Mitochondria play a central role in controlling apoptosis, and activation of the caspase cascade appears to be crucial event during the apoptotic process. Human B lymphoma Raji cells are resistant to nuclear apoptosis induced by various stimuli. Using this cell line, we have asked whether reduction of the mitochondrial transmembrane potential and activation of caspase-3 are sufficient to induce DNA fragmentation during the apoptotic process. After stimulation with cell-permeable C2-ceramide or mitochondrial permeability transition (PT) inducers, not only apoptosis-sensitive cell lines (HL-60, Jurkat, and Daudi cells), but also Raji cells showed reduction of the mitochondrial transmembrane potential (▵ψm), activation of caspase-3, and loss of clonogenic potential. However, Raji cells did not show detectable levels of nuclear apoptosis (DNA degradation). In a cell-free system, cell lysates from tetra-butylhydroperoxide (t-BHP)–treated HL-60 cells induced DNA degradation of Raji nuclei, whereas cell lysates from t-BHP–treated Raji cells failed to induce DNA degradation in either apoptosis-sensitive cell lines or apoptosis-resistant Raji cells. Cleavage of DFF-45, which is a downstream target molecule for caspase-3, was observed in Raji cells as well as in apoptosis-sensitive Daudi cells. These results indicate that there is a defective apoptotic pathway in the cytoplasm downstream of caspase-3 in Raji cells.
APOPTOSIS IS A CRUCIAL pathway not only for physiological mammalian development, but also for tumor cell killing by cytotoxic T cells, natural killer cells,1irradiation or cytotoxic drugs.2-5 Although chemoradiotherapy or immunological therapy has been successful in the treatment of several human cancers, such as hematopoietic malignancies, and rarely in melanoma, these approaches have been mostly unsuccessful in most other malignant tumors. Resistance to apoptosis has been established as one important mechanism of treatment failure, in addition to altered drug metabolism in cancer cells conferred through the P-glycoprotein encoded by MDR1, redox detoxifying action of glutathione, enhanced hepatic P450 activity, and mutation of topoisomerase II.6 Comparing the apoptotic process in apoptosis-sensitive with that in apoptosis-resistant cancer cells may provide new insights into treatment resistance in malignant tumors.
The apoptotic process can be divided into the cell-type–specific induction phase, which is death-stimulus–dependent, and the common effector/degradation phase.7 Mitochondria play a central role in determining whether to die during the common apoptotic signaling pathway, and the decision to die is transduced via the caspase cascade, subsequently leading to DNA degradation. Disruption of the mitochondrial transmembrane potential (Δψm) and subsequent DNA fragmentation occur in various cell types after distinct types of apoptosis-inducing stimulation.8-10 In addition, disruption of the mitochondrial transmembrane potential has been shown to lead to an irreversible apoptotic process.8
We have previously reported that cell-permeable ceramide inhibits the growth of B-lymphoma Raji cells by inducing G0/G1 cell cycle arrest but not apoptosis.11 Since completing this report, we have also found that antitumor agents such as doxorubicin and etoposide could not induce DNA fragmentation in Raji cells.
We have been addressing the question of what is required for the apoptotic process by asking what is missing in the apoptotic pathways of this cell line. In this report, we demonstrate that reduction of mitochondrial transmembrane potential and activation of caspase-3 are not sufficient to induce DNA fragmentation in Raji cells and show evidence that certain molecules downstream of caspase-3 appear to be lacking or not functioning properly in this particular cell line. We also discuss the role of DNA fragmentation factor (DFF) during the apoptotic process in this cell line.
MATERIALS AND METHODS
Cell lines.
HL-60 (acute myelogenous leukemia), U937 (monoblastic leukemia), Jurkat (T-cell leukemia), NALM-6 (B-cell leukemia), K562 (chronic myelogenous leukemia), Ramos (Epstein-Barr virus [EBV]-negative Burkitt’s lymphoma), Daudi (EBV-positive Burkitt’s lymphoma), and Raji (EBV-positive Burkitt’s lymphoma) cells were provided by Fujisaki Cell Center (Okayama, Japan) and were maintained in RPMI 1640 medium supplemented with mycoplasma-free heat-inactivated 10% fetal calf serum (FCS; Hyclone, Logan, UT), 2 mmol/L L-glutamine, 100 U/mL penicillin and 100 μg/mL streptomycin at 37°C. Exponentially growing cells were used in this study, and viability was determined by trypan blue dye exclusion test.
Reagents.
Mitochondrial transmembrane permeability transition (PT) inducers, tetra-butylhydroperoxide (t-BHP) and diamide, were purchased from Sigma (St Louis, MO). A cell-permeable ceramide analog, C2-ceramide, was purchased from Matreya (Pleasant Gap, PA). Rhodamine 123 (Rh123) was purchased from Sigma. Purified anti-Fas (CD95) monoclonal antibody (MoAb; IgM, clone CH-11) and fluorescein isothiocyanate (FITC)-conjugated anti-Fas MoAb (IgG1, clone DX2) were obtained from MBL (Nagoya, Japan) and Becton Dickinson (San Jose, CA), respectively.
Measurement of the mitochondrial transmembrane potential.
A cationic lipophilic fluorochrome, Rh123, was used to measure the mitochondrial transmembrane potential (Δψm), as previously reported.12 Briefly, cells were treated with mitochondrial transmembrane PT inducers or C2-ceramide and were then incubated with Rh123 at a final concentration of 10 μmol/L for 30 minutes. After washing twice with phosphate-buffered saline (PBS), fluorescence intensity was determined by a flow cytometer. Fluorescence intensity was also monitored by fluorescence microscopy.
DNA fragmentation.
To determine the extent of DNA fragmentation, agarose gel electrophoresis and cell-cycle analysis using a flow cytometer were used in this study. The details of the procedures have been described elsewhere.11 In some experiments, TdT-mediated dUTP nick end labeling (TUNEL) assay was performed using an In situ Apoptosis Detection Kit (TaKaRa, Osaka, Japan). For DNA electrophoresis, 2 × 106 cells were washed twice with PBS and were lysed in 200 μL of lysis buffer (50 mmol/L Tris-HCl [pH 8.0]/10 mmol/L EDTA/0.5% lauroylsarcosinate). Cell lysates were incubated with proteinase K (0.5 mg/mL) for 1 hour at 50°C. RNase A (0.25 mg/mL; Nippongene, Osaka, Japan) was then added and incubated for an additional 1 hour at 50°C. Extracted DNA was loaded in 2% agarose gels and electrophoresed in 1× TBE buffer. DNA was visualized by soaking the gels in TBE buffer containing 1 μg/mL ethidium bromide for 30 minutes.
Flow cytometric analysis of DNA loss was performed as follows. After treatment with the various stimuli, cells were washed twice with PBS and were then fixed with 70% ethanol/PBS on ice. After fixation, the medium was removed by centrifugation, 500 μL of PBS was added to each sample, and the cells were treated by DNase-free RNase A (20 μg/mL) for 45 minutes at 37°C. The cells were then stained with 500 μL of propidium iodide (PI; 50 μg/mL) for 30 minutes at room temperature and subjected to flow cytometry. Flow cytometric analysis was performed using a Cytron (Ortho Diagnostics, Tokyo, Japan) with a cell cycle program (Version 1.4). DNA degradation in each cell was defined as a nucleus containing less than 2N DNA.
In some experiments, cell viability was determined by vital staining with PI.
Isolation of intact nuclei.
Intact nuclei were prepared according to the method previously reported.13 Untreated HL-60 and Raji cells (5 × 106) were washed twice with cold PBS and pelleted by centrifugation at 800g for 5 minutes. The cells were incubated in STKM buffer (0.25 mol/L sucrose/50 mmol/L Tris-HCl [pH 7.5]/25 mmol/L KCl/5 mmol/L MgCl2/0.25% Triton X-100) for 30 minutes on ice. After incubation, nuclei were pelleted by centrifugation at 800g for 10 minutes. Sedimental nuclei were washed once in STKM buffer. The release of cytosol-free nuclei was monitored by phase contrast microscopy.
Preparation of cell lysates.
HL-60 and Raji cells (1 × 107) were exposed to 500 μmol/L t-BHP for 4 hours at a density of 1 × 106/mL. The cells were then washed once with cold PBS and were incubated in STKM buffer for 30 minutes on ice. After incubation, nuclei were pelleted by centrifugation at 800g for 10 minutes. Supernatants were then centrifuged at 2,600g for 15 minutes and were used as cell lysates in a cell-free system. Nuclei-free cytosol was confirmed by phase contrast microscopy.
Analysis of DNA degradation in a cell-free system.
Intact nuclei from untreated HL-60 or Raji cells (5 × 105) were suspended in 150 μL of each cell lysate derived from untreated or t-BHP–treated cells (1 × 106 cell equivalent). After adding EGTA at a final concentration of 5 mmol/L, the reaction mixtures were incubated at 37°C for the indicated periods. After incubation, the reaction mixtures were centrifuged at 800g for 5 minutes. Pellets were washed once with STKM buffer and were fixed with cold 70% ethanol/PBS. The medium was removed by centrifugation, and 500 μL of STKM buffer was added to each sample. The cells were treated by DNase-free RNase A (20 μg/mL) for 45 minutes. The nuclei were then stained with 500 μL of PI (50 μg/mL) for 30 minutes at room temperature and subjected to flow cytometry.
Immunoblotting for caspase-3 and DFF-45.
After washing twice with PBS, 1 × 106 cells were resuspended in 20 μL of RIPA buffer (1% NP40/0.1% sodium deoxycholate/150 mmol/L NaCl/50 mmol/L Tris-HCl [pH 7.5]/1 mmol/L phenylmethylsulfonyl fluoride [PMSF]/20 U/mL aprotinin) on ice. After centrifugation at 10,000g at 4°C for 10 minutes, supernatants were harvested and were subjected to 12.5% sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) under reducing conditions. Transfer of the resolved proteins and visualization of the proteins were previously described in detail.14 Mouse antihuman caspase-3 and mouse antihuman DFF-45 MoAbs were purchased from MBL. Polyclonal rabbit anti–caspase-3 antibody reacting with both the 32-kD unprocessed pro–caspase-3 and the 17-kD subunit of the active caspase-3 was obtained from PharMingen (San Diego, CA).
Assay for caspase-3 activity.
Activity of intracellular caspase-3 was determined using a PhiPhiLux-G6D2 Kit15 16 (OncoImmunin, Inc, College Park, MD) according to the manufacturer’s instructions. Cells (5 × 105) were exposed to 500 μmol/L t-BHP for the indicated intervals at 37°C at a density of 1 × 105/mL. After incubation, 50 μL of 10 μmol/L fluorogenic caspase-3 substrate solution and 5 μL of FCS were added to each cell pellet and centrifuged at 800g for 5 minutes. The cells were then incubated at 37°C for 60 minutes. After incubation, 500 μL of ice-cold flow cytometry dilution buffer was added to this reaction mixture, and the cells were subjected to flow cytometry.
Reverse transcriptase-polymerase chain reaction (RT-PCR) for DFF-45.
Total cellular RNA was prepared according to the method previously reported.17 cDNA synthesis was performed using a First-Strand Synthesis Kit (Pharmacia, Uppsala, Sweden). The primers 5′-TGTGGCATTGGCTAGTAATGAGA-3′ and 5′-ACTAGATAAGCTCAGCTCTGGAG-3′ were used to amplify the DFF-45 cDNA. The PCR products include the 2 cleavage sites of caspase-3, DETD (aa117) and DAVD (aa224). Thirty PCR cycles were performed at 94°C for denaturation, 56°C for annealing, and 72°C for extension reaction. PCR products were loaded onto 2% agarose gels and electrophoresed in 1× TBE buffer. Gels were stained with ethidium bromide and visualized under UV light.
Sequencing of DFF-45 cDNA.
The PCR products of DFF-45 cDNA were directly sequenced using a Dye Terminator Cycle Sequencing Kit (PE Applied Biosystems, Foster City, CA). PCR products were extensively purified using Centricon-100 concentrator columns, and the same forward and reverse primers were used for sequencing chemistry. Sequence analysis was performed using an Applied Biosystems 377A automatic DNA sequencer (Applied Biosystems, Foster City, CA).
Assay for clonogenic cell growth of tumor lines.
Cells (1,000 cells/mL) were cultured in a 35-mm petri dish in semisolid culture medium consisting of 0.8% methylcellulose/RPMI 1640/20% FCS at 37°C. After 7 days of culture, a cell cluster containing more than 50 cells was counted as a colony under an inverted microscope.
RESULTS
Cell-permeable ceramide and DNA-damaging agents failed to induce nuclear apoptosis (DNA fragmentation) in Raji cells.
We have previously reported that a cell-permeable ceramide analogue can inhibit the growth of Raji cells but not induce nuclear apoptosis.11 We have extended this observation to learn whether other apoptosis-inducing stimuli cause DNA fragmentation in this particular cell line. As shown in Table 1, not only C2-ceramide, but also antitumor agents, such as doxorubicin and etoposide, failed to induce DNA degradation in Raji cells, whereas in other cell lines these reagents induced apoptosis. Moreover, Raji cells did not undergo apoptosis in response to anti-Fas MoAb treatment, which could induce apoptosis in Jurkat cells. To define the mechanisms of resistance to apoptosis in Raji cells, we first focused on the alteration of the mitochondrial transmembrane potential and the activation of caspase-3.
Induction of DNA Fragmentation in Lympho-Hematopoietic Tumor Lines by Various Apoptosis Inducers
| Reagent . | Dose . | DNA Degradation* . | |||||||
|---|---|---|---|---|---|---|---|---|---|
| HL-60 . | U937 . | Jurkat . | K562 . | NALM-6 . | Ramos . | Daudi . | Raji . | ||
| DNR | 1 μg/mL | + | + | − | − | ND | − | − | − |
| VP-16 | 10 μg/mL | + | + | + | − | ND | + | + | − |
| TNF-α | 20 ng/mL | + | + | ND | ND | ND | ND | ND | − |
| C2-ceramide | 20 μmol/L | + | + | + | + | + | + | + | − |
| Diamide | 1 mmol/L | + | + | + | + | + | + | + | − |
| t-BHP | 50 μmol/L | + | + | + | + | + | + | + | − |
| Anti-Fas† | 1 μg/mL | ND | ND | + | ND | ND | − | − | − |
| Irradiation‡ | 5,000 rad | + | + | + | ND | ND | + | + | − |
| Reagent . | Dose . | DNA Degradation* . | |||||||
|---|---|---|---|---|---|---|---|---|---|
| HL-60 . | U937 . | Jurkat . | K562 . | NALM-6 . | Ramos . | Daudi . | Raji . | ||
| DNR | 1 μg/mL | + | + | − | − | ND | − | − | − |
| VP-16 | 10 μg/mL | + | + | + | − | ND | + | + | − |
| TNF-α | 20 ng/mL | + | + | ND | ND | ND | ND | ND | − |
| C2-ceramide | 20 μmol/L | + | + | + | + | + | + | + | − |
| Diamide | 1 mmol/L | + | + | + | + | + | + | + | − |
| t-BHP | 50 μmol/L | + | + | + | + | + | + | + | − |
| Anti-Fas† | 1 μg/mL | ND | ND | + | ND | ND | − | − | − |
| Irradiation‡ | 5,000 rad | + | + | + | ND | ND | + | + | − |
Abbreviation: ND, not determined.
Cells were cultured in 2% FCS/RPMI1640 in the presence of varying reagents for 18 hours at 37°C. DNA degradation was determined by the presence of a hypodiploid peak in cell-cycle analysis.
Cells were incubated with apoptogenic anti-Fas MoAb (IgM, clone CH-11) for 18 hours at 37°C.
Cells were irradiated with an x-ray source.
Loss of the mitochondrial transmembrane potential did not result in DNA fragmentation in Raji cells.
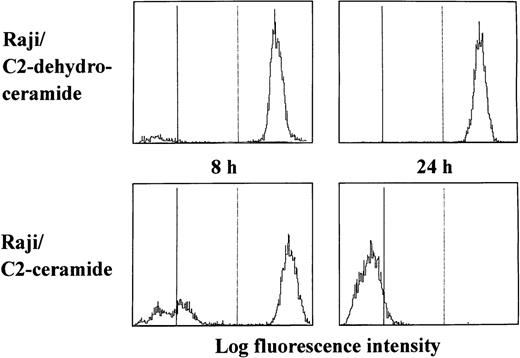
We first examined whether a cell-permeable ceramide analogue might induce reduction of the mitochondrial transmembrane potential in apoptosis-resistant Raji cells. After treatment with C2-ceramide, disruption of the mitochondrial transmembrane potential and subsequent apoptosis were observed in HL-60, U937, Jurkat, and Daudi cells. Interestingly, C2-ceramide reduced the mitochondrial transmembrane potential in Raji cells, although it did not induce DNA degradation (Fig 1). The MTT assay showed that Raji cells were more resistant to ceramide treatment than were other cell lines (Table 2), but approximately 20% of the C2-ceramide-treated Raji cells were found to be dead after 48 hours of culture by trypan blue staining (data not shown). As shown in Table 3, Raji cells lost their clonogenic potential after treatment with cell-permeable ceramide, suggesting that Δψm disruption is the point of no return for cell death, irrespective of death type.
Reduction of the mitochondrial transmembrane potential by a cell-permeable ceramide in Raji cells. Cells were cultured with either C2-ceramide (20 μmol/L) or C2-dehydroceramide (20 μmol/L) for the indicated periods. After incubation, cells were harvested for measurement of the mitochondrial transmembrane potential.
Reduction of the mitochondrial transmembrane potential by a cell-permeable ceramide in Raji cells. Cells were cultured with either C2-ceramide (20 μmol/L) or C2-dehydroceramide (20 μmol/L) for the indicated periods. After incubation, cells were harvested for measurement of the mitochondrial transmembrane potential.
Effect of Ceramide on Cell Viability
| Cells . | Ceramide . | MTT Assay*(OD570-620) . | |
|---|---|---|---|
| 24 h . | 48 h . | ||
| Ramos | C2-dehydroceramide | 0.166 ± 0.014 | 0.248 ± 0.031 |
| C2-ceramide | 0.106 ± 0.008 | 0.064 ± 0.004 | |
| Daudi | C2-dehydroceramide | 0.172 ± 0.005 | 0.296 ± 0.017 |
| C2-ceramide | 0.100 ± 0.005 | 0.080 ± 0.006 | |
| Raji | C2-dehydroceramide | 0.260 ± 0.007 | 0.361 ± 0.018 |
| C2-ceramide | 0.249 ± 0.003 | 0.219 ± 0.004 | |
| HL-60 | C2-dehydroceramide | 0.210 ± 0.013 | 0.379 ± 0.038 |
| C2-ceramide | 0.039 ± 0.001 | 0.038 ± 0.002 | |
| Cells . | Ceramide . | MTT Assay*(OD570-620) . | |
|---|---|---|---|
| 24 h . | 48 h . | ||
| Ramos | C2-dehydroceramide | 0.166 ± 0.014 | 0.248 ± 0.031 |
| C2-ceramide | 0.106 ± 0.008 | 0.064 ± 0.004 | |
| Daudi | C2-dehydroceramide | 0.172 ± 0.005 | 0.296 ± 0.017 |
| C2-ceramide | 0.100 ± 0.005 | 0.080 ± 0.006 | |
| Raji | C2-dehydroceramide | 0.260 ± 0.007 | 0.361 ± 0.018 |
| C2-ceramide | 0.249 ± 0.003 | 0.219 ± 0.004 | |
| HL-60 | C2-dehydroceramide | 0.210 ± 0.013 | 0.379 ± 0.038 |
| C2-ceramide | 0.039 ± 0.001 | 0.038 ± 0.002 | |
Cells (20 × 104/mL) were cultured in 2% FCS/RPMI1640 in the presence of 20 μmol/L cell-permeable ceramide for the indicated periods. C2-dehydroceramide was used as a negative control.
MTT assay was performed according to the method described by Mosmann.41 Data are expressed as the mean ± SD of triplicate cultures.
Loss of Clonogenic Potential After Ceramide Treatment
| Cells . | Pretreatment3-150 . | Concentration . | No. of Colonies (/1,000 cells plated)3-151 . |
|---|---|---|---|
| Daudi | C2-dehydroceramide | 20 μmol/L | 637 ± 41 |
| C2-ceramide | 20 μmol/L | 19 ± 2 | |
| Raji | C2-dehydroceramide | 20 μmol/L | 799 ± 61 |
| C2-ceramide | 20 μmol/L | 38 ± 9 |
| Cells . | Pretreatment3-150 . | Concentration . | No. of Colonies (/1,000 cells plated)3-151 . |
|---|---|---|---|
| Daudi | C2-dehydroceramide | 20 μmol/L | 637 ± 41 |
| C2-ceramide | 20 μmol/L | 19 ± 2 | |
| Raji | C2-dehydroceramide | 20 μmol/L | 799 ± 61 |
| C2-ceramide | 20 μmol/L | 38 ± 9 |
Cells were cultured in the presence of the indicated concentration of cell-permeable ceramide. After 48 hours of culture, cells were washed twice with 2% FCS/RPMI1640 and trypan blue-unstained viable cells (1,000 cells/mL) were plated in a semisolid culture medium.
Data are expressed as the mean ± SD of triplicate cultures.
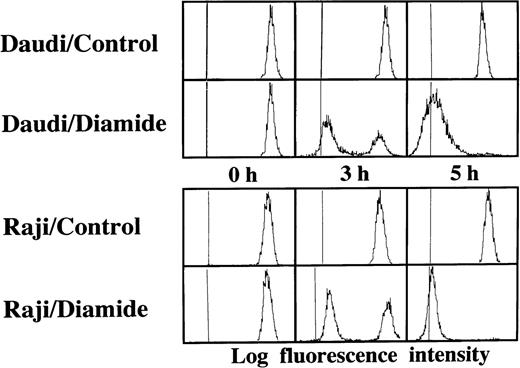
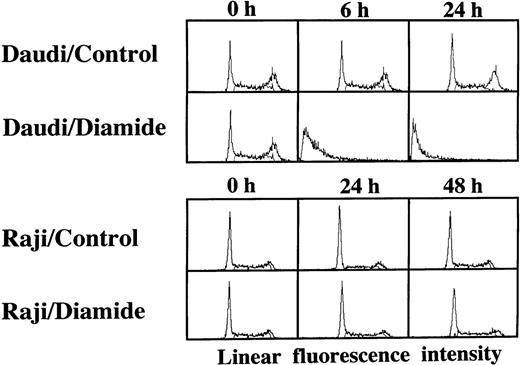
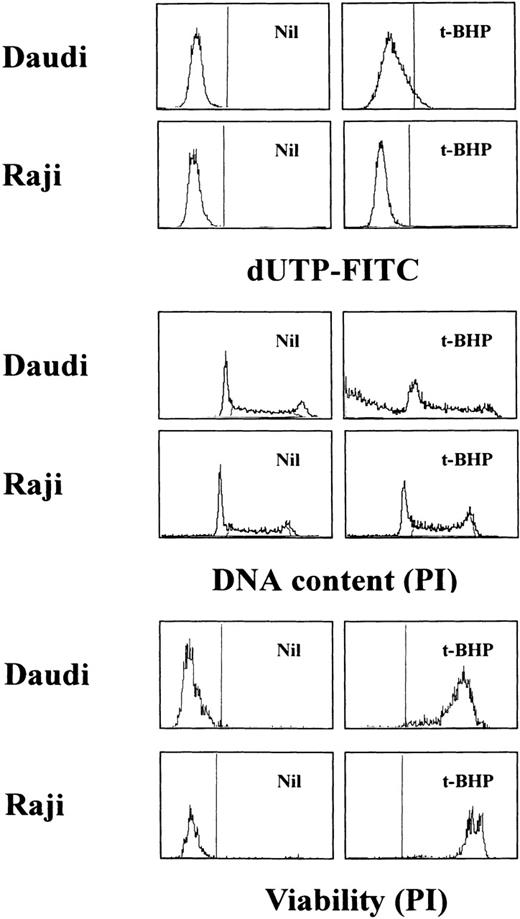
Loss of the mitochondrial transmembrane potential has been shown to result in mitochondrial permeability transition, which then causes mitochondrial release of apoptogenic proteins.17 Thus, we asked whether forced mitochondrial permeability transition would cause DNA fragmentation in Raji cells. We used Daudi cells as a control in this experiment, because the EBV genome was integrated in both Raji and Daudi cells. After the treatment with diamide, Raji cells showed a decrease of incorporation of a cationic lipophilic fluorochrome, Rh123, to the same degree as apoptosis-inducible Daudi cells (Fig 2). Other apoptosis-sensitive cells, such as HL-60, U937, and Jurkat cells, showed similar results (data not shown). Apoptosis, which was judged by the presence of cells with hypodiploid DNA content on flow cytometry, was observed in Daudi cells as early as 6 hours after treatment with diamide, and almost all cells showed DNA degradation at 24 hours of incubation with diamide (Fig 3). Other apoptosis-sensitive cells, such as HL-60, U937, Jurkat, K562, NALM-6, and Ramos cells, also showed similar results (Table 1). In contrast, the ratio of apoptotic cells was not significant, even 48 hours after stimulation with diamide in Raji cells (Fig 3). This observation was confirmed by using another PT inducer, t-BHP, and DNA electrophoresis analysis. t-BHP failed to induce DNA fragmentation in Raji cells, whereas t-BHP induced apoptosis in HL-60 and Jurkat cells (Fig 4). Moreover, we have performed TUNEL assay to confirm that Raji cells are resistant to nuclear apoptosis (Fig 5). However, vital staining of Raji cells with PI showed that Raji cells died after t-BHP. These results indicate that mitochondrial dysfunction induced by PT inducers does not lead to DNA degradation, but rather leads to cell death in Raji cells.
Disruption of the mitochondrial transmembrane potential by PT inducers. Raji and Daudi cells were cultured in the presence or absence of 1 mmol/L diamide for the indicated periods. Reduction of the mitochondrial transmembrane potential was observed in both Daudi and Raji cells. Similar results were obtained with t-BHP (data not shown).
Disruption of the mitochondrial transmembrane potential by PT inducers. Raji and Daudi cells were cultured in the presence or absence of 1 mmol/L diamide for the indicated periods. Reduction of the mitochondrial transmembrane potential was observed in both Daudi and Raji cells. Similar results were obtained with t-BHP (data not shown).
Failure to induce DNA degradation in Raji cells by PT inducers. After treatment with 1 mmol/L diamide, the cells were fixed, stained with PI, and subjected to flow cytometry. This experiment was repeated 3 times with similar results. The use of t-BHP also gave similar results (data not shown).
Failure to induce DNA degradation in Raji cells by PT inducers. After treatment with 1 mmol/L diamide, the cells were fixed, stained with PI, and subjected to flow cytometry. This experiment was repeated 3 times with similar results. The use of t-BHP also gave similar results (data not shown).
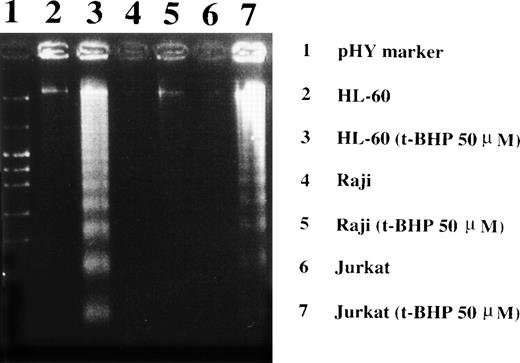
Induction of DNA fragmentation in apoptosis-sensitive HL-60 and Jurkat cells but not in apoptosis-resistant Raji cells. The cells were treated with 50 μmol/L t-BHP for 4 hours at 37°C and were then harvested for DNA isolation. Characteristic apoptotic DNA ladders were observed in HL-60 and Jurkat cells but not in Raji cells. This experiment was repeated more than 3 times with similar results.
Induction of DNA fragmentation in apoptosis-sensitive HL-60 and Jurkat cells but not in apoptosis-resistant Raji cells. The cells were treated with 50 μmol/L t-BHP for 4 hours at 37°C and were then harvested for DNA isolation. Characteristic apoptotic DNA ladders were observed in HL-60 and Jurkat cells but not in Raji cells. This experiment was repeated more than 3 times with similar results.
TUNEL assay and cell-cycle analysis to determine the presence of DNA fragmentation. Daudi and Raji cells were stimulated with 50 μmol/L t-BHP for 6 hours at 37°C. Apoptosis-permissive Daudi cells, but not apoptosis-resistant Raji cells, were nick-end labeled by TdT. Viability was determined by staining the unfixed cells with PI.
TUNEL assay and cell-cycle analysis to determine the presence of DNA fragmentation. Daudi and Raji cells were stimulated with 50 μmol/L t-BHP for 6 hours at 37°C. Apoptosis-permissive Daudi cells, but not apoptosis-resistant Raji cells, were nick-end labeled by TdT. Viability was determined by staining the unfixed cells with PI.
t-BHP can induce activation of caspase-3 in nuclear apoptosis-resistant Raji cells.
Recently, it has been suggested that reduction of the mitochondrial transmembrane potential and activation of the caspase cascade are common events leading to DNA degradation during the apoptotic process.7,18-23 Apoptogenic proteins released from mitochondria have been shown to activate the caspase cascade.24,25 We asked whether t-BHP could induce the activation of caspase after the induction of mitochondrial transmembrane potential loss in Raji cells. Caspase-3 is a protease that is activated downstream of caspase-1 during apoptosis25 and cleaves substrates such as poly (ADP-ribose) polymerase, actin, fodrin, and lamin.26 27Therefore, we determined the expression of caspase-3 protease in Raji cells. Immunoblotting analysis showed that Raji cells expressed a level of procaspase-3 protein similar to that of other apoptosis-sensitive cells (data not shown).
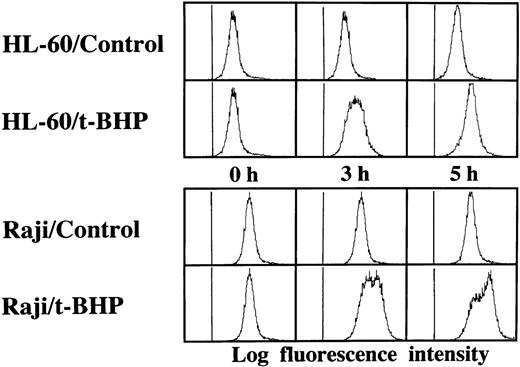
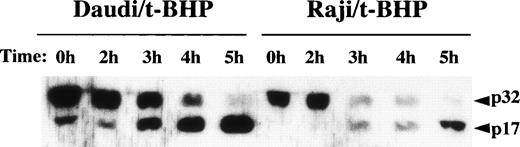
We then asked whether PT inducers induce the activation of caspase-3 in Raji cells. To examine the activity of caspase-3, we performed flow cytometric analysis using a fluorogenic substrate for caspase-3 that released fluorescence in proportion to caspase-3 activity in living cells. As shown in Fig 6, activation of caspase-3 was observed in t-BHP–treated Raji cells. To confirm this finding, we also performed immunoblotting to detect cleavage of procaspase-3 in Raji cells. After treatment with t-BHP, a decrease of p32 procaspase-3 protein expression was observed and simultaneously p17 active caspase-3 was detected in Raji cells as well as Daudi cells (Fig 7).
Activation of caspase-3 in Raji cells by t-BHP. HL-60 and Raji cells were cultured in the presence or absence of 500 μmol/L t-BHP for the indicated periods. Intracellular caspase-3 activity was assessed by flow cytometric analysis using a PhiPhiLux-G6D2 kit.
Activation of caspase-3 in Raji cells by t-BHP. HL-60 and Raji cells were cultured in the presence or absence of 500 μmol/L t-BHP for the indicated periods. Intracellular caspase-3 activity was assessed by flow cytometric analysis using a PhiPhiLux-G6D2 kit.
Cleavage of procaspase-3 in Raji cells. Daudi and Raji cells were stimulated with 500 μmol/L t-BHP for the indicated periods, and then cell lysates were prepared. Samples were loaded onto 13% SDS-PAGE gels. Transferred proteins on polyvinylidene difluoride (PVDF) membranes were reacted with polyclonal rabbit anti–caspase-3 antibody (PharMingen). This experiment was repeated 3 times with similar results.
Cleavage of procaspase-3 in Raji cells. Daudi and Raji cells were stimulated with 500 μmol/L t-BHP for the indicated periods, and then cell lysates were prepared. Samples were loaded onto 13% SDS-PAGE gels. Transferred proteins on polyvinylidene difluoride (PVDF) membranes were reacted with polyclonal rabbit anti–caspase-3 antibody (PharMingen). This experiment was repeated 3 times with similar results.
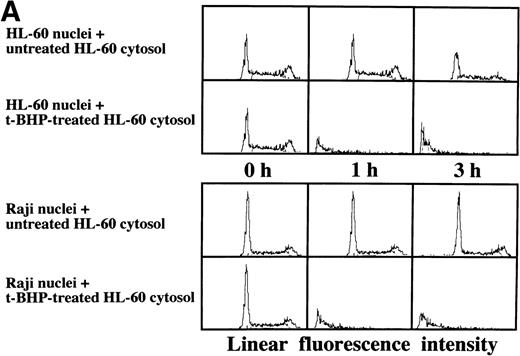
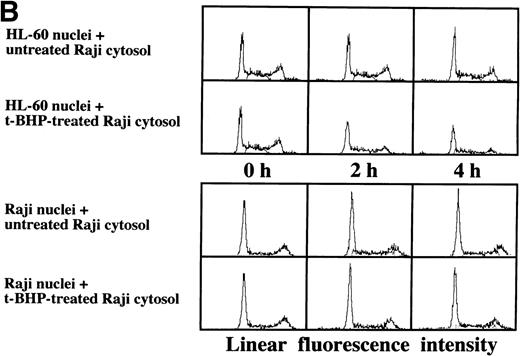
Induction of DNA degradation of isolated Raji nuclei with a cell-free cytosolic extract from t-BHP–treated HL-60 cells.
As described above, the activation of caspase-3 did not lead to DNA degradation in Raji cells. We raised the question of whether there might be some defects in Raji nuclei that prevent induction of DNA degradation. We employed a cell-free system using isolated nuclei and cytosolic extracts. The cell-free cytosolic extract from t-BHP–treated HL-60 cells induced DNA degradation not only in HL-60 nuclei, but also in Raji nuclei (Fig 8A). In contrast, the cytosolic extract from t-BHP–treated Raji cells did not induce DNA degradation in either Raji or HL-60 nuclei (Fig 8B). These results indicate that sufficient apoptotic signals generated in the cytoplasm of apoptosis-permissive cells induce DNA degradation in Raji nuclei and that apoptotic signals in the cytoplasm of Raji cells treated with t-BHP are not sufficient to induce DNA degradation in nuclei of apoptosis-permissive cells. These data suggest that cytoplasmic downstream target molecules for caspase-3, which should be activated during the apoptotic process, might be missing or functionally defective in Raji cells.
Induction of DNA degradation in Raji nuclei by the t-BHP–treated HL-60 cytosolic extract. (A) The t-BHP–treated HL-60 cytosolic extract induced DNA degradation in Raji nuclei as well as in HL-60 nuclei. (B) The cell-free cytosolic extract from t-BHP–treated Raji cells did not induce DNA degradation in either HL-60 or Raji nuclei.
Induction of DNA degradation in Raji nuclei by the t-BHP–treated HL-60 cytosolic extract. (A) The t-BHP–treated HL-60 cytosolic extract induced DNA degradation in Raji nuclei as well as in HL-60 nuclei. (B) The cell-free cytosolic extract from t-BHP–treated Raji cells did not induce DNA degradation in either HL-60 or Raji nuclei.
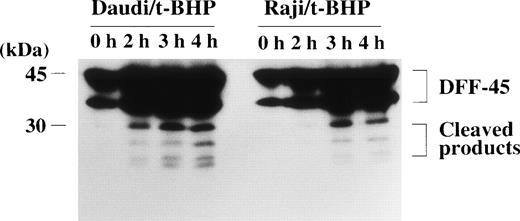
Expression and cleavage of DFF-45 in Raji cells.
The DFF, which induces DNA fragmentation after it is activated by caspase-3, has been identified in human HeLa and U937 cytoplasm.28 The DFF has been reported to be a heterodimeric protein of 40- and 45-kD subunits, of which the 45-kD subunit (DFF-45) can be cleaved by caspase-3 into smaller polypeptides of 30 and 11 kD.28 The 40-kD subunit (DFF-40) remains intact after stimulation by active caspase-3. Caspase-3 cleaves DFF-45 at the 2 cleavage sites to generate an active factor that produces DNA fragmentation without further requirement for caspase-3 or other cytosolic proteins.28 The amino acid sequence of DFF-45 was identified as a novel protein, of which an open reading frame of 331 amino acids was predicted from its cDNA sequence.28Furthermore, Liu et al28 reported that the sequences of the 2 cleavage sites of DFF-45 for active caspase-3 were DETD (aa177) and DATD (aa224), respectively, in HeLa and U937 cells. We tested the possibility that DFF-45 might be deficient or mutated in Raji cells. To determine the expression of DFF-45 in Raji cells, we first performed RT-PCR using primers that were designed to amplify the sequence spanning the 2 caspase-3 cleavage sites. Agarose gel electrophoresis of these PCR-amplified cDNAs showed that expression of DFF-45 mRNA was detected in Raji cells with the same migration pattern as in other cells (data not shown). The cDNA sequence of DFF-45 in Raji cells was identical to that of U937 cells, and it included the same sequences of the 2 known cleavage sites for caspase-3 reported in HeLa and U937 cells (data not shown). In fact, the cleavage of DFF-45 was confirmed by immunoblotting analysis in Raji cells as well as apoptosis-sensitive Daudi cells (Fig 9).
Cleavage of the DFF-45 protein after mitochondrial dysfunction. After treatment with t-BHP (500 μmol/L), smaller protein bands, which were reactive with anti-DFF45 MoAb, were detected in both Daudi and Raji cells.
Cleavage of the DFF-45 protein after mitochondrial dysfunction. After treatment with t-BHP (500 μmol/L), smaller protein bands, which were reactive with anti-DFF45 MoAb, were detected in both Daudi and Raji cells.
DISCUSSION
Mitochondria play a crucial role in apoptosis.7 During the apoptotic process, mitochondria release several apoptogenic molecules to activate the downstream apoptotic signal transduction pathways, including apoptosis-inducing factor (AIF)23 and cytochrome C.29 AIF is released from mitochondria after the reduction of the mitochondrial transmembrane potential and subsequent membrane permeability transition.12,18 AIF then activates caspases.24 Cytochrome C is released during the early phase of apoptosis before the occurrence of mitochondrial membrane potential loss,30,31 which appears to be regulated independently from mitochondrial dysfunction. Cytochrome C also activates caspases.29 Caspase-3 is a protease that cleaves various cellular proteins, such as poly (ADP-ribose) polymerase, actin, fodrin, and lamin during apoptosis,26,27 and has been postulated to be an “executioner” protein activated by the caspase cascade.31
In this report, we have demonstrated that reduction of mitochondrial transmembrane potential and activation of caspase-3 do not lead to DNA fragmentation in human B-lymphoma Raji cells. The results from experiments using a cell-free system suggest that there is a defective apoptotic pathway in the cytoplasm downstream of caspase-3 in this particular cell line. Disruption of the mitochondrial transmembrane potential has been detected in various cell types by distinct apoptosis-inducing stimuli, and subsequently occurring nuclear apoptosis cannot be dissociated from this biochemical event in each system.7,9,18,32,33 To our knowledge, this is the first report describing the model in which loss of the mitochondrial transmembrane potential and activation of caspase-3 do not lead to DNA degradation in living cells. However, Raji cells lose their proliferative capacity after ceramide treatment, suggesting that Δψm disruption is an irreversible step of cell death programs. There is an interesting report that Raji and Ramos cells show distinct types of cell death in response to polyunsaturated fatty acids.34 Raji and Ramos cells die by necrosis and apoptosis, respectively. We are currently investigating the cell death mechanisms in Raji cells in response to ceramide or mitochondrial permeability transition inducers.
The signal transduction pathway between the caspase cascade and the DNA degradation phase in humans has not been fully elucidated. Human DFF has recently been identified as a protein that is activated by caspase-3 and induces nuclear DNA fragmentation without any requirement for other cytosolic proteins.28 DFF is a heterodimeric protein consisting of 45- and 40-kD subunits (designated DFF-45 and DFF-40). DFF-45 has been cloned, and it is cleaved by caspase-3 at the sequences DETD (aa 117) and DAVD (aa 224).28 We performed partial sequencing of DFF-45 cDNA in Raji and U937 cells and could not find any difference in the sequence, which is identical to that previously reported in HeLa and U937 cells.28 We also confirmed the cleavage of DFF-45 in Raji cells. These results suggest that certain apoptotic pathways after the cleavage of DFF-45 are defective in this cell line.
Murine caspase-activated deoxyribonuclease (CAD) and its inhibitor (ICAD) have recently been identified and cloned. 35The amino acid sequence of human DFF-45 is highly homologous to that of murine ICAD,35 suggesting that human DFF-45 could be a counterpart of murine ICAD. Murine CAD exists as a complex with ICAD in the cytoplasm and caspase-3 cleaved ICAD at the 2 cleavage sites to allow CAD to enter the nuclei and induce DNA fragmentation.35,36 Human DFF-40 has been recently cloned,37 and the homology between DFF-40 and murine CAD has been reported to be 71%.37 It is possible that there might be structural and/or functional defects in DFF-40 in Raji cells, and this issue should be addressed in the near future.
p53 and Bcl-2 are extensively investigated molecules that modulate apoptotic cell death, and their functional alteration is thought to be involved in resistance to apoptosis in human cancers.38-40In our previous report, it was demonstrated that the expression of p53 protein was normal, and the expression of Bcl-2 was even less in Raji cells compared with HL-60 or U937 cells, which are known to be sensitive to apoptosis.11 Because the apoptotic machinery in the nuclei of Raji cells was normal, alterations in p53 do not appear to be the cause of apoptosis resistance. Although the structural and/or functional defects downstream of caspase-3 in Raji cells remain to be elucidated, exploration of the mechanisms will lead to a new model of resistance to nuclear apoptosis.
ACKNOWLEDGMENT
The authors thank Fujisaki Cell Center for providing us with cell lines.
Supported in part by grants from the Japan Research Foundation for Clinical Pharmacology and the Ministry of Welfare, Science and Culture, Japan to M.H.
The publication costs of this article were defrayed in part by page charge payment. This article must therefore be hereby marked “advertisement” in accordance with 18 U.S.C. section 1734 solely to indicate this fact.
REFERENCES
Author notes
Address reprint requests to Makoto Hirokawa, MD, PhD, Department of Internal Medicine III, Akita University School of Medicine, 1-1-1 Hondo, Akita 010-8543, Japan; e-mail: hirokawa@med.akita-u.ac.jp.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal