp product internal to KS330233. BMSCs from 24 of 26 (92%) patients with MM and 1 of 4 normal donors were KSHV PCR+. DNA sequence analyses showed interpatient specific mutations (2 to 3 bp). Both Southern blot and sequence analyses confirmed the specificity of PCR results. The presence of the KSHV gene sequences was further confirmed by amplifying T 1.1 (open reading frame [ORF] K7) and viral cyclin D (ORF 72), two other domains within the KSHV genome. Immunohistochemical studies of KSHV PCR+ MM BMSCs demonstrate expression of dendritic cell (DC) lineage markers (CD68, CD83, and fascin). Serological studies for the presence of KSHV lytic or latent antibodies were performed using sera from 53 MM patients, 12 normal donors, and 5 human immunodeficiency virus (HIV)/KSHV+ patients. No lytic or latent antibodies were present in sera from either MM patients or normal donors. Taken together, these findings show that KSHV DNA sequences are detectable in BMSCs from the majority of MM patients, but that serologic responses to KSHV are not present. Ongoing studies are defining whether the lack of antibody response is caused by the absence of ongoing infection, the presence of a novel viral strain associated with MM, or underlying immunodeficiency in these patients.
WHETHER KAPOSI’S SARCOMA herpesvirus (KSHV) is associated with multiple myeloma (MM), and if present, whether it is of pathophysiologic significance or merely an epiphenomenon, remains controversial. Although early studies did not detect KSHV in MM cells,1 Rettig et al2 showed that bone marrow stromal cells (BMSCs), but not MM cells themselves, are infected with KSHV.2 These studies are consistent with prior observations that MM cells themselves are not infected with KSHV,1 and suggest the possibility that KSHV may promote the growth and/or survival of tumor cells in a paracrine mechanism by secretion of cytokines, ie, viral interleukin-6 (vIL-6),3 from infected BMSCs. However, to date only three groups have shown KSHV DNA within BM biopsy specimens from MM patients,4-6 whereas multiple other groups have not confirmed an association of KSHV with MM. Specifically, investigators from France, England, Italy, and the United States have not found antibodies to KSHV in sera from MM patients despite humoral responses to other herpesviruses,7-12 and other investigators using polymerase chain reaction (PCR) have failed to detect KSHV DNA in either freshly isolated bone marrow mononuclear cells (BMMCs),9,13 BMSCs,10 or BM biopsy samples14 from patients with MM.
In the present study, we assayed for KSHV DNA sequences in long-term BMSCs from 26 patients with MM and 4 normal donors. PCR using primers that detect KSHV gene sequence (KS330233)15 was negative; however, subsequent nested PCR using primers that amplify sequences internal to KS330233 to yield a final PCR product of 186 bp was positive in 24 of 26 (92%) patients with MM and 1 of 4 normal donors. Southern blotting analyses and gene sequence analysis confirmed specificity of PCR results. Moreover, PCR amplification of T 1.1 and viral cyclin D regions in the KSHV genome further confirmed the presence of KSHV gene sequences in MM BMSCs. However, serological analyses showed lack of either KSHV lytic or latent antibodies in MM patients as well as normal donors. Therefore, our studies demonstrate that KSHV gene sequences are detectable in MM BMSCs in the absence of a serological response.
MATERIALS AND METHODS
Preparation of long-term BMSC cultures.
After appropriate informed consent and under the auspices of an Insitutional Review Board approved protocol, Ficoll Hypaque (Pharmacia Biotech, Inc, Piscataway, NJ) BMMCs were freshly obtained from 26 patients with MM and 4 normal donors, obtained at the time of BM harvest for allografting. BMMCs were cultured for 2 to 6 weeks in Iscove’s media (Sigma Diagnostics, St Louis, MO) with 10% fetal bovine serum (FBS, Sigma), 10% horse serum (Sigma), and penicillin/streptomycin (pen/strep; GIBCO-BRL, Gaithersburg, MD) to generate long-term BMSC cultures, as previously described.2Adherent layers were trypsinized and assayed for presence of KSHV DNA, as well as cell-surface phenotype.
PCR assays for KSHV sequences.
DNA was prepared using the Easy-DNA kit per manufacturer’s instructions (Invitrogen, Carlsbad, CA). Genomic DNA was extracted from freshly isolated MM BMMCs and normal BMMCs, from 3- to 6-week-old MM BMSC and normal BMSC cultures. To increase the specificity of PCR, the DNA was amplified in a two-step PCR, first with outer primers and then with inner primers nested within the first primers.16,17PCR with nested primers has been used to detect HIV-1 in tumor cell lines and in clinical samples.16,18 We first performed PCR using primers that recognize and amplify a region within the KSHV gene sequence to yield a 233-bp fragment (KS330233), as previously described.15 An aliquot from the first PCR amplification product was used as template in a subsequent nested PCR using primers (forward primer, 5′-CTC GAA TCC AAC GGA TTT GA-3′; reverse primer, 5′-ATA TGT GCG CCC CAT AAA TG-3′) that recognize sequences internal to this 233-bp fragment to yield a final PCR product of 186 bp. The thermal cycling conditions, similar for both the initial and nested PCR reactions, were as follows: 95°C for 3 minutes (1 cycle); 94°C for 1 minute, 60°C for 1 minute, 72°C for 1 minute (45 cycles); 72°C extension for 5 minutes (1 cycle). Each PCR reaction uses 500 ng to 800 ng of genomic DNA, and 30 pmol of each primer. The PCR reactions were performed in Ready-To-Go PCR beads (Pharmacia Biotech AB, Uppsala, Sweden) with a final volume of 25 μL reaction mixture, and amplifications performed in a Perkin-Elmer 480 Thermocycler (Applied Biosystems, Foster City, CA). Fifteen microliters of the PCR product was analyzed on 1.5% agarose gels containing ethidium bromide and photographed under UV light. Genomic DNA from body cavity–based lymphoma (BCBL)-1 cell line infected with KSHV19 served as a positive control for the detection of KSHV gene sequences. PCR with β-actin primers was performed on all samples to assure quality of genomic DNA.
A similar nested PCR approach was used to amplify viral cyclin D (open reading frame [ORF] 72) with previously described primer sets and thermal cycling conditions.20 In addition, genomic DNA from MM patients was also assayed for another gene unique to KSHV, designated ORF K7 or T1.1. For T1.1 (ORF K7), the outer primer set was 5′ CTT GCC GCT TCT GGT TTT CA 3′ as forward primer and 5′ CAC CAG TGG GCG CTG CTT CTT TC 3′ as backward primer; the inner primer set was 5′ CTT GCC GCT TCT GGT TTT CA 3′ as forward primer and 5′ GGC GCT GCT TTC CTT TCA CA 3′ as backward primer.21-23 The thermal cycling conditions were as follows: 95°C for 3 minutes (1 cycle); 94°C for 1 minute, 62°C for 1 minute, and 72°C for 1 minute (45 cycles); and 72°C extension for 5 minutes (1 cycle).
Southern blot analyses and DNA sequencing of PCR products.
Southern blot analyses were performed to confirm the specificity of PCR. The PCR products observed in the agarose gels were transferred onto nitrocellulose filter membranes which were then hybridized to a32P end-labeled 25-bp oligomer probe internal to the KS330233 sequence (5′ TGC AGC AGC TGT TGG TGT ACC ACA T), as previously described.15 Filters were washed and exposed to Kodak X-O mat XAR film (Eastman Kodak, Rochester, NY) using an intensifying screen. The autoradiograms were scanned using the LKB Produkter (LKB, Bromma, Sweden) ultrascan XL laser densitometer and analyzed with a Gelscan XL software package (LKB). DNA sequencing of the 186-bp nested PCR product was performed on 7 KSHV+ MM samples. The nested PCR products obtained after amplification of viral cyclin D and T1.1 regions were also purified and sequenced from 4 and 2 MM patients, respectively.
Phenotypic analysis.
Serologic assays.
RESULTS AND DISCUSSION
KSHV DNA sequence is detectable in MM BMSCs.
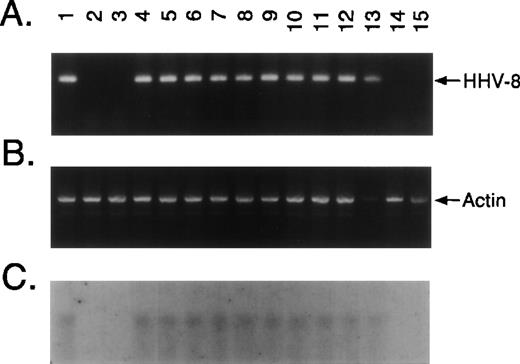
Ficol-Hypaque BMMCs were freshly obtained from 26 patients with MM and 4 normal donors and cultured for 2 to 6 weeks in Iscove’s media. Adherent layers were trypsinized and assayed for presence of KSHV DNA sequence by PCR amplification of extracted DNA with primers that detect KSHV gene sequences (KS330233).15 No KS330233 was detectable in these samples. Aliquots from these PCR amplification products were used as templates for nested PCR with a second set of primers that recognize sequences internal to KS330233 to yield a final PCR product of 186 bp. KSHV-specific amplicons were demonstrable in BMSCs from 24 of 26 (92%) patients with MM and 1 of 4 normal donors, but were not detected in freshly isolated BMMCs from 4 MM patients. Representative results in BMSCs from 12 MM patients and 2 normal donors are shown in Fig1A. Genomic DNA from BCBL-1 cell line infected with KSHV19 served as a positive control for the detection of KSHV gene sequences (Fig 1A, lane 1). KSHV DNA was evident in long-term BMSCs from 10 of 12 patients with MM (lanes 4 through 13), including those 4 cases (lanes 7, 8, 12, and 13) in which fresh MM BMMCs were negative for KSHV by nested PCR. In contrast, KSHV DNA was absent in BMSCs from 2 MM patients (lanes 14 and 15) and 2 normal donors (lanes 2 and 3). Amplification with β-actin–specific primers confirmed DNA in each lane (Fig 1B).
KSHV DNA sequence (ORF 26) is detectable in MM BMSCs. Ficoll Hypaque BMMCs were freshly obtained from 26 patients with MM and 4 normal donors and cultured for 2 to 6 weeks in Iscove’s media with 10% FBS, 10% horse serum, and penicillin/streptomycin to generate long-term BMSC cultures. Adherent layers were trypsinized and assayed for presence of KSHV DNA sequence. (A) DNA from long-term BMSCs from 12 patients with MM and 2 normal donors were assayed by PCR using primers that amplify KSHV gene sequences (KS330233) followed by nested PCR using primers designed to recognize sequences internal to KS330233 to yield a final PCR product of 186 bp. DNA from BCBL-1 cell line (lane 1) served as a positive control for the detection of KSHV gene sequences and DNA obtained from normal BMSCs (lanes 2 and 3) as a negative control; MM patient BMSCs are shown in lanes 4 through 15. (B) Amplification with β-actin specific primers confirmed DNA in each lane. (C) To confirm the specificty of PCR, nested PCR products were transferred onto nitrocellulose filters. Southern blot analysis was performed by hybridizing the filters with a32P end-labeled 25-bp oligomer probe internal to the KS330233 sequence.
KSHV DNA sequence (ORF 26) is detectable in MM BMSCs. Ficoll Hypaque BMMCs were freshly obtained from 26 patients with MM and 4 normal donors and cultured for 2 to 6 weeks in Iscove’s media with 10% FBS, 10% horse serum, and penicillin/streptomycin to generate long-term BMSC cultures. Adherent layers were trypsinized and assayed for presence of KSHV DNA sequence. (A) DNA from long-term BMSCs from 12 patients with MM and 2 normal donors were assayed by PCR using primers that amplify KSHV gene sequences (KS330233) followed by nested PCR using primers designed to recognize sequences internal to KS330233 to yield a final PCR product of 186 bp. DNA from BCBL-1 cell line (lane 1) served as a positive control for the detection of KSHV gene sequences and DNA obtained from normal BMSCs (lanes 2 and 3) as a negative control; MM patient BMSCs are shown in lanes 4 through 15. (B) Amplification with β-actin specific primers confirmed DNA in each lane. (C) To confirm the specificty of PCR, nested PCR products were transferred onto nitrocellulose filters. Southern blot analysis was performed by hybridizing the filters with a32P end-labeled 25-bp oligomer probe internal to the KS330233 sequence.
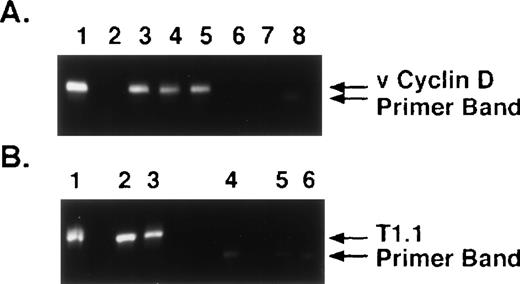
The specificty of nested PCR product was determined by Southern blot analyses. The results showed reactivity in lanes 4 through 13 and lack of reactivity in lanes 2, 3, 14, and 15, confirming our PCR results (Fig 1C). To confirm KSHV gene sequences, we purified the 186-bp nested PCR product from 7 MM patient samples and performed DNA sequencing, using either forward or reverse primers. The DNA sequences from all 7 patients demonstrated 92% to 96% homology to sequences (1030 bp to 1177 bp) present within the KS330 Bam fragment of KSHV genome.15 Interpatient specific mutations were observed among these patients, which reaffirms the lack of PCR contamination. In addition, two different regions of the KSHV genome, ORF 72 (viral cylin D) and ORF K7 (T 1.1), were also amplified by nested PCR in DNA of BMSCs from MM patients and normal donors (Fig 2A and 2B, respectively). ORF 72 encodes for a functional cyclin D homolog with 31% identity to cellular cyclin D which is also found in Herpesvirus saimiri. Viral cyclin D is capable of phosphorylating the retinoblastoma protein and is known to be expressed in latency. ORF K7 or T1.1 is among the 15 different genes (K1-15) that are unique to KSHV. The results showed PCR positivity for both of these genes in MM patient DNA samples, whereas normal BMSCs were PCR negative. Moreover, sequence analyses of PCR products showed 97% homology to known KSHV sequence and demonstrated patient specific mutations.
KSHV DNA sequences viral cyclin D (ORF 72) and T1.1 (ORF K7) are detectable in MM BMSCs. (A) DNA from long-term BMSCs from 4 patients with MM (lanes 2 through 5) and 2 normal donors (lanes 6 and 7) were assayed by nested PCR to amplify viral cyclin D (ORF 72) KSHV gene sequence. DNA from BCBL-1 cell line (lane 1) served as a positive control and water (H2O) (lane 8) served as control for nested PCR contamination. (B) DNA from long-term BMSCS from 2 patients with MM (lanes 2 and 3) and 2 normal donors (lanes 5 and 6) were assayed by nested PCR to amplify T1.1 (ORF K7). DNA from BCBL-1 cell line (lane 1) served as a positive control and water (H2O) (lane 4) served as a control for nested PCR contamination.
KSHV DNA sequences viral cyclin D (ORF 72) and T1.1 (ORF K7) are detectable in MM BMSCs. (A) DNA from long-term BMSCs from 4 patients with MM (lanes 2 through 5) and 2 normal donors (lanes 6 and 7) were assayed by nested PCR to amplify viral cyclin D (ORF 72) KSHV gene sequence. DNA from BCBL-1 cell line (lane 1) served as a positive control and water (H2O) (lane 8) served as control for nested PCR contamination. (B) DNA from long-term BMSCS from 2 patients with MM (lanes 2 and 3) and 2 normal donors (lanes 5 and 6) were assayed by nested PCR to amplify T1.1 (ORF K7). DNA from BCBL-1 cell line (lane 1) served as a positive control and water (H2O) (lane 4) served as a control for nested PCR contamination.
Our data are consistent with the prior reports2,4,5 that MM BMSCs are KSHV PCR+. Moreover, our results are in concert with other studies showing that MM cells themselves are not infected with KSHV,1,2,9 since KSHV DNA was not detected in freshly isolated MM BMMCs. In contrast, our results differ from those of Masood et al,10 who failed to detect KSHV DNA in long-term BMSCs from patients with MM. It is possible that MM BMSCs examined in this study were uninfected, or alternatively, that negative PCR results were due to a lack of sensitivity of the single-step PCR methodology. In the present study, we also did not detect KSHV DNA by PCR using primers to amplify KS330233,15 and therefore used aliquots from the first PCR amplification product as templates for nested PCR with internal primers. With this nested PCR strategy, KSHV sequences were demonstrable in the majority (24 of 26) of MM BMSCs.
Morphologic and phenotypic profile of MM BMSCs with detectable KSHV gene sequences.
To identify the lineage of the KSHV PCR+ cell within BMSCs, immunohistochemical staining was performed on cytosmears prepared from these long-term MM BMSCs using MoAbs reactive with B (CD20, κ, λ, and CD38), T (CD3), monocytoid (CD14 and CD68), hematopoietic progenitor (CD34), and dendritic (CD83 and fascin) lineage cells. Immunohistochemical studies (indirect immunoperoxidase or immunoalkaline phosphatase techniques) of the dominant large BMSCs are summarized in Table 1. These cells strongly expressed CD68, CD83, and fascin, but lacked κ and λ chain, CD20, CD3, CD14, and CD34 expression. Ill-defined cytoplasmic staining for CD38 of uncertain significance was observed. The population of smaller mononuclear cells, generally less than 5% to 10% of the cell population, included rare cells reactive for CD3 or CD14 and rare or small numbers of cells reactive for κ or λ light chains, CD20, and CD38; these cells lacked CD68, CD83, and fascin. Double-marker studies for CD20 and fascin verified the presence of two distinct cell populations: a dominant fascin-positive population consistent with DCs, and a very minor population of cells of B lineage by virtue of CD20 staining. However, it is important to note that fascin is expressed on Reed-Sternberg cells25 and can be induced by EBV on B cells.28 These findings are in concert with prior reports2 that KSHV PCR+ MM BMSCs are of DC lineage.
Immunohistochemical Analysis of MM BMSCs With Detectable KSHV Sequences
| Kappa . | Lambda . | CD3 . | CD20 . | CD14 . | CD34 . | CD38 . | CD68 . | CD83 . | Fascin . |
|---|---|---|---|---|---|---|---|---|---|
| 0/5* | 0/5 | 0/9 | 0/9 | 0/9 | 0/9 | 0/5† | 5/5 | 9/9 | 9/9 |
| Kappa . | Lambda . | CD3 . | CD20 . | CD14 . | CD34 . | CD38 . | CD68 . | CD83 . | Fascin . |
|---|---|---|---|---|---|---|---|---|---|
| 0/5* | 0/5 | 0/9 | 0/9 | 0/9 | 0/9 | 0/5† | 5/5 | 9/9 | 9/9 |
Cases reactive/cases evaluated; reactivity observed for predominant population of large mononuclear cells with abundant cytoplasm with pseudopods or cytoplasmic blebs.
Ill-defined cytoplasmic staining; no surface reactivity.
Although MM cells do not appear to be infected in the present and previous studies,1,2 it is nonetheless of interest to note that KSHV has been isolated from CD19+ B cells of normal blood donors.29 In MM, several lines of evidence show the existence of CD19+ clonotypic B cells30; however, these CD19+ cells comprise a minor population in which detection of viral infection may be difficult. Importantly, we and others have shown in prior studies that CD40 ligand (CD40L) stimulation of MM cells expands these CD19+ MM cells and triggers IL-6 secretion.31-33 Therefore, in ongoing studies we are activating MM cells in vitro using CD40L to facilitate assay of CD19+ malignant cells for the presence of KSHV sequences by nested PCR. Moreover, because human IL-6 was previously used to activate normal CD19+ B cells and thereby enhance sensitivity of KSHV detection,29 induction of IL-6 related to CD40 activation of MM cells may also aid in detection of virus in tumor cells. Thus, the precise lineage(s) of cells with detectable KSHV sequences within MM BMSCs remains under investigation.
Seroprevalence of KSHV antibodies in MM patient and normal donor sera.
Serological studies were performed by using KSHV ELISA and immunofluorescent assay to latent antigens (LANA), as previously described.26,27 The ELISA studies for the presence of KSHV lytic antibodies were performed in the laboratory of Dr D.V. Ablashi (Advanced Biotechnologies, Columbia, MD) and the LANA in the laboratory of Dr C. Boshoff (Chester Beatty Laboratory Cancer Research Institute, London, UK) on sera from 53 MM patients, 12 normal donors, and 5 HIV/KSHV+ patients. No lytic or latent antibodies were detectable in sera of MM patients or normal donors, whereas sera from HIV/KSHV patients showed detectable antibody titers to KSHV. Multiple groups have failed to demonstrate a serological response to KSHV in patients with MM, as is true in our studies, and concluded on this basis that KSHV is not associated with MM.7-12 Based on the lack of serological response to KSHV and the absence of an epidemiological link between MM and KS, these investigators have concluded that there is no association between KSHV and MM. However, given the studies demonstrating KSHV gene sequences in MM BMSCs now emanating from UCLA,2,4 France,5,6and our laboratory, this conclusion may be premature. First, the absence of humoral response to KSHV may truly reflect a lack of infection in some MM patients. Secondly, it may be that assays used in our and other negative studies do not adequately detect antibodies. In this regard, a recent report used ORF 65 and latent nuclear antigen (LNA) immunoblotting to show antibodies to ORF 65 and to LNA in 81% and 52% of MM patients, respectively.34 These results need to be confirmed using these assays to study larger numbers of MM patients. Alternatively, a specific lack of humoral response to KSHV with preserved response to other herpesviruses in KSHV may occur in KSHV infected MM patients. Finally and most likely, it may be that epitopes on KSHV which are immunogenic in other settings, ie, KS patients, do not trigger detectable immune response in MM due to the presence of different strains of KSHV in these clinical settings. In this regard, a recent report has amplified ORF 26 in MM BMSCs and some normal donor BMSCs using nested PCR, but failed to amplify ORF 72 and ORF 75, suggesting the presence of a related (KS330-containing) virus.35
In summary, the present results show (1) that KSHV gene sequences are detectable in the majority of MM BMSCs; (2) that KSHV+ MM BMSCs express DC lineage antigens, and (3) a lack of KSHV seropostivity despite the presence of KSHV gene sequences in MM patient BMSCs. Although these data show that KSHV gene sequences are evident in the majority of MM BMSCs and less commonly in normal BMSCs, whether there is infection with BMSC in MM, and whether biologically active KSHV gene products play a role in MM pathogenesis, is presently unknown and the object of ongoing studies.
Supported by National Institutes of Health Grants No. CA50947 and CA78378, and the Kraft Family Research Fund.
The publication costs of this article were defrayed in part by page charge payment. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. section 1734 solely to indicate this fact.
REFERENCES
Author notes
Address reprint requests to Kenneth C. Anderson, MD, Dana-Farber Cancer Institute, 44 Binney St, Boston, MA 02215; e-mail:kenneth_anderson@dfci.harvard.edu.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal