The B-cell antigen receptor (BCR) comprises membrane Igs (mIgs) and a heterodimer of Ig (CD79a) and Igβ (CD79b) transmembrane proteins, encoded by the mb-1 and B29 genes, respectively. These accessory proteins are required for surface expression of mIg and BCR signaling. B cells from chronic lymphocytic leukemia (B-CLL) frequently express low to undetectable surface Ig, as well as CD79b protein. Recent work described genetic aberrations affecting B29 expression and/or function in B-CLL. Because the prevalence of CLL is increased among first degree relatives, we analyzed the B29 gene in 10 families including 2 affected members each. A few silent or replacement mutations were observed at the genomic level, which never lead to truncated CD79b protein. Both members of the same family did not harbor the same mutations. However, a single silent base change in the B29 extracellular domain, corresponding to a polymorphism, was detected on 1 allele of most patients. These results indicate that the few mutations observed in the B29 gene in these patients do not induce structural abnormalities of the CD79b protein and thus do not account for its low surface expression in B-CLL. Furthermore, genetic factors were not implicated, because identical mutations were not observed among 2 members of the same family.
B-CELL CHRONIC LYMPHOCYTIC leukemia (B-CLL), the leukemia with highest incidence in western countries, is a neoplastic disease characterized by a progressive accumulation of functionally incompetent, long-lived, small, mature lymphocytes in the blood, bone marrow, and lymphoid tissues.1,2B-CLL lymphocyte is characterized by (1) expression of CD5 antigen3; (2) constant expression of low amounts of surface Ig (sIg)4,5 and CD79b6,7; (3) inability to adequately respond when stimulated through the B-cell receptor (BCR) pathway8,9; (4) resistance to infection by the Epstein-Barr Virus (EBV), in contrast with normal CD5+ B cells, despite expressing the CD21 molecule10; (5) autoreactive binding specificities10,11; (6) a prolonged G0 phase; and (7) overexpression of the bcl-2 protein.12
In B-CLL, B lymphocytes express low amounts of sIg. sIg on B cells bind noncovalently with a heterodimer Igα (CD79a)/Igβ (CD79b) to constitute the B-cell antigen receptor (BCR).13,14 CD79a and CD79b are encoded by the mb-1 and B29 genes, respectively. Both proteins contain an extracellular Ig-like domain, a transmembrane segment, and a cytoplasmic tail, which contains a motif sequence including 2 tyrosine residues. Phosphorylation of these tyrosine residues upon Ag receptor stimulation initiates signal transduction cascades.15 In addition, association of Ig with CD79a and CD79b is essential for intracellular transport and subsequent surface expression of this BCR16,17 and plays a major role in antigen internalization and induction of apoptosis through the BCR pathway.18
Defects in the BCR may account for functional impairment in BCR signaling. In addition to low expression of sIg, it was also reported that the vast majority of B-CLL patients also express low to undetectable amounts of CD79b extracellular domain.6 7
A study of mRNA in B-CLL by Thompson et al7 has recently shown that, whereas B29 mRNA was undetectable in half of their B-CLL samples, it was detected in the other samples at a level identical to that of normal B cells. However, B-CLL cells with a normal level of B29 mRNA had point mutations, insertions, or deletions in the B29 transmembrane and cytoplasmic domains, which could account for the low CD79b surface expression.
The possibility that genetic or familial factors may predispose to CLL is suggested by reports of multiple instances of the disease in some families, indicating a 2- to 7-fold excess risk in first degree relatives.19,20 In addition, the low incidence of CLL among individuals of Japanese origin, including those having migrated to Hawaii, suggests that genetic influences may play a stronger role than environmental factors in the pathogenesis of the disease.21However, the nature of the genetic predisposition remains unknown.
It is presently unclear whether the BCR could play a major role in the pathogenesis of the disease and whether the above-mentioned genetic defects observed in CD79b gene could constitute a primary event in B-CLL leukemogenesis. The study of familial cases should help address this question. We report a study at the genomic level of the B29 gene in 10 families, each with 2 siblings manifesting typical CLL. We found only a few mutations leading to amino acid replacement, none of which would be predicted to result in structural alterations of the CD79b protein. Because identical mutations of the B29 gene were not found in both affected siblings in the same family, genetics factors involving CD79b may not play a major role in CLL pathogenesis.
MATERIALS AND METHODS
Patients.
Total blood samples from 10 different Italian families, including 2 members affected with CLL, were stored at −20°C in the Clinica de la Sapienza (Rome, Italy). Families 1 through 8 have been studied for the VH gene expression of their Ig.21a Families 13 and 14 have been added in this study. All of the patients studied demonstrated typical CD5+ B-CLL. They comprised 9 men and 11 women aged from 35 to 95 years who were staged at diagnosis following Binet’ staging22 (stage A, 14 patients; stage B, 4; stage C, 1; in 1 case, stage could not be determined). Two patients progressed to stage B during evolution. At the time of the study, 5 patients had received previous treatment, whereas 15 patients did not receive any previous treatment.
DNA extraction.
Total DNA was purified from up to 10 mL of whole citrated blood with the QIAamp Blood Maxi kit (QIAGEN, Hilden, Germany). The purified DNA ranged in size up to 50 kb, with fragments of approximately 30 kb predominating. DNA yields ranged from 15 to 600 μg according to the cell densities.
Polymerase chain reaction (PCR) amplification.
PCR amplification was performed with genomic DNA using Taq DNA polymerase (GIBCO-BRL, Bethesda, MD) and 2 sets of oligonucleotides for the B29 gene. The first one, designed to amplify the Ig domain and the transmembrane region, consisted of the forward primer 5′GTAGTGCTTGTTCGCGGATC3′ (corresponding to nucleotides 2006-2025 in the DNA sequence23) and the reverse primer 5′CTTGTCCAGCAGCAGGAAGA3′ (corresponding to nucleotides 2824-2805). The second one, designed to amplify the intracytoplasmic domain, consisted of the forward primer 5′GATGACAGCAAGGCTGGCAT3′ (nucleotides 3122-3141) and the reverse primer 5′CTCCTGGCCTGGGTGCTCA3′ (3368-3350).
Thirty cycles of amplification were performed under the following conditions: 95°C for 30 seconds, 60°C for 30 seconds (for Ig domain and transmembrane segment) or 65°C for 30 seconds (for intracytoplasmic domain), and 72°C for 1 minute.
Amplification of VH Ig genes was performed from genomic DNA using 5′ primers designed to anneal to the leader sequence (LH) of VH1 to VH6 families, and a consensus JH 3′ primer. The LHprimers consisted of the following: LH1: 5′CCATGGACTGGACCTGG3′; LH2: 5′ATGGACACACTTTGCTMCAC3′; LH3: 5′CCATGGAGTTTGGGCTGAGC3′; LH4: 5′ATGAAACACCTGTGGTTCTTCC3′; LH5: 5′ATGGGGTCAACCGCCATCC3′; and LH6: 5′ATGTCTGTCTCCTTCCTCATC3′. The consensus JHprimer consisted of 5′ACCTGAGGAGACGGTGACCAGGGT3′. In all cases, 30 cycles of amplification (95°C for 30 seconds, 65°C for 30 seconds, and 72°C for 1 minute) were performed.
Cloning and sequencing of B29 and VH Ig genes.
PCR products were gel purified and subcloned into a TA cloning vector, followed by transformation into INVαF′ cells (Invitrogen, San Diego, CA). Recombinant plasmids were purified and sequence analysis was performed.
RESULTS
Clinical and biological data in 10 families affected with CLL.
Ten families, each with 2 members affected with CLL, were studied. Clinical and biological data concerning these patients are reported in Table 1. Except for 2 patients (1A and 14B), they all had lymphocytosis, whether treated or not. It can be observed in the 6 families with vertical transmission of the disease that the mean age of the parental generation was 81 years as compared with a mean of 50 years for the second generation. This earlier appearance of CLL in the second generation suggested that genetic factors could influence the development of the disease.
Clinical and Biological Data in Members With CLL From 10 Different Italian Families
| Family . | Sex/Age . | Relationship . | Stage . | Treatment . | LY . |
|---|---|---|---|---|---|
| 1A | F/81 | MO | B | CB | 1 |
| 1B | M/57 | SO | A | NO | 30 |
| 2A | M/43 | BR | A | NO | 30 |
| 2B | F/52 | SI | C | NO | 120 |
| 3A | F/85 | MO | A | CB | 40 |
| 3B | F/63 | DA | A | NO | 8 |
| 4A | F/68 | MO | A | NO | 20 |
| 4B | M/35 | SO | A-B | FLU | 7 |
| 5A | M/62 | BR | A | NO | 45 |
| 5B | F/56 | SI | A | NO | 15 |
| 6A | F/95 | MO | A | NO | 40 |
| 6B | F/56 | DA | B | NO | 28 |
| 7A | F/57 | SI | B | CB | 8 |
| 7B | M/60 | BR | A-B | NO | 75 |
| 8A | M/72 | FA | A | NO | 15 |
| 8B | M/43 | SO | NO | 40 | |
| 13A | F/57 | SI | A | NO | 18 |
| 13B | F/59 | SI | A | NO | 25 |
| 14A | M/88 | FA | A | NO | 58 |
| 14B | M/48 | SO | B | FLU | 4 |
| Family . | Sex/Age . | Relationship . | Stage . | Treatment . | LY . |
|---|---|---|---|---|---|
| 1A | F/81 | MO | B | CB | 1 |
| 1B | M/57 | SO | A | NO | 30 |
| 2A | M/43 | BR | A | NO | 30 |
| 2B | F/52 | SI | C | NO | 120 |
| 3A | F/85 | MO | A | CB | 40 |
| 3B | F/63 | DA | A | NO | 8 |
| 4A | F/68 | MO | A | NO | 20 |
| 4B | M/35 | SO | A-B | FLU | 7 |
| 5A | M/62 | BR | A | NO | 45 |
| 5B | F/56 | SI | A | NO | 15 |
| 6A | F/95 | MO | A | NO | 40 |
| 6B | F/56 | DA | B | NO | 28 |
| 7A | F/57 | SI | B | CB | 8 |
| 7B | M/60 | BR | A-B | NO | 75 |
| 8A | M/72 | FA | A | NO | 15 |
| 8B | M/43 | SO | NO | 40 | |
| 13A | F/57 | SI | A | NO | 18 |
| 13B | F/59 | SI | A | NO | 25 |
| 14A | M/88 | FA | A | NO | 58 |
| 14B | M/48 | SO | B | FLU | 4 |
Abbreviations: MO, mother; FA, father; SO, son; SI, sister; BR, brother; DA, daughter; CB, chlorambucil; FLU, fludarabin; NO, no treatment; LY, lymphocyte count ×10−9/L; 1A, 1B, two members of family 1.
Analysis of mutations in B29 DNA from B-CLL cells.
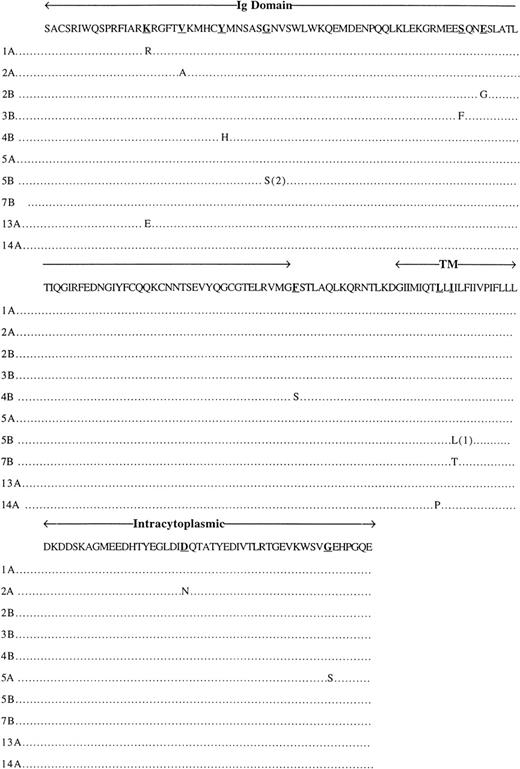
DNA from 20 patients corresponding to the 10 families described above had the B29 gene sequenced. For each B-CLL sample analyzed, 2 classes of B29 DNA clones were identified in the PCR-generated clones corresponding to both B29 alleles. Mutations identified on the B29 DNA sequence are described in Tables 2 and3. In all patients (except patient 3B), 1 allele had the silent mutation TGT→TGC at amino acid position 122 (Cys) in the Ig domain (Table 2), corresponding to a polymorphism of the B29 sequence. In the Ig domain, only 1 other silent mutation was observed (patient 1A), whereas 7 replacement mutations producing an amino acid change were noted in 7 patients: patient 1A at position 56 (Lys → Arg), 13A at position 56 (Lys → Glu), 2A at position 61 (Val → Ala), 4B at position 66 (Tyr → His), 5B at position 72 (Gly → Ser), 3B at position 99 (Ser → Phe), and 2B at position 102 (Glu → Gly). These mutations do not affect known important sites of this domain, namely the 5 extracellular cysteine residues allowing for intrachain and interchain (with Igα) S-S bonds or the 4 glycosylation sites on the Asn residues. Concerning the transmembrane portion, 1 silent mutation was observed in patient 4A at amino acid position 145, whereas 4 replacement mutations at positions 145 (Phe → Ser), 167 (Leu → Pro), 169 (Ile → Leu), and 169 (Ile → Thr) were noted in patients 4B, 14A, 5B, and 7B, respectively. Mutations in the intracytoplasmic domain were detected in only 4 patients (Table 3): 2 silent mutations at amino acid position 192 in patient 6A and 193 in patient 14B and 2 replacement mutations at amino acid position 202 (Asp → Asn) and 223 (Gly →Ser) in patients 2A and 5A, respectively. The Tyr residues, substrates for phosphorylation, were not affected by the mutations. All of the replacement mutations of the B29 gene are described in Fig 1.
Mutations in B29 Ig and Transmembrane Domains
| Patient . | Allele . | Nucleotide Change . | Amino Acid Change . | B29 Domain Affected . |
|---|---|---|---|---|
| 1A | 1 | 2254 AAA → AGA | 56 Lys → Arg | Ig |
| 2377 GAA → GAG | ||||
| 2453 TGT → TGC | ||||
| 2 | None | |||
| 1B | 1 | None | ||
| 2 | 2453 TGT → TGC | |||
| 2A | 1 | 2453 TGT → TGC | ||
| 2 | 2269 GTG → GCG | 61 Val → Ala | Ig | |
| 2453 TGT → TGC | ||||
| 2B | 1 | 2453 TGT → TGC | ||
| 2 | 2392 GAA → GGA | 102 Glu → Gly | Ig | |
| 3A | 1 | None | ||
| 2 | 2453 TGT → TGC | |||
| 3B | 1 | 2383 TCC → TTC | 99 Ser → Phe | Ig |
| 2 | None | |||
| 4A | 1 | None | ||
| 2 | 2453 TGT → TGC | |||
| 2711 TTC → TTT | ||||
| 4B | 1 | 2453 TGT → TGC | ||
| 2 | 2283 TAC → CAC | 66 Tyr → His | Ig | |
| 2710 TTC → TCC | 145 Phe → Ser | TM | ||
| 5A | 1 | None | ||
| 2 | 2453 TGT → TGC | |||
| 5B | 1 | 2453 TGT → TGC | ||
| 2780 ATC → CTC | 169 Ile → Leu | TM | ||
| 2 | 2301 GGC → AGC | 72 Gly → Ser | Ig | |
| 6A | 1 | None | ||
| 2 | 2453 TGT → TGC | |||
| 6B | 1 | None | ||
| 2 | 2453 TGT → TGC | |||
| 7A | 1 | 2453 TGT → TGC | ||
| 2 | ||||
| 7B | 1 | 2453 TGT → TGC | ||
| 2781 ATC → ACC | 169 Ile → Thr | TM | ||
| 2 | 2453 TGT → TGC | |||
| 8A | 1 | 2453 TGT → TGC | ||
| 2 | ||||
| 8B | 1 | None | ||
| 2 | 2453 TGT → TGC | |||
| 13A | 1 | 2453 TGT → TGC | ||
| 2 | 2453 TGT → TGC | |||
| 2253 AAA → GAA | 56 Lys → Glu | Ig | ||
| 13B | 1 | None | ||
| 2453 TGT → TGC | ||||
| 14A | 1 | 2453 TGT → TGC | ||
| 2 | 2453 TGT → TGC | |||
| 2775 CTG → CCG | 167 Leu → Pro | TM | ||
| 14B | 1 | None | ||
| 2 | 2453 TGT → TGC |
| Patient . | Allele . | Nucleotide Change . | Amino Acid Change . | B29 Domain Affected . |
|---|---|---|---|---|
| 1A | 1 | 2254 AAA → AGA | 56 Lys → Arg | Ig |
| 2377 GAA → GAG | ||||
| 2453 TGT → TGC | ||||
| 2 | None | |||
| 1B | 1 | None | ||
| 2 | 2453 TGT → TGC | |||
| 2A | 1 | 2453 TGT → TGC | ||
| 2 | 2269 GTG → GCG | 61 Val → Ala | Ig | |
| 2453 TGT → TGC | ||||
| 2B | 1 | 2453 TGT → TGC | ||
| 2 | 2392 GAA → GGA | 102 Glu → Gly | Ig | |
| 3A | 1 | None | ||
| 2 | 2453 TGT → TGC | |||
| 3B | 1 | 2383 TCC → TTC | 99 Ser → Phe | Ig |
| 2 | None | |||
| 4A | 1 | None | ||
| 2 | 2453 TGT → TGC | |||
| 2711 TTC → TTT | ||||
| 4B | 1 | 2453 TGT → TGC | ||
| 2 | 2283 TAC → CAC | 66 Tyr → His | Ig | |
| 2710 TTC → TCC | 145 Phe → Ser | TM | ||
| 5A | 1 | None | ||
| 2 | 2453 TGT → TGC | |||
| 5B | 1 | 2453 TGT → TGC | ||
| 2780 ATC → CTC | 169 Ile → Leu | TM | ||
| 2 | 2301 GGC → AGC | 72 Gly → Ser | Ig | |
| 6A | 1 | None | ||
| 2 | 2453 TGT → TGC | |||
| 6B | 1 | None | ||
| 2 | 2453 TGT → TGC | |||
| 7A | 1 | 2453 TGT → TGC | ||
| 2 | ||||
| 7B | 1 | 2453 TGT → TGC | ||
| 2781 ATC → ACC | 169 Ile → Thr | TM | ||
| 2 | 2453 TGT → TGC | |||
| 8A | 1 | 2453 TGT → TGC | ||
| 2 | ||||
| 8B | 1 | None | ||
| 2 | 2453 TGT → TGC | |||
| 13A | 1 | 2453 TGT → TGC | ||
| 2 | 2453 TGT → TGC | |||
| 2253 AAA → GAA | 56 Lys → Glu | Ig | ||
| 13B | 1 | None | ||
| 2453 TGT → TGC | ||||
| 14A | 1 | 2453 TGT → TGC | ||
| 2 | 2453 TGT → TGC | |||
| 2775 CTG → CCG | 167 Leu → Pro | TM | ||
| 14B | 1 | None | ||
| 2 | 2453 TGT → TGC |
Two kinds of B29 DNA clones were detected in B-CLL cell samples, corresponding to 2 alleles (1 and 2). Numbering of the nucleotides begins with the first base of the complete DNA sequence described in Hashimoto et al.23
Abbreviations: Ig, Ig domain; TM, transmembrane domain.
Mutations in B29 Intracytoplasmic Domain
| Patient . | Allele . | Nucleotide Change . | Amino Acid Change . |
|---|---|---|---|
| 2A | 1 | None | |
| 2 | 3285 GAC → AAC | 202 Asp → Asn | |
| 5A | 1 | None | |
| 2 | 3348 GGT → AGT | 223 Gly → Ser | |
| 6A | 1 | None | |
| 2 | 3148 GAA → GAG | ||
| 14B | 1 | None | |
| 2 | 3151 GAT → GAC |
| Patient . | Allele . | Nucleotide Change . | Amino Acid Change . |
|---|---|---|---|
| 2A | 1 | None | |
| 2 | 3285 GAC → AAC | 202 Asp → Asn | |
| 5A | 1 | None | |
| 2 | 3348 GGT → AGT | 223 Gly → Ser | |
| 6A | 1 | None | |
| 2 | 3148 GAA → GAG | ||
| 14B | 1 | None | |
| 2 | 3151 GAT → GAC |
Only 4 patients of 20 had silent or replacement mutations. Numbering of the nucleotides begins with the first base of the complete DNA sequence described in Hashimoto et al.23
Replacement mutations in B29 genes. Among 20 patients affected with CLL, 10 express replacement mutations in 1 allele of their B29 gene; 1 patient, 5B, expresses replacement mutation on both alleles (1) and (2). Ig domain, Ig-like domain; TM, transmembrane segment; intracytoplasmic, intracytoplasmic domain. Underlined amino acids correspond to the replacement mutations.
Replacement mutations in B29 genes. Among 20 patients affected with CLL, 10 express replacement mutations in 1 allele of their B29 gene; 1 patient, 5B, expresses replacement mutation on both alleles (1) and (2). Ig domain, Ig-like domain; TM, transmembrane segment; intracytoplasmic, intracytoplasmic domain. Underlined amino acids correspond to the replacement mutations.
The only identical mutation observed between 2 members of the same family was the silent TGT → TGC (amino acid position 122) in the Ig domain. We never observed another identical silent or replacement mutation between both members from the 10 different families.
As we used whole blood for DNA extraction because blood samples were stored as pellets at −20°C and fresh cells were not available, the possibility exists that the amplified B29 gene could be derived from nontumoral cells such as T cells, neutrophils, and monocytes. However, the majority of patients included in this study displayed high numbers of monoclonal B cells. In these conditions, most patients had a remarkable excess of neoplastic DNA used for the PCR amplification and sequencing; thus, any important deletion or insertion affecting the B29 gene should not escape detection.
VH gene usage.
The composition of the H chain variable domain sequences obtained from the 20 patients with familial clustering of CLL is summarized in Table 4. The VH distribution (VH1, 10%; VH2, 0%; VH3, 4%; VH4, 30%; VH5, 10%; and VH6, 5%) is close to that corresponding to the normal complexity of the system (VH1, 22%; VH2, 6%; VH3, 43%; VH4, 22%; VH5, 6%; and VH6, 2%), except for VH2, which is not represented in our familial cases. Among the 18 patients whose VH genes have been sequenced, 14 cases expressed less than 98% similarity with the germinal counterpart, and 11 of 18 had less than 95% similarity. Among these 11 cases, 8 presented a CDR/FR rate of mutation greater than 0.3, which could suggest, at least for some cases, the existence of an antigen-driven selection process.21a
Characteristics of the Twenty Sequenced Genes
| Patients . | VH Family . | Germinal Counterpart . | Homology % . | FR Mutation . | RS . | CDR Mutation . | RS . | CDR/FR . | D . | JH . |
|---|---|---|---|---|---|---|---|---|---|---|
| CLL-1A | VH5 | V5-a | 98.3 | 3 | 0.5 | 2 | 0 | 1.5 | 2-15 | JH5b |
| CLL-1B | VH3 | V3-15 | 100 | 0 | 0 | 0 | 0 | 0 | 3-09 | JH4b |
| CLL-2A | VH6 | V6-01 | 95.4 | 11 | 0.8 | 3 | 2 | 0.27 | 2-15 | JH4d |
| CLL-2B | VH3 | V3-73 | 90.3 | 16 | 2.2 | 13 | 3.3 | 0.81 | 2-02 | JH6b |
| CLL-3A | VH3 | V3-21 | 93.3 | 11 | 1.2 | 9 | 2 | 0.82 | 2-21 | JH4b |
| CLL-3B | VH3 | V3-23 | 93.3 | 14 | 2.5 | 6 | 2 | 0.43 | 3-22 | JH4b |
| CLL-4A | VH5 | V5-51 | 92.2 | 17 | 2.4 | 6 | 5 | 0.35 | ND | JH5b |
| CLL-4B | VH4 | V4-34 | 96.9 | 5 | 4 | 4 | >4 | 0.8 | 5-05 | JH6b |
| CLL-5A | VH1 | V1-02 | 90.1 | 21 | 1.33 | 8 | 3 | 0.38 | 2-21 | JH4b |
| CLL-5B | VH4 | V4-34 | 94.8 | 13 | 2.25 | 2 | >2 | 0.15 | 2-21 | JH4b |
| CLL-6A | VH3 | V3-23 | 90.8 | 22 | 0.83 | 5 | 4 | 0.22 | 6-19 | JH4b |
| CLL-6B | VH1 | V1-18 | 93.2 | 16 | 3 | 4 | 1 | 0.25 | ND | JH4b |
| CLL-7A | VH3 | V3-13 | 99.0 | 2 | 1 | 1 | >1 | 0.5 | 2-15 | JH4b |
| CLL-7B | VH4 | V4-34 | 92.4 | 16 | 0.78 | 6 | 1 | 0.37 | 3-22 | JH4b |
| CLL-8A | VH3 | V3-23 | 93.2 | 12 | 0.71 | 8 | >8 | 0.67 | 3-03 | JH4b |
| CLL-8B | VH3 | V3-30.3 | 98.3 | 5 | 0.67 | 0 | 0 | 0 | 2-02 | JH4b |
| CLL-13A | VH4 | V4-39 | 95.6 | 10 | 4 | 3 | >3 | 0.3 | 6-19 | JH4a |
| CLL-13B | VH3 | ND | ND | JH5a | ||||||
| CLL-14A | VH4 | ND | 2-02 | JH4a | ||||||
| CLL-14B | VH4 | V4-59 | 94.8 | 10 | 1 | 5 | 0.6 | 0.5 | 3-22 | JH1 |
| Patients . | VH Family . | Germinal Counterpart . | Homology % . | FR Mutation . | RS . | CDR Mutation . | RS . | CDR/FR . | D . | JH . |
|---|---|---|---|---|---|---|---|---|---|---|
| CLL-1A | VH5 | V5-a | 98.3 | 3 | 0.5 | 2 | 0 | 1.5 | 2-15 | JH5b |
| CLL-1B | VH3 | V3-15 | 100 | 0 | 0 | 0 | 0 | 0 | 3-09 | JH4b |
| CLL-2A | VH6 | V6-01 | 95.4 | 11 | 0.8 | 3 | 2 | 0.27 | 2-15 | JH4d |
| CLL-2B | VH3 | V3-73 | 90.3 | 16 | 2.2 | 13 | 3.3 | 0.81 | 2-02 | JH6b |
| CLL-3A | VH3 | V3-21 | 93.3 | 11 | 1.2 | 9 | 2 | 0.82 | 2-21 | JH4b |
| CLL-3B | VH3 | V3-23 | 93.3 | 14 | 2.5 | 6 | 2 | 0.43 | 3-22 | JH4b |
| CLL-4A | VH5 | V5-51 | 92.2 | 17 | 2.4 | 6 | 5 | 0.35 | ND | JH5b |
| CLL-4B | VH4 | V4-34 | 96.9 | 5 | 4 | 4 | >4 | 0.8 | 5-05 | JH6b |
| CLL-5A | VH1 | V1-02 | 90.1 | 21 | 1.33 | 8 | 3 | 0.38 | 2-21 | JH4b |
| CLL-5B | VH4 | V4-34 | 94.8 | 13 | 2.25 | 2 | >2 | 0.15 | 2-21 | JH4b |
| CLL-6A | VH3 | V3-23 | 90.8 | 22 | 0.83 | 5 | 4 | 0.22 | 6-19 | JH4b |
| CLL-6B | VH1 | V1-18 | 93.2 | 16 | 3 | 4 | 1 | 0.25 | ND | JH4b |
| CLL-7A | VH3 | V3-13 | 99.0 | 2 | 1 | 1 | >1 | 0.5 | 2-15 | JH4b |
| CLL-7B | VH4 | V4-34 | 92.4 | 16 | 0.78 | 6 | 1 | 0.37 | 3-22 | JH4b |
| CLL-8A | VH3 | V3-23 | 93.2 | 12 | 0.71 | 8 | >8 | 0.67 | 3-03 | JH4b |
| CLL-8B | VH3 | V3-30.3 | 98.3 | 5 | 0.67 | 0 | 0 | 0 | 2-02 | JH4b |
| CLL-13A | VH4 | V4-39 | 95.6 | 10 | 4 | 3 | >3 | 0.3 | 6-19 | JH4a |
| CLL-13B | VH3 | ND | ND | JH5a | ||||||
| CLL-14A | VH4 | ND | 2-02 | JH4a | ||||||
| CLL-14B | VH4 | V4-59 | 94.8 | 10 | 1 | 5 | 0.6 | 0.5 | 3-22 | JH1 |
Abbreviations: GC, germline counterpart; FR, framework; RS, replacement/silent mutation ratio; CDR, complementary determining region; CDR/FR, CDR/FR mutation ratio; when all mutations induced replacements, results are expressed as > number of replacements observed; ND, not determined.
DISCUSSION
The B-CLL lymphocyte differs from the normal CD5+ B cell by constant expression of low amounts of surface Ig and CD79b4,5; resistance to infection by the EBV virus, despite expressing the CD21 molecule10; inability to adequately respond when stimulated through the BCR pathway8,9; and overexpression of the bcl-2 protein.12 It remains unknown whether some of the particular characteristics of the B-CLL lymphocyte could be malignancy-related.
Because genetic or familial factors have been suggested in the etiology of CLL19,20 and genetic aberrations were reported to exist at the level of the B29 gene,7 we were interested in studying the B29 gene in several families who had 2 affected members each. The excess risk of CLL for individuals with affected first degree relatives suggests that familial CLL might constitute a useful model to study the pathogenesis of this disease, as has been the case in other neoplastic disorders. However, previous studies in familial CLL cases failed to show the presence of consistent chromosomal or proto-oncogene abnormalities.24,25 We could not perform such karyotypic studies on our patients, because the material we studied here consisted of blood samples frozen at −20°C. Furthermore, the number of cases reporting common HLA phenotypes among affected CLL siblings is small.19,26,27 The study of family histories or family trees from our series shows evidence favoring a vertical transmission, which may be consistent with the expression of an autosomal dominant gene. Anticipation as defined by worsening severity or earlier age at onset with each generation for an inherited disease has been reported to occur in familial cases of CLL. In a recent series of 9 parent-child pairs with CLL,28 8 showed younger ages at onset in the child and showed a mean decline in age at onset of 21 years, with very similar results found in a British study.29 This suggests that dynamic mutation of unstable DNA sequence repeats could be a common mechanism of inherited hematopoietic malignancy, with implications for the role of somatic mutation in the more frequent sporadic cases. In the 6 families from our series in which a vertical transmission was observed, the mean age of the parental generation was 81 years as compared with a mean age of 50 years for the second generation, which is lower than the mean age of 64 years found in French series.30 Because sampling bias is unlikely to explain these findings, these results could point to the existence among familial CLL patients of another genetic mechanism for developing disease.
In B cells, the CD79b protein is supposed to be essential in the transport of sIg to the membrane,16,17 initiation of signal transduction through the BCR,15 and apoptosis.18 Thus, a defect in expression of this protein may play a major role in the pathophysiology of the disease. Recent work indicating the presence of genetic aberrations leading to affected expression and/or function in B-CLL patients suggests that these alterations could have a genetic origin.7 To better define whether these aberrations could play a primary role in CLL leukemogenesis, we have sequenced the B29 gene at the genomic level in 10 different families including 2 affected members each.
Our results demonstrated that some mutations were present either in the Ig, transmembrane, or cytoplasmic domains of the CD79b protein, but we never observed any insertion or deletion in the B29 gene leading to a truncated protein. Indeed, we have not screened normal genomic DNA for mutations in B29 gene, but the few published sequences derived from normal subjects showed the presence of some polymorphism such as the TGT or TGC in AA position 12231 that we frequently found in our series. Thus, because the few mutations described in the present work have not been reported before in normal B29 gene sequence, it is difficult to conclude whether they correspond to new polymorphisms or whether they are CLL related. However, they are not genetically determined, because they never occured at the same position among 2 members of the same family.
Our results are in accordance with those reported by Rassenti et al,32 who studied unrelated patients affected with CLL and also failed to observe deletion or insertion in Ig, transmembrane, or cytoplasmic domains, but only base substitutions. Thus, our familial CLL did not differ from these common CLL cases. However, both our series and that from Rassenti et al32 differ with the results reported by Thompson et al,7 who demonstrated by sequencing cDNA clones generated from B-CLL cell B29 mRNA, that point mutations, insertions, or deletions, largely located in the B29 transmembrane and cytoplasmic domains, affected B29 expression and/or function. This discrepancy with our data could result from a different origin of the 10 families studied here, but the present findings exclude, at least in our ethnic group, the presence of causal mutations. Both alleles were sequenced and genetic alterations were not detected in paternal and maternal alleles. The few mutations observed here did not induce structural abnormalities of the protein, which could explain its low expression in CLL; furthermore, these mutations are not coding for amino acids involved in the intrachain or interchain bonds between the CD79b and CD79a proteins, namely the Cys of the Ig domain, glycosylation sites, or intracytoplasmic sites for phosphorylation. Thus, the function of the CD79b protein would not be predicted to be affected by these mutations.
Analysis of the VH gene usage among these 10 families affected with CLL showed a higher level of mutation than that described in previous studies concerning unrelated patients,33,34 and the vast majority of the CLL variable domains contained a high degree of somatic mutation and exhibited an excess of replacement mutations in the CDR intervals. These findings suggest that familial CLL cases may preferentially derive from B-cell progenitors that have responded to antigen.21a
Because our results appear to exclude the possibility that a genetic defect could account for the low expression of the CD79b molecule in B-CLL, it would be interesting to study the possibility that the low expression of the CD79b protein on the B-CLL could result from a mechanism of alternative splicing. This event could result in overexpression of a truncated CD79 transcript form, which lacks exon 3, encoding for the extracellular domain of this molecule, and low if any expression of the complete CD79b transcript. Previous work has shown that both transcripts exist in B cells in normal conditions,35 although activated B cells inverse relative expression of the truncated form.36 Thus, the possibility exists that low expression of CD79b in CLL could result from preferential transcription of the truncated CD79b form. Although this possibility needs to be tested, results from Thompson el al7 indicate that transcripts coding for the complete protein are found in consistent amounts in at least half of the CLL patients they have studied. Most of these patients were expressing low to undetectable amounts of the CD79b membrane protein, indicating that CD79b expression may not be exclusively explained by the absence of complete transcripts.
In these conditions, the possibility that a defect at the posttranscriptional level accounting for the discrepancy between the presence of adequate transcripts and very low expression of CD79b and sIg needs to be considered. This defect could occur at the level of intracellular synthesis, could consist of accelerated proteolysis of CD79b and sIg proteins, or in inadequate transport of the BCR to the cell membrane as described on tolerant B lymphocytes.37
Supported by Grant No. 98003512 from the Fondation contre la Leucémie and Grant No. 9734 from the Association pour la Recherche sur le Cancer.
The publication costs of this article were defrayed in part by page charge payment. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. section 1734 solely to indicate this fact.
REFERENCES
Author notes
Address reprint requests to Béatrice Payelle-Brogard, PhD, Unité d’Immunohématologie et d’Immunopathologie, Institut Pasteur, 25 rue du Dr Roux, 75724 Paris Cedex 15, France.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal